Genome-Wide Identification and Functional Analysis of RF2 Gene Family and the Critical Role of GhRF2-32 in Response to Drought Stress in Cotton
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Property Analysis of RF2 Gene Family in Cotton
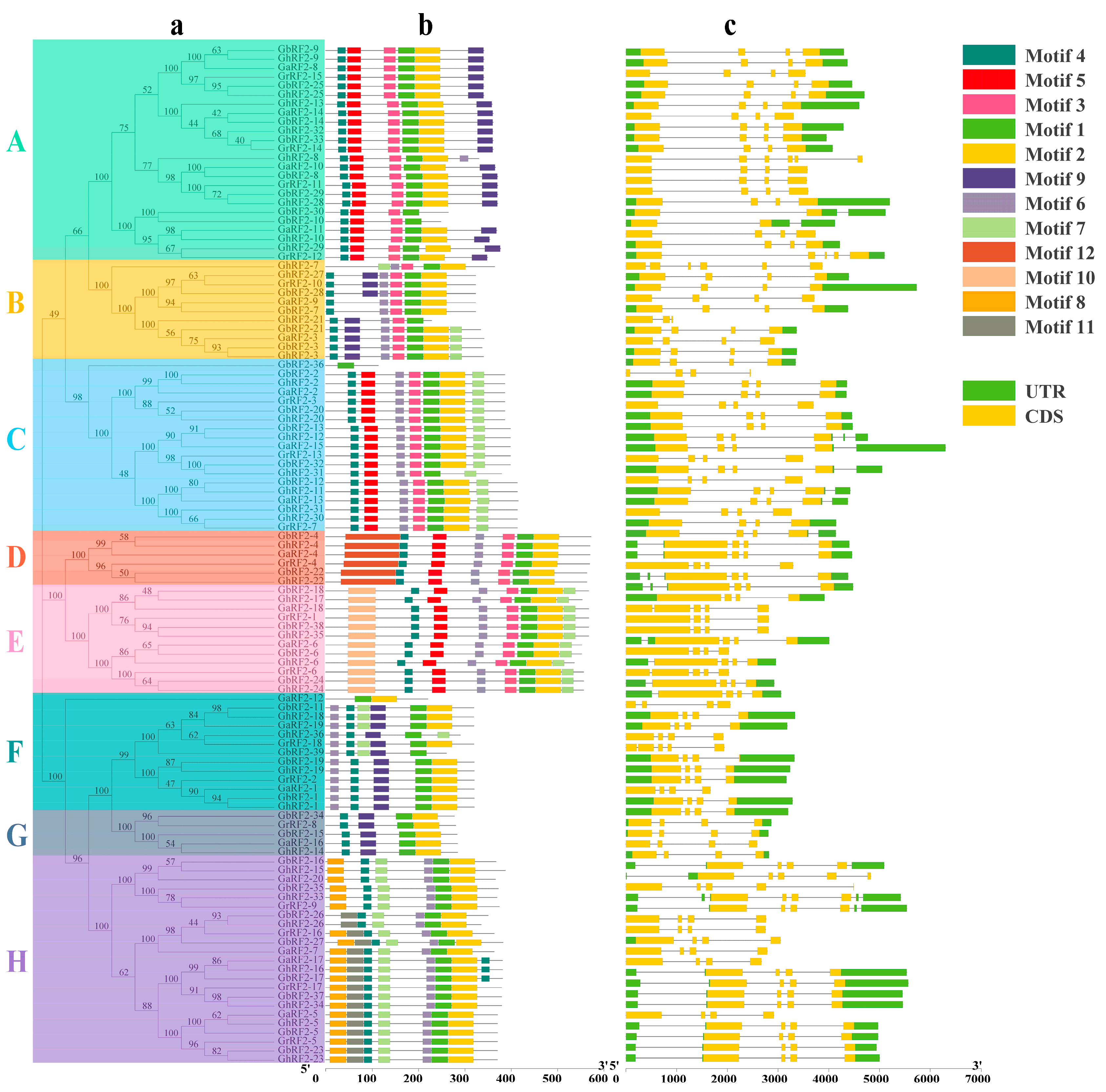
2.2. Construction of Phylogenetic Tree of RF2 Genes in Cotton
2.3. Analysis of RF2 Gene Conserved Motif and Exon–Intron Distribution Pattern in Cotton
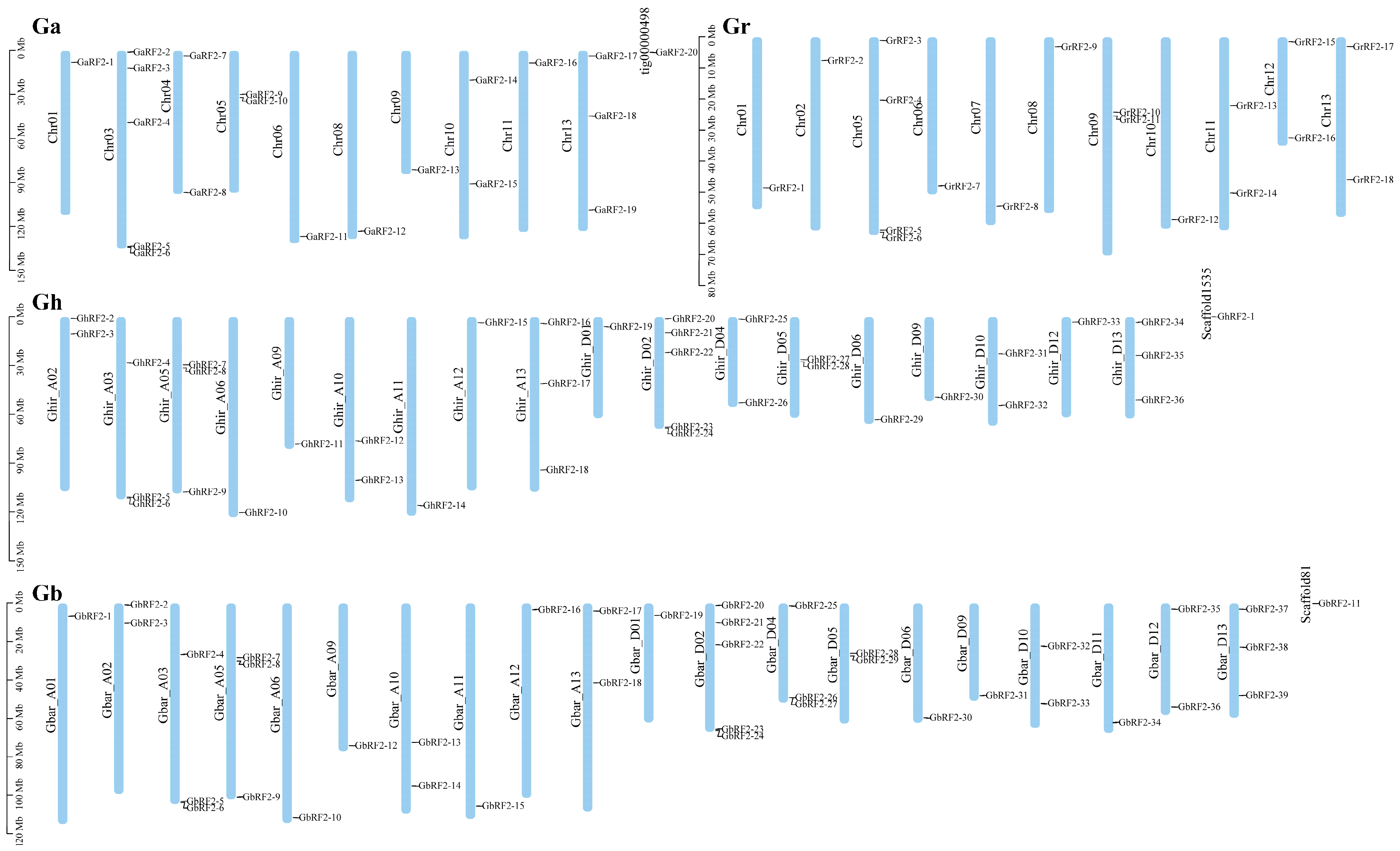
2.4. Distribution of RF2 Gene Family on Chromosomes in Cotton
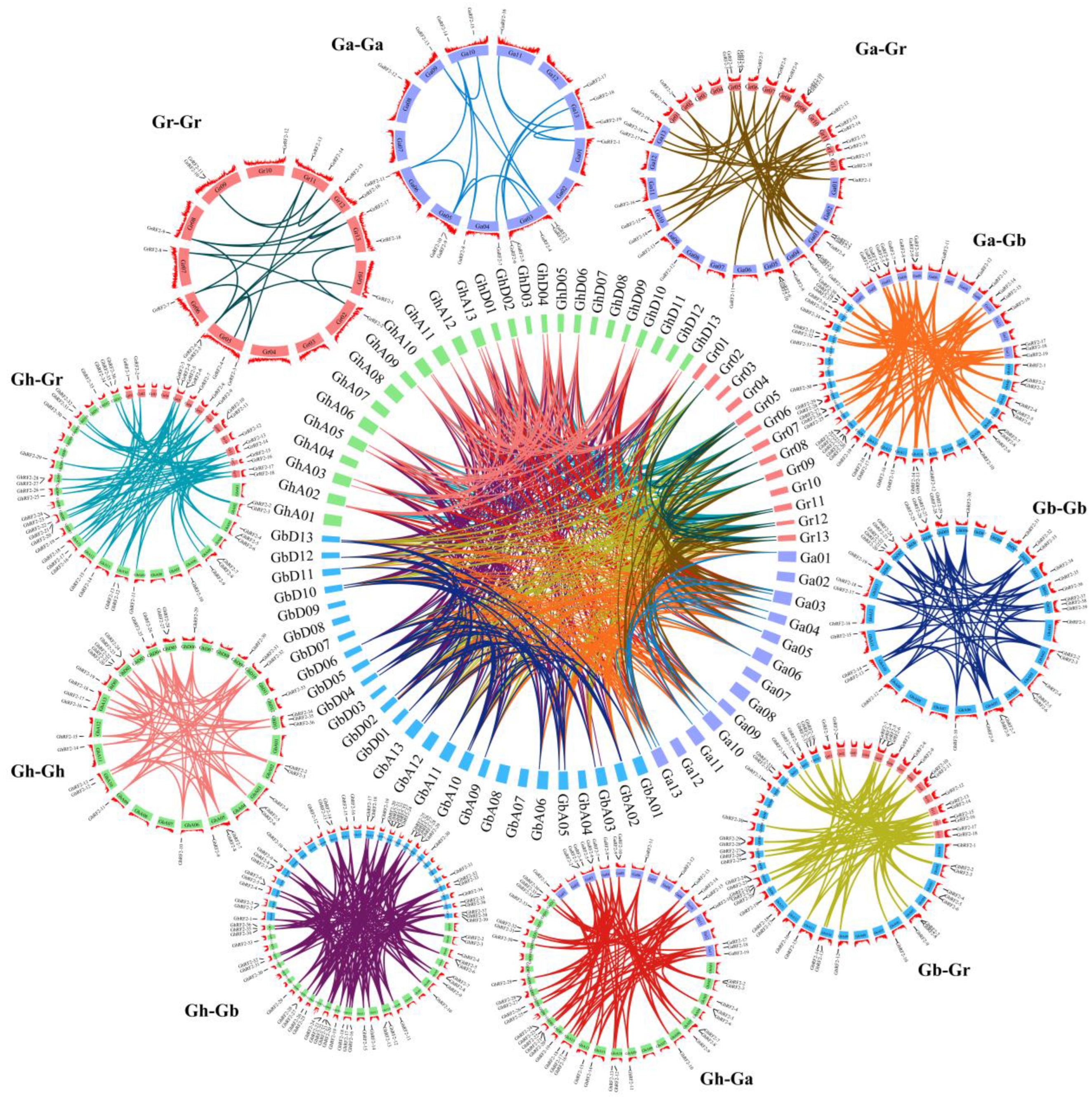
2.5. Intraspecific and Interspecific Collinearity Analysis of RF2 Gene in Cotton
2.6. Analysis of Cis Elements of RF2 Gene Promoters in Cotton
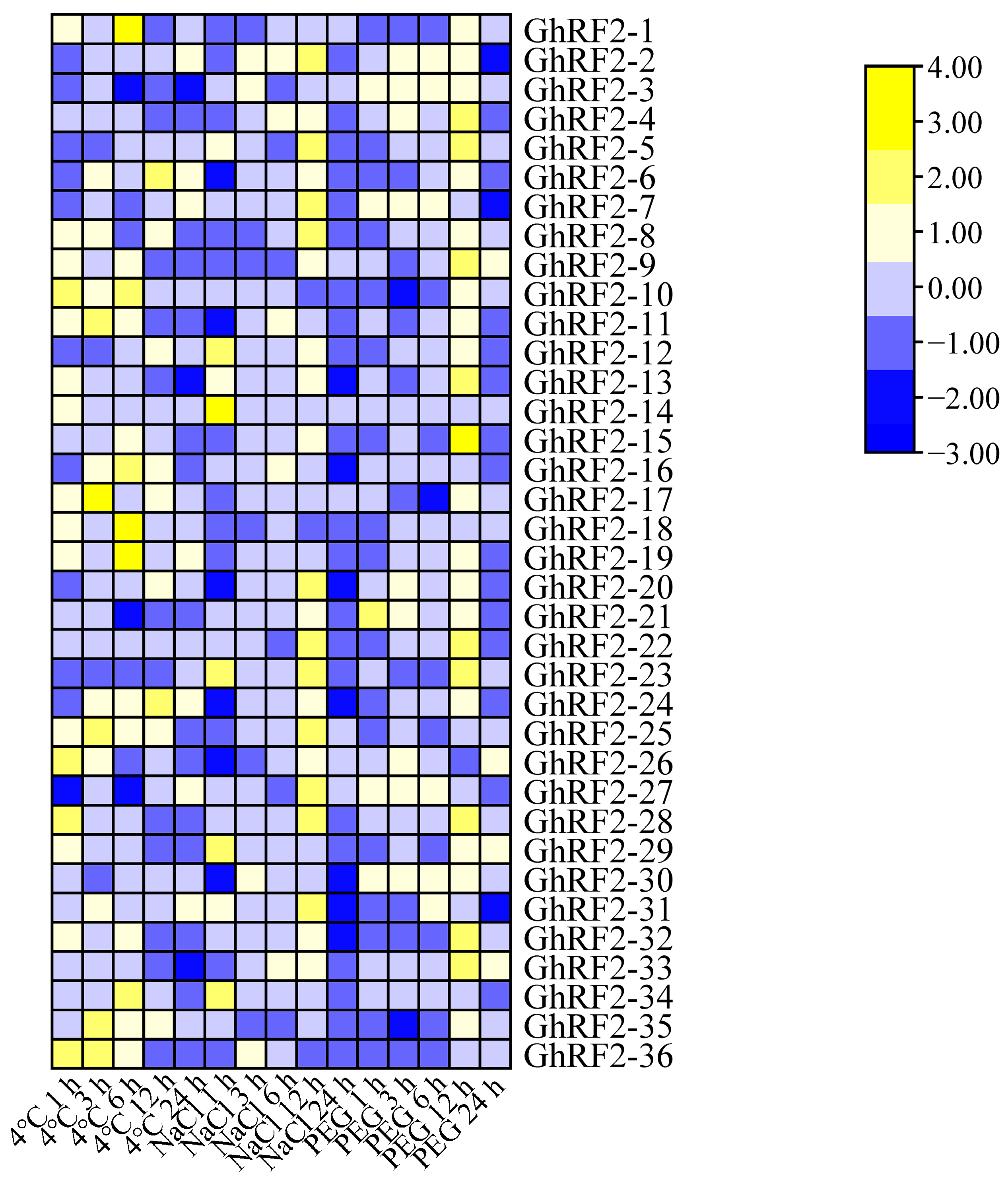
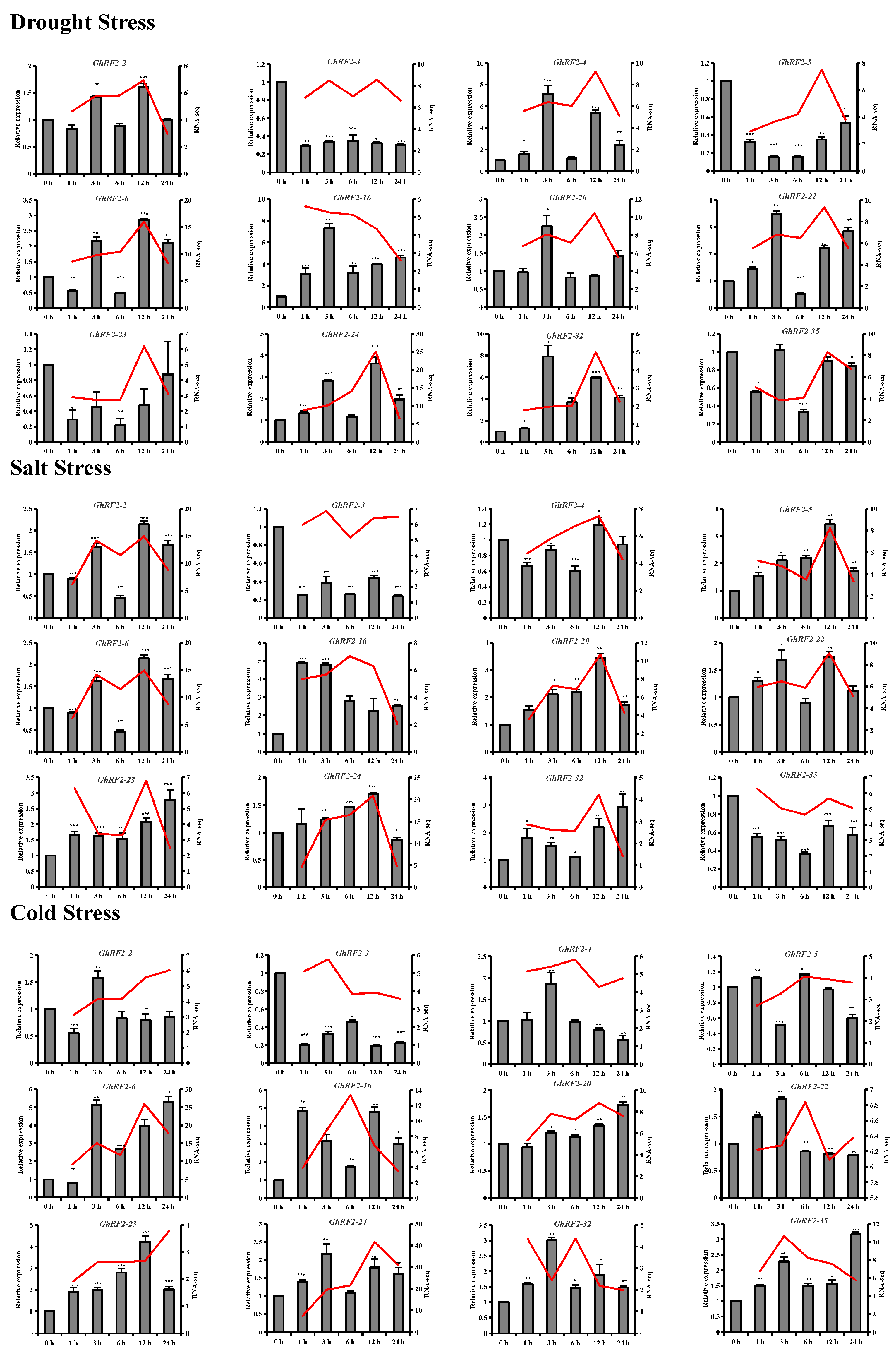
2.7. RF2 Gene Expression Patterns in Cotton under Abiotic Stress
2.8. qRT-PCR Analysis of RF2 Genes in Upland Cotton under Different Abiotic Stresses
2.9. Analysis of the Subcellular Localization of GhRF2-32
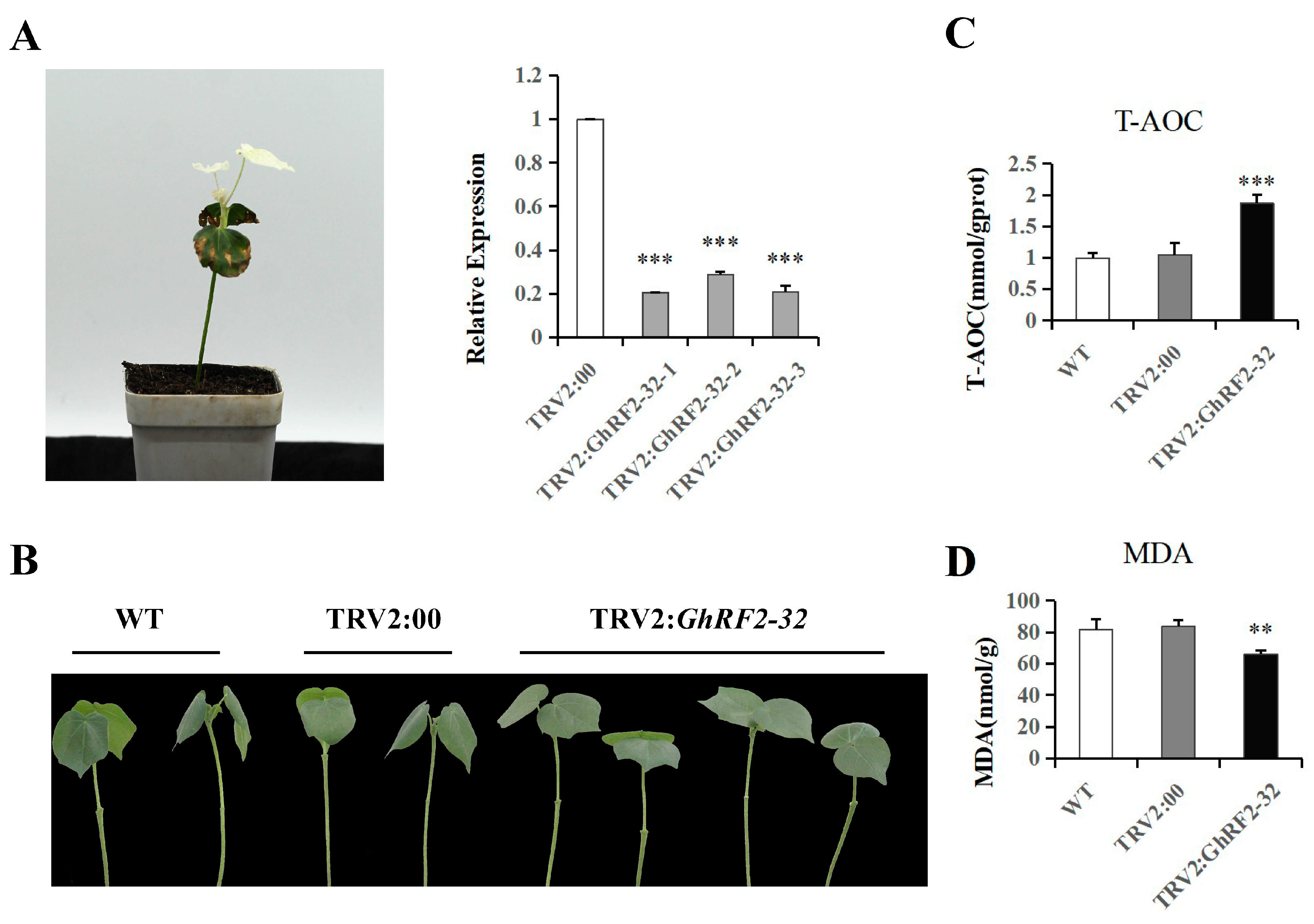
2.10. Virus-Induced Gene Silencing (VIGS) of GhRF2-32 in Upland Cotton
3. Materials and Methods
3.1. Identification and Physicochemical Property Analysis of RF2 Gene Family in Cotton
3.2. Phylogenetic Analysis of RF2 Gene in Cotton
3.3. Analysis of Conserved Motifs and Gene Structures of RF2 Genes in Cotton
3.4. Mapping RF2 Genes in Cotton Chromosome
3.5. Intraspecific and Interspecific Collinearity Analysis of RF2 Genes in Cotton
3.6. Prediction of Cis Elements of RF2 Gene in Cotton
3.7. Expression Pattern of RF2 under Abiotic Stress
3.8. Stress Treatments and qRT-PCR Analysis
3.9. Subcellular Localization of GhRF2-32
3.10. Using Virus-Induced Gene Silencing (VIGS) to Test RF2 Gene Function
3.11. Determination of Enzyme Activity in Gene-Silencing Cotton after Drought Stress Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Valliyodan, B.; Nguyen, H.T. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 2006, 9, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Fu, B.; Liu, Y.; Li, Y.; Wang, S.; Zhan, T.; Wang, Y.; Gao, D. Evaluation of ecosystem resilience to drought based on drought intensity and recovery time. Agric. For. Meteorol. 2022, 314, 108809. [Google Scholar] [CrossRef]
- Liang, W.; Cui, W.; Ma, X.; Wang, G.; Huang, Z. Function of wheat Ta-UnP gene in enhancing salt tolerance in transgenic Arabidopsis and rice. Biochem. Biophys. Res. Commun. 2014, 450, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; James, R.A.; Läuchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef]
- Theocharis, A.; Clément, C.; Barka, E.A. Physiological and molecular changes in plants grown at low temperatures. Planta 2012, 235, 1091–1105. [Google Scholar] [CrossRef]
- Corrêa, L.G.; Riaño-Pachón, D.M.; Schrago, C.G.; dos Santos, R.V.; Mueller-Roeber, B.; Vincentz, M. The role of bZIP transcription factors in green plant evolution: Adaptive features emerging from four founder genes. PLoS ONE 2008, 3, e2944. [Google Scholar] [CrossRef]
- Wang, X.; Lu, X.; Malik, W.A.; Chen, X.; Wang, J.; Wang, D.; Wang, S.; Chen, C.; Guo, L.; Ye, W. Differentially expressed bZIP transcription factors confer multi-tolerances in Gossypium hirsutum L. Int. J. Biol. Macromol. 2020, 146, 569–578. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.; Shi, H.; Guo, M.; Chai, M.; He, Q.; Yan, M.; Cao, D.; Zhao, L.; Cai, H.; et al. Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genom. 2018, 19, 159. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, D.; Jia, L.; Huang, X.; Ma, G.; Wang, S.; Zhu, M.; Zhang, A.; Guan, M.; Lu, K.; et al. Genome-Wide Identification and Structural Analysis of bZIP Transcription Factor Genes in Brassica napus. Genes 2017, 8, 288. [Google Scholar] [CrossRef] [PubMed]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Petruccelli, S.; Ordiz, M.I.; Zhang, Z.; Chen, S.; Beachy, R.N. Functional analysis of RF2a, a rice transcription factor. J. Biol. Chem. 2003, 278, 36396–36402. [Google Scholar] [CrossRef]
- Dai, S.; Zhang, Z.; Chen, S.; Beachy, R.N. RF2b, a rice bZIP transcription activator, interacts with RF2a and is involved in symptom development of rice tungro disease. Proc. Natl. Acad. Sci. USA 2004, 101, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Yang, Y.; Cao, L.; Liu, W.; Bi, C. RF2 basic leucine zipper transcription factor TabZIP3 involved in salt stress response in wheat. J. Agric. Sci. Technol. 2019, 21, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Xu, Y.; Tian, S.; Unver, T.; Liu, Z.; Zhou, Z.; Cai, X.; Wang, K.; Wei, Y.; Liu, Y.; et al. Evolutionary divergence of duplicated genomes in newlydescribed allotetraploid cottons. Proc. Natl. Acad. Sci. USA 2022, 119, e220849611. [Google Scholar]
- Peng, R.; Jones, D.C.; Liu, F.; Zhang, B.H. From sequencing to genome editing for cotton improvement. Trend Biotech 2021, 39, 221–224. [Google Scholar] [CrossRef]
- Zhao, Z.; Shuang, J.; Li, Z.; Xiao, H.; Liu, Y.; Wang, T.; Wei, Y.; Hu, S.; Wan, S.; Peng, R. Identification of the Golden-2-like transcription factors gene family in Gossypium hirsutum. PeerJ 2021, 9, e12484. [Google Scholar] [CrossRef]
- Prakash, S.; Kumar, M.; Radha; Kumar, S.; Jaconis, S.; Parameswari, E.; Sharma, K.; Dhumal, S.; Senapathy, M.; Deshmukh, V.P.; et al. The resilient cotton plant: Uncovering the effects of stresses on secondary metabolomics and its underlying molecular mechanisms. Funct. Integr. Genom. 2023, 23, 183. [Google Scholar] [CrossRef]
- Pourabed, E.; Ghane Golmohamadi, F.; Soleymani Monfared, P.; Razavi, S.M.; Shobbar, Z.S. Basic leucine zipper family in barley: Genome-wide characterization of members and expression analysis. Mol. Biotechnol. 2015, 57, 12–26. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J.; Chen, X.; Lu, X.; Wang, D.; Wang, J.; Wang, S.; Chen, C.; Guo, L.; Malik, W.A.; et al. Genome-wide identification and characteristic analysis of the downstream melatonin metabolism gene GhM2H in Gossypium hirsutum L. Biol. Res. 2021, 54, 36. [Google Scholar] [CrossRef]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019, 51, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Huang, G.; He, S.; Yang, Z.; Sun, G.; Ma, X.; Li, N.; Zhang, X.; Sun, J.; Liu, M.; et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 2018, 50, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Udall, J.A.; Long, E.; Hanson, C.; Yuan, D.; Ramaraj, T.; Conover, J.L.; Gong, L.; Arick, M.A.; Grover, C.E.; Peterson, D.G.; et al. De Novo Genome Sequence Assemblies of Gossypium raimondii and Gossypium turneri. G3 2019, 9, 3079–3085. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, J.; Huang, Y.; Wang, S.; Wei, L.; Liu, D.; Weng, Y.; Xiang, J.; Zhu, Q.; Yang, Z.; et al. CottonMD: A multi-omics database for cotton biological study. Nucleic Acids Res. 2023, 51, D1446–D1456. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Hao, X.D.; Liu, Y.; Li, B.W.; Wu, W.; Zhao, X.W. Exome sequencing analysis identifies novel homozygous mutation in ABCA4 in a Chinese family with Stargardt disease. Int. J. Ophthalmol. 2020, 13, 671–676. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Ghosh, S.; Chan, C.K. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods Mol. Biol. 2016, 1374, 339–361. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B.H. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F.; Udall, J.A. Duplicate gene evolution, homoeologous recombination, and transcriptome characterization in allopolyploid cotton. BMC Genom. 2012, 13, 302. [Google Scholar] [CrossRef]
- Senchina, D.S.; Alvarez, I.; Cronn, R.C.; Liu, B.; Rong, J.; Noyes, R.D.; Paterson, A.H.; Wing, R.A.; Wilkins, T.A.; Wendel, J.F. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 2003, 20, 633–643. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F.; bZIP Research Group. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Qiu, S.R.; Li, H.; Li, S.J.; Li, X.; Xiang, H.; Zeng, S.H.; Liu, Y.J. Identification of bZIP gene family and gene expression analysis of its subgroup A under ABA treatment in Nicotiana tabacum. Fenzi Zhiwu Yuzhong (Mol. Plant Breed.) 2020, 18, 5607–5621. [Google Scholar]
- Hajibarat, Z.; Saidi, A.; Hajibarat, Z. Genome-wide identification of 14-3-3 gene family and characterization of their expression in developmental stages of Solanum tuberosum under multiple biotic and abiotic stress conditions. Funct. Integr. Genom. 2022, 22, 1377–1390. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhang, H.; Hu, J.; Nie, G.; Khan, I.; Feng, G.; Zhang, X.; Wang, X.; Huang, L. Genome-wide identification and characterization of bHLH family genes from orchardgrass and the functional characterization of DgbHLH46 and DgbHLH128 in drought and salt tolerance. Funct. Integr. Genom. 2022, 22, 1331–1344. [Google Scholar] [CrossRef]
- Xing, L.; Peng, K.; Xue, S.; Yuan, W.; Zhu, B.; Zhao, P.; Wu, H.; Cheng, Y.; Fang, M.; Liu, Z. Genome-wide analysis of zinc finger-homeodomain (ZF-HD) transcription factors in diploid and tetraploid cotton. Funct. Integr. Genom. 2022, 22, 1269–1281. [Google Scholar] [CrossRef]
- Duan, P.; Wei, M.; Zhang, R.; Zhao, S.; Wang, Y.; Gou, B.; Yang, N.; Zhang, T.; Zhang, G.; Wei, B. Identification and bioinformatic analysis of the CaCesA/Csls family members and the expression of the CaCslD1 in the flower buds of CMS/Rf system in pepper. Funct. Integr. Genom. 2022, 22, 1411–1431. [Google Scholar] [CrossRef]
- Mallick, B.; Kumari, M.; Pradhan, S.K.; Acharya, G.C.; Naresh, P.; Das, B.; Shashankar, P. Genome-wide analysis and characterization of heat shock transcription factors (Hsfs) in common bean (Phaseolus vulgaris L.). Funct. Integr. Genom. 2022, 22, 743–756. [Google Scholar] [CrossRef]
- Cheng, Y.-Z.; He, G.-Q.; Yang, S.-D.; Ma, S.-H.; Ma, J.-P.; Shang, F.-H.-Z.; Li, X.-F.; Jin, H.-Y.; Guo, D.-L. Genome-wide identification and expression analysis of JmjC domain–containing genes in grape under MTA treatment. Funct. Integr. Genom. 2022, 22, 783–795. [Google Scholar] [CrossRef]
- Huang, G.; Wu, Z.; Percy, R.G.; Bai, M.; Li, Y.; Frelichowski, J.E.; Hu, J.; Wang, K.; Yu, J.Z.; Zhu, Y. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat. Genet. 2020, 52, 516–524. [Google Scholar] [CrossRef]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2014, 84, 19–36, Erratum in Plant Mol. Biol. 2018, 97, 467–468. [Google Scholar] [CrossRef]
- Luang, S.; Sornaraj, P.; Bazanova, N.; Jia, W.; Eini, O.; Hussain, S.S.; Kovalchuk, N.; Agarwal, P.K.; Hrmova, M.; Lopato, S. The wheat TabZIP2 transcription factor is activated by the nutrient starvation-responsive SnRK3/CIPK protein kinase. Plant Mol. Biol. 2018, 96, 543–561. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Zhang, D.F.; Fu, J.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 2012, 235, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Yang, Y.; Wang, W.; Guo, G.; Liu, W.; Bi, C. Overexpression of a wheat (Triticum aestivum L.) bZIP transcription factor gene, TabZIP6, decreased the freezing tolerance of transgenic Arabidopsis seedlings by down-regulating the expression of CBFs. Plant Physiol. Biochem. 2018, 124, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cao, H.; Qian, W.; Yao, L.; Hao, X.; Li, N.; Yang, Y.; Wang, X. Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic arabidopsis. Ann. Bot. 2017, 119, 1195–1209. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, B.H. The landscape of genome sequencing and assembling in plants. Funct. Integr. Genom. 2022, 22, 1147–1152. [Google Scholar] [CrossRef]
- Li, C.; Brant, E.; Budak, H.; Zhang, B.H. CRISPR/Cas: A Nobel Prize award-winning precise genome editing technology for gene therapy and crop improvement. J. Zhejiang Univ. Sci. B 2021, 22, 253–284. [Google Scholar] [CrossRef]
- Li, C.; Chu, W.; Gill, R.A.; Sang, S.; Shi, Y.; Hu, X.; Yang, Y.; Zaman, Q.U.; Zhang, B.H. Computational tools and resources for CRISPR/Cas genome editing. Genom. Proteom. Bioinform. 2022, in press. [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, H.; Zhao, Z.; Wei, Y.; Li, P.; Lu, Q.; Liu, Y.; Wang, T.; Hu, N.; Wan, S.; Zhang, B.; et al. Genome-Wide Identification and Functional Analysis of RF2 Gene Family and the Critical Role of GhRF2-32 in Response to Drought Stress in Cotton. Plants 2023, 12, 2613. https://doi.org/10.3390/plants12142613
Gu H, Zhao Z, Wei Y, Li P, Lu Q, Liu Y, Wang T, Hu N, Wan S, Zhang B, et al. Genome-Wide Identification and Functional Analysis of RF2 Gene Family and the Critical Role of GhRF2-32 in Response to Drought Stress in Cotton. Plants. 2023; 12(14):2613. https://doi.org/10.3390/plants12142613
Chicago/Turabian StyleGu, Haonan, Zilin Zhao, Yangyang Wei, Pengtao Li, Quanwei Lu, Yuling Liu, Tao Wang, Nan Hu, Sumei Wan, Baohong Zhang, and et al. 2023. "Genome-Wide Identification and Functional Analysis of RF2 Gene Family and the Critical Role of GhRF2-32 in Response to Drought Stress in Cotton" Plants 12, no. 14: 2613. https://doi.org/10.3390/plants12142613
APA StyleGu, H., Zhao, Z., Wei, Y., Li, P., Lu, Q., Liu, Y., Wang, T., Hu, N., Wan, S., Zhang, B., Hu, S., & Peng, R. (2023). Genome-Wide Identification and Functional Analysis of RF2 Gene Family and the Critical Role of GhRF2-32 in Response to Drought Stress in Cotton. Plants, 12(14), 2613. https://doi.org/10.3390/plants12142613






