The Key Role of Glutamate Dehydrogenase 2 (GDH2) in the Control of Kernel Production in Maize (Zea mays L.)
Abstract
1. Introduction
2. Results
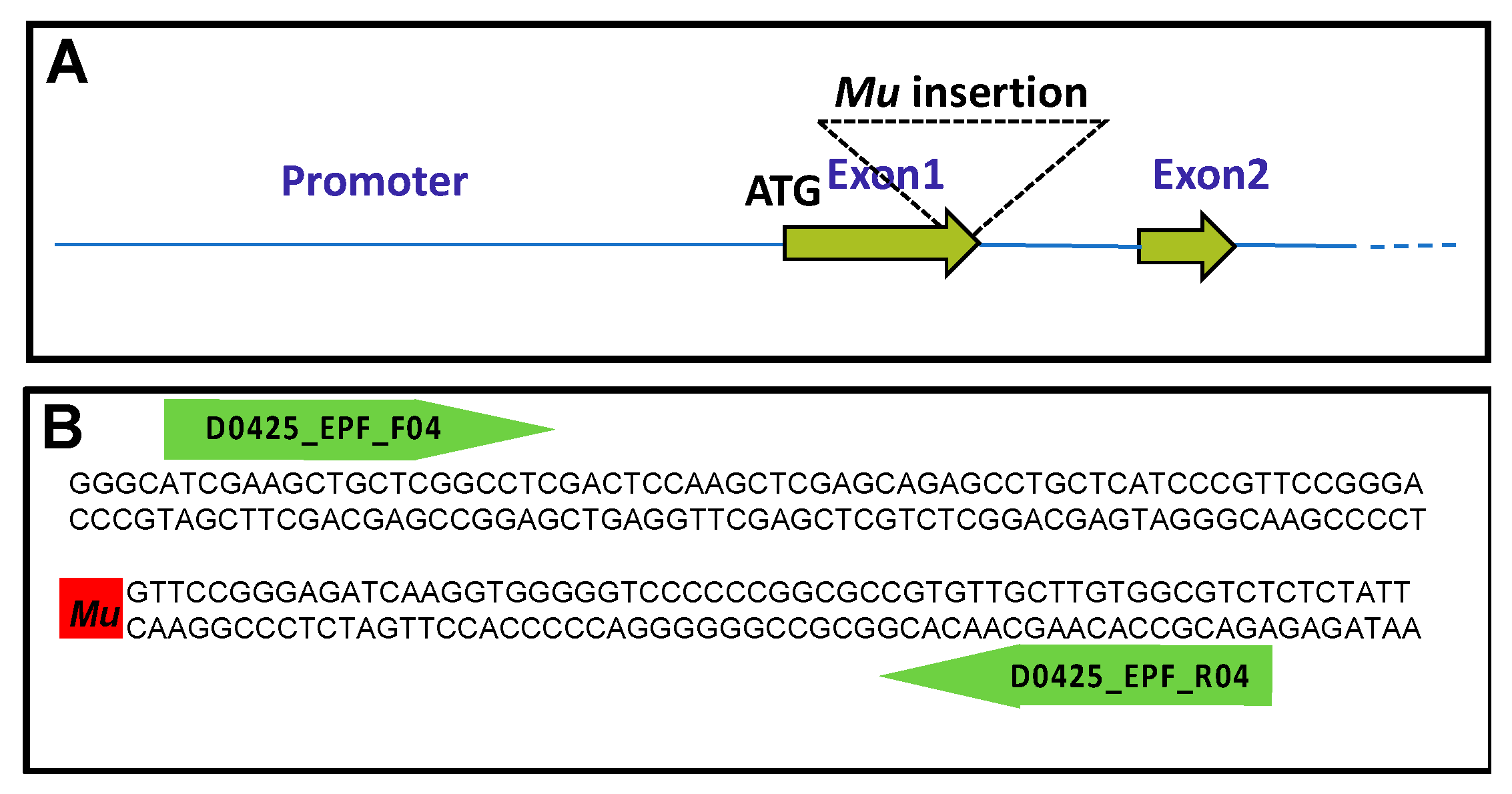
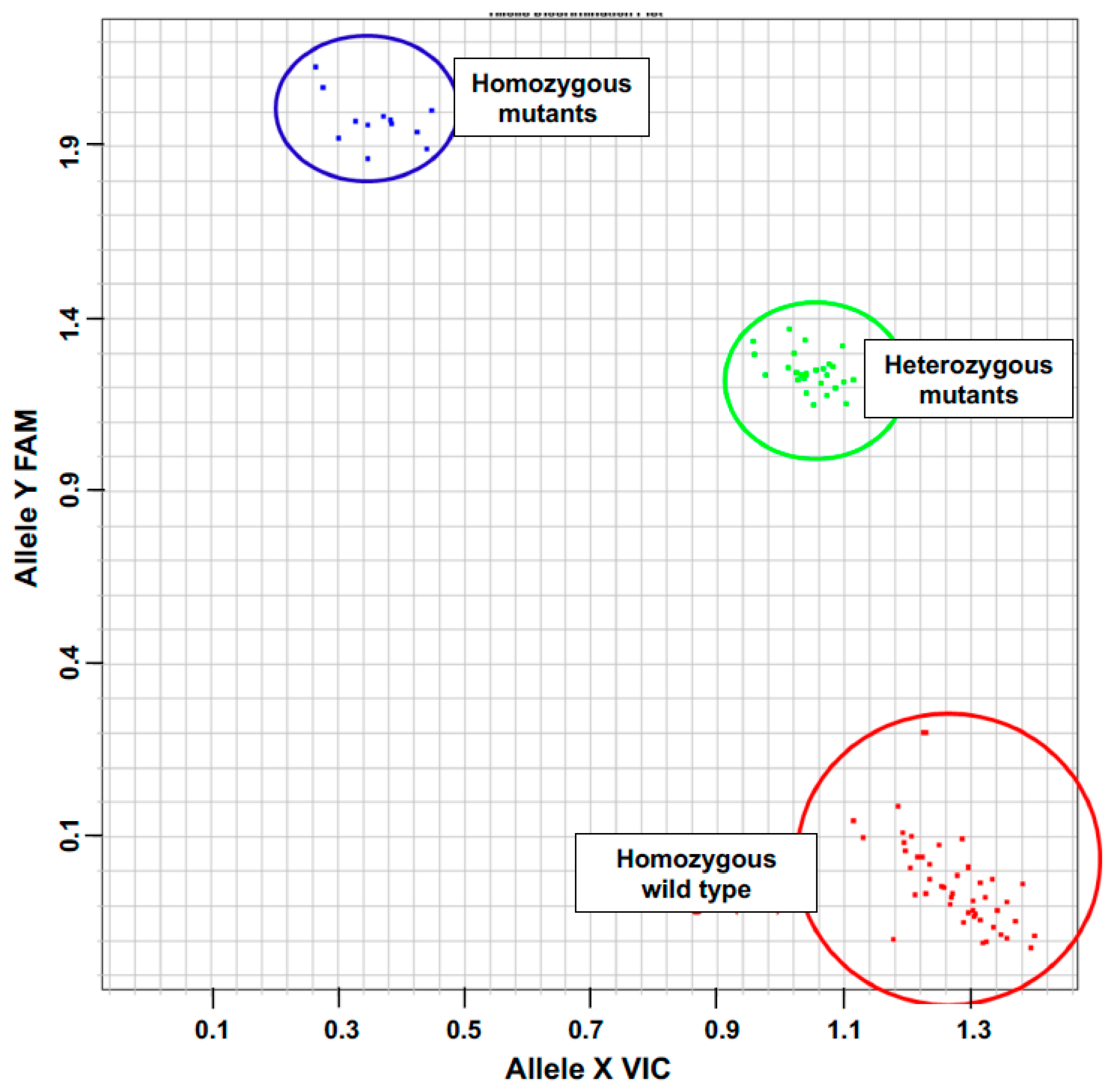
2.1. Isolation and Characterization of a gdh2-Deficient Mutant
2.2. Glutamate Dehydrogenase Protein Content and Isoenzyme Composition in the gdh2 Mutant
2.3. Metabolic Profile of the gdh2 Mutant
2.4. Mutant Phenotype and Biomass Production
3. Discussion
4. Materials and Methods
4.1. Isolation and Characterization of the gdh2 Mutant
4.2. Plant Material for Molecular, Physiological and Agronomic Studies
4.3. Enzymatic In Vitro and In-Gel Assay, Determination of Total Soluble Protein and Protein Gel Blot Analysis
4.4. Metabolome Analysis
4.5. Statistics
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hirel, B.; Lea, P.J. Ammonia assimilation. In Plant Nitrogen; Lea, P.J., Morot-Gaudry, J.F., Eds.; INRA Editions; Springer: Berlin/Heidelberg, Germany, 2001; pp. 79–99. [Google Scholar]
- Dubois, F.; Tercé-Laforgue, T.; Gonzalez-Moro, M.B.; Estavillo, M.B.; Sangwan, R.; Gallais, A.; Hirel, B. Glutamate dehydrogenase in plants: Is there a new story for an old enzyme? Plant Physiol. Biochem. 2003, 41, 565–576. [Google Scholar] [CrossRef]
- Skopelitis, D.; Paranychiankis, N.V.; Paschalidis, K.A.; Plianokis, E.D.; Delis, I.D.; Yakoumakis, D.I.; Kouvarakis, A.; Papadakis, A.; Stephanou, E.G.; Roubelakis-Angelakis, K.A. Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenase to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 2006, 18, 2767–2781. [Google Scholar] [CrossRef] [PubMed]
- Bittsánsky, A.; Pilinsky, K.; Gyulai, G.; Komives, T. Overcoming ammonium toxicity. Plant Sci. 2015, 231, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, Y.; Good, A.G. NAD (H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J. Exp. Bot. 2008, 59, 667–680. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, J.X.; Tercé-Laforgue, T.; Armengaud, P.; Clément, G.; Renou, J.P.; Pelletier, S.; Catterou, M.; Azzopardi, M.; Gibon, Y.; Lea, P.J.; et al. Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell 2012, 24, 4044–4065. [Google Scholar] [CrossRef]
- Robinson, S.A.; Stewart, G.R.; Phillips, R. Regulation of glutamate dehydrogenase activity in relation to carbon limitation and protein catabolism in carrot cell suspension cultures. Plant Physiol. 1992, 98, 1190–1195. [Google Scholar] [CrossRef]
- Labboun, S.; Tercé-Laforgue, T.; Roscher, A.; Bedu, M.; Restivo, F.M.; Velanis, C.N.; Skopelitis, D.S.; Moshou, P.N.; Roubelakis-Angelakis, K.A.; Suzuki, A.; et al. Resolving the role of plant glutamate dehydrogenase: I. In vivo real time nuclear magnetic resonance spectroscopy experiments. Plant. Cell Physiol. 2009, 50, 1761–1773. [Google Scholar] [CrossRef]
- Fontaine, J.X.; Tercé-Laforgue, T.; Bouton, S.; Pageau, K.; Lea, P.J.; Dubois, F.; Hirel, B. Further insights into the isoenzyme composition and activity of glutamate dehydrogenase in Arabidopsis thaliana. Plant Signal. Behav. 2013, 8, e233291-5. [Google Scholar] [CrossRef]
- Qiu, X.; Xie, W.; Lian, X.; Zhang, Q. Molecular analyses of the rice glutamate dehydrogenase gene family and their response to nitrogen and phosphorous deprivation. Plant Cell Rep. 2009, 28, 1115–1126. [Google Scholar] [CrossRef]
- Turano, F.J.; Thakkar, S.S.; Fang, T.; Weisemann, J.M. Characterization and expression of NAD (H)-dependent glutamate dehydrogenase genes in Arabidopsis. Plant Physiol. 1997, 113, 1329–1341. [Google Scholar] [CrossRef]
- Marchi, L.; Degola, F.; Polverini, E.; Tercé-Laforgue, T.; Dubois, F.; Hirel, B.; Restivo, F. Glutamate dehydrogenase isoenzyme 3 (GDH3) of Arabidopsis thaliana is regulated by a combined effect of nitrogen and cytokinin. Plant Physiol. Biochem. 2013, 73, 368–374. [Google Scholar] [CrossRef]
- Miflin, B.J.; Habash, D.Z. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 2002, 53, 979–987. [Google Scholar] [CrossRef]
- Tercé-Laforgue, T.; Bedu, M.; Dargel-Graffin, C.; Dubois, F.; Gibon, Y.; Restivo, F.M.; Hirel, B. Resolving the role of plant glutamate dehydrogenase: II. Physiological Characterization of plants overexpressing individually or simultaneously the two enzyme subunits. Plant. Cell Physiol. 2013, 54, 1634–1647. [Google Scholar] [CrossRef]
- Forde, B.G.; Lea, P.J. Glutamate in plants: Metabolism, regulation, and signaling. J. Exp. Bot. 2007, 58, 2339–2358. [Google Scholar] [CrossRef] [PubMed]
- Tercé-Laforgue, T.; Dubois, F.; Ferrario-Mery, S.; Pou de Crecenzo, M.A.; Sangwan, R.; Hirel, B. Glutamate dehydrogenase of tobacco (Nicotiana tabacum L.) is mainly induced in the cytosol of phloem companion cells when ammonia is provided either externally or released during photorespiration. Plant Physiol. 2004, 136, 4308–4317. [Google Scholar] [CrossRef] [PubMed]
- Cañas, R.A.; Yesbergenova-Cuny, Z.; Belanger, L.; Rouster, J.; Brulé, L.; Gilard, F.; Quilleré, I.; Sallaud, C.; Hirel, B. NADH-GOGAT overexpression does not improve maize (Zea mays L.) performance even when pyramiding with NAD-IDH, GDH and GS. Plants 2020, 9, 130. [Google Scholar] [CrossRef]
- Schmidt, R.R.; Miller, P. Polypeptides and Polynucleotides Relating to the α and β Subunits of a Glutamate Dehydrogenase and Methods of Use. U.S. Patent 5,879,941, 9 March 1999. [Google Scholar]
- Lightfoot, D.A.; Mungur, R.; Ameziane, R.; Nolte, S.; Long, L.; Bernhard, K.; Colter, A.; Jones, K.; Iqbal, M.J.; Varsa, E.; et al. Improved drought tolerance of transgenic Zea mays plants that express the glutamate dehydrogenase gene (gdhA) of E. coli. Euphytica 2007, 156, 103–116. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, H.; Zhou, X.; Yan, Y.; Du, C.; Li, Y.; Liu, D.; Zhang, C.; Deng, X.; Tang, D.; et al. Over-expression of a fungal NADP (H)-dependent glutamate dehydrogenase Pc GDH improves nitrogen assimilation and growth quality in rice. Mol. Breed. 2014, 34, 335–349. [Google Scholar] [CrossRef]
- Gallais, A.; Hirel, B. An approach to the genetics of nitrogen use efficiency in maize. J. Exp. Bot. 2004, 55, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Purnell, M.P.; Stewart, G.R.; Botella, J.R. Cloning and characterisation of a glutamate dehydrogenase cDNA from tomato (Lycopersicon esculentum L.). Gene 1997, 186, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Restivo, F.M. Molecular cloning of glutamate dehydrogenase genes of Nicotiana plumbaginifolia: Structure analysis and regulation of their expression by physiological and stress conditions. Plant Sci. 2004, 166, 971–982. [Google Scholar] [CrossRef]
- Taranto, F.; D’Agostino, N.; Rodriguez, M.; Pavan, S.; Minervini, A.P.; Pecchioni, N.; Papa, R.; De Vita, P. Whole genome scan reveals molecular signatures of divergence and selection related to important traits in durum wheat germplasm. Front. Plant Sci. 2020, 11, 217. [Google Scholar] [CrossRef] [PubMed]
- Purcino, A.A.C.; Arellano, C.; Athwal, G.S.; Huber, S.C. Nitrate effect on carbon and nitrogen assimilating enzymes of maize hybrids representing seven eras of breeding. Maydica 1998, 43, 83–94. [Google Scholar]
- Pryor, A.J.A. Maize glutamic dehydrogenase null mutant is cold temperature sensitive. Maydica 1990, 35, 367–372. [Google Scholar]
- Hanley, S.; Edwards, D.; Stevenson, D.; Haines, S.; Hegarty, M.; Schuch, W.; Edwards, K.J. Identification of transposon-tagged genes by the random sequencing of Mutator-tagged DNA fragments from Zea mays. Plant J. 2000, 23, 557–566. [Google Scholar] [CrossRef]
- Jiao, Y.; Peluso, P.; Shi, J.; Liang, T.; Stitzer, M.C.; Wang, B.; Campbell, M.S.; Stein, J.C.; Wei, X.; Chin, C.S.; et al. Improved maize reference genome with single-molecule technologies. Nature 2017, 546, 524–527. [Google Scholar] [CrossRef]
- Sakakibara, H.; Fujii, K.; Sugiyama, T. Isolation and characterization of a cDNA that encodes maize glutamate dehydrogenase. Plant Cell Physiol. 1995, 36, 789–797. [Google Scholar] [CrossRef]
- Loulakakis, K.A.; Roubelakis-Angelakis, K.A. Immunocharacterization of NADH-glutamate dehydrogenase from Vitis vinifera L. Plant Physiol. 1990, 94, 109–113. [Google Scholar] [CrossRef]
- Eprintsev, A.T.; Anokhina, G.B. The Role of Methylation of Glutamate Dehydrogenase Gene Promoters (GDH1 and GDH2) in the Regulation of Their Expression in Corn Leaves under Hypoxia. Russ. J. Plant Physiol. 2023, 70, 1–9. [Google Scholar] [CrossRef]
- Osuji, G.O.; Mangaroo, A.S.; Reyes, J.; Bulgin, A.; Wright, V. Biomass enhancement in maize and soybean in response to glutamate dehydrogenase isomerization. Biol. Plant. 2003, 47, 45–52. [Google Scholar] [CrossRef]
- Bado, S.; Forster, B.P.; Nielen, S.; Ali, A.M.; Lagoda, P.J.; Till, B.J.; Laimer, M. Plant mutation breeding: Current progress and future assessment. Plant Breed. Rev. 2015, 39, 23–88. [Google Scholar]
- Pandit, R.; Bhusal, B.; Regmi, R.; Neupane, P.; Bhattarai, K.; Maharjan, B.; Acharya, S.; Bigyan, K.C.; Poudel, M.R. Mutation breeding for crop improvement: A review. Review Food Agr. 2021, 2, 31–35. [Google Scholar] [CrossRef]
- Fox, T.; DeBruin, J.; Haug Collet, K.; Trimnell, M.; Clapp, J.; Leonard, A.; Li, B.; Scolaro, E.; Collinson, S.; Glassman, K.; et al. A single point mutation in Ms44 results in dominant male sterility and improves nitrogen use efficiency in maize. Plant Biotechnol. J. 2017, 15, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Alfatih, A.; Wu, J.; Zhang, Z.S.; Xia, J.Q.; Jan, S.U.; Yu, L.H.; Xiang, C.B. Rice NIN-LIKE PROTEIN 1 rapidly responds to nitrogen deficiency and improves yield and nitrogen use efficiency. J. Exp. Bot. 2020, 71, 6032–6042. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Li, S.; Li, J.; Yan, L.; Xia, L. Increasing yield potential through manipulating of an ARE1 ortholog related to nitrogen use efficiency in wheat by CRISPR/Cas9. J. Integr. Plant Biol. 2021, 63, 1649–1663. [Google Scholar] [CrossRef]
- Mungur, R.; Glass, A.D.M.; Goodenow, D.B.; Lightfoot, D.A. Metabolite fingerprinting in transgenic Nicotiana tabacum altered by the Escherichia coli glutamate dehydrogenase gene. J. Biomed. Biotechnol. 2005, 2005, 198. [Google Scholar] [CrossRef]
- Cañas, R.A.; Quilleré, I.; Lea, P.J.; Hirel, B. Analysis of amino acid metabolism in the ear of maize mutants deficient in two cytosolic glutamine synthetase isoenzymes highlights the importance of asparagine for nitrogen translocation within sink organs. Plant Biotechnol. J. 2010, 8, 966–978. [Google Scholar] [CrossRef]
- Boller, T. Peptide signalling in plant development and self/non-self-perception. Curr. Opin. Cell Biol. 2005, 17, 116–122. [Google Scholar] [CrossRef]
- Yu, Y.; Qian, Y.; Jiang, M.; Xu, J.; Yang, J.; Zhang, T.; Gou, L.; Pi, E. Regulation mechanisms of plant basic leucine zippers to various abiotic stresses. Front. Plant Sci. 2020, 11, 1258. [Google Scholar] [CrossRef]
- Cañas, R.A.; Yesbergenova-Cuny, Z.; Simons, M.; Chardon, F.; Armengaud, P.; Quilleré, I.; Cukier, C.; Gibon, Y.; Limami, A.M.; Nicolas, S.; et al. Exploiting the genetic diversity of maize using a combined metabolomic, enzyme activity profiling, and metabolic modeling approach to link leaf physiology to kernel yield. Plant Cell 2017, 29, 919–943. [Google Scholar] [CrossRef]
- Urrutia, M.; Blein-Nicolas, M.; Fernandez, O.; Bernillon, S.; Maucourt, M.; Deborde, C.; Balliau, T.; Rabier, D.; Bénard, C.; Prigent, S.; et al. Identification of Metabolic and Protein Markers Representative of the Impact of Mild Nitrogen Deficit on Agronomic Performance of Maize Hybrids. 2023. Available online: https://www.researchsquare.com/article/rs-2591494/v1 (accessed on 4 June 2023).
- Broyart, C.; Fontaine, J.X.; Molinié, R.; Cailleu, D.; Tercé-Laforgue, T.; Dubois, F.; Hirel, B.; Mesnard, F. Metabolic profiling of maize mutants deficient for two glutamine synthetase isoenzymes using 1H-NMR-based metabolomics. Phytochemical Analysis: An International. J. Plant Chem. Biochem. Technol. 2010, 21, 102–109. [Google Scholar]
- Yang, X.S.; Wu, J.; Ziegler, T.E.; Yang, X.; Zayed, A.; Rajani, M.S.; Zhou, D.; Basra, A.; Schachtman, D.; Peng, M.; et al. Gene expression biomarkers provide sensitive indicators of in planta nitrogen status in maize. Plant Physiol. 2011, 1574, 1841–1852. [Google Scholar] [CrossRef] [PubMed]
- Amiour, N.; Imbaud, S.; Clément, G.; Agier, N.; Zivy, M.; Valot, B.; Balliau, T.; Armengaud, P.; Quilleré, I.; Cañas, R.A.; et al. The use of metabolomics integrated with transcriptomic and proteomic studies for identifying key steps involved in the control of nitrogen metabolism in crops such as maize. J. Exp. Bot. 2012, 63, 5017–5033. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.Q.; Deng, F.; Zhou, P.; Yan, J.B.; Wang, Q.F.; Yang, R.C.; Li, X.Q. QTL mapping for the tocopherols at milk stage of kernel development in sweet corn. Euphytica 2013, 193, 409–417. [Google Scholar] [CrossRef]
- Bailey, D.W.; Attia, Z.; Reinert, S.; S Hulke, B.; Kane, N.C. Effective strategies for isolating DNA from members of Asteraceae with high concentrations of secondary metabolites. BioTechniques 2022, 72, 85–89. [Google Scholar] [CrossRef]
- Bertin, P.; Gallais, A. Physiological and genetic basis of nitrogen use efficiency in maize. I. Agrophysiological results. Maydica 2000, 45, 53–66. [Google Scholar]
- Bertin, P.; Gallais, A. Physiological and genetic basis of nitrogen use efficiency in maize. II. QTL detection and coincidences. Maydica 2001, 46, 53–68. [Google Scholar]
- Turano, F.J.; Dashner, R.; Upadhayaya, A.; Caldwell, C.R. Purification of mitochondrial glutamate dehydrogenase from dark-grown soybean seedlings. Plant Physiol. 1996, 112, 1357–1364. [Google Scholar] [CrossRef]
- Loulakakis, K.A.; Roubelakis-Angelakis, K.A. Characterization of Vitis vinifera L. glutamine synthetase and molecular cloning of cDNAs for the cytosolic enzyme. Plant Mol. Biol. 1996, 31, 983–992. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolite profiling in Arabidopsis. In Methods in Molecular Biology, Arabidopsis Protocols, 2nd ed.; Salinas, J., Sanchez-Serrano, J.J., Totowa, N.J., Eds.; Humana Press: Totowa, NJ, USA, 2006; pp. 439–447. [Google Scholar]



| Heterozygous | Homozygous | ||||||
|---|---|---|---|---|---|---|---|
| Metabolite | WT | m | FC | t-Test | mm | FC | t-Test |
| Alanine | 0.3140 | 0.2356 | 0.75 | 0.0002 | 0.2430 | 0.77 | 0.0403 |
| Allantoin | 0.0019 | 0.0006 | 0.57 | 0.0313 | nc | nc | nc |
| Arginine | 0.0437 | 0.0217 | 0.51 | 0.0000 | 0.0246 | 0.58 | 0.0014 |
| Asparagine | 0.1336 | 0.0692 | 0.59 | 0.0062 | nc | nc | nc |
| Aspartate | 0.2034 | 0.1643 | 0.81 | 0.0018 | nc | nc | nc |
| Glutamate | 0.6834 | 0.5697 | 0.84 | 0.0343 | nc | nc | nc |
| Glutamine | 0.4184 | 0.3329 | 0.82 | 0.0495 | 0.3310 | 0.81 | 0.0426 |
| Homoserine | 0.0002 | 0.0001 | 0.85 | 0.0593 | nc | nc | nc |
| Leucine | 0.0314 | 0.0275 | 0.87 | 0.0046 | nc | nc | nc |
| Lysine | 0.0091 | 0.0058 | 0.69 | 0.0230 | nc | nc | nc |
| Phenylalanine | 0.0174 | 0.0146 | 0.84 | 0.0278 | nc | nc | nc |
| Threonine | 0.0467 | 0.0393 | 0.85 | 0.0205 | 0.0393 | 0.85 | 0.0512 |
| Tryptophan | 0.0052 | 0.0042 | 0.84 | 0.0698 | nc | nc | nc |
| Tyrosine | 0.0500 | 0.0427 | 0.86 | 0.0133 | nc | nc | nc |
| Galactose | 0.0081 | 0.0064 | 0.81 | 0.0220 | 0.0065 | 0.82 | 0.0399 |
| Myo-Inositol | 0.0209 | 0.0136 | 0.66 | 0.0001 | nc | nc | nc |
| Ribose | 0.0274 | 0.0226 | 0.84 | 0.0421 | 0.0219 | 0.82 | 0.0251 |
| Digalactosylglycerol | 0.0010 | 0.0013 | 1.27 | 0.0078 | 0.0015 | 1.51 | 0.0129 |
| Heterozygous | Homozygous | ||||||
|---|---|---|---|---|---|---|---|
| Metabolite | WT | m | FC | t-Test | mm | FC | t-Test |
| Arabitol | 0.0052 | 0.0039 | 0.76 | 0.0014 | nc | nc | nc |
| Asparagine | 0.1695 | 0.0308 | 0.45 | 0.0501 | nc | nc | nc |
| α-Amyrin | 0.0020 | 0.0011 | 0.64 | 0.0293 | nc | nc | nc |
| Erythronate | 0.0004 | 0.0002 | 0.78 | 0.0424 | nc | nc | nc |
| Ethanolamine | 0.0315 | 0.0235 | 0.75 | 0.0004 | 0.0260 | 0.83 | 0.0260 |
| Galactosylglycerol | 0.0008 | 0.0005 | 0.63 | 0.0027 | 0.0006 | 0.76 | 0.0399 |
| 3-P-Glycerate | 0.0013 | 0.0008 | 0.65 | 0.0055 | nc | nc | nc |
| Mannitol | 0.0030 | 0.0021 | 0.69 | 0.0002 | nc | nc | nc |
| Myo-Inositol | 0.0041 | 0.0032 | 0.83 | 0.0529 | 0.4543 | 1.22 | 0.0341 |
| Sorbitol | 0.0039 | 0.0030 | 0.80 | 0.0629 | nc | nc | nc |
| 1-3-Diaminopropane | 0.0002 | 0.0003 | 1.34 | 0.0490 | nc | nc | nc |
| 3-trans-Caffeoylquinate- | 0.0003 | 0.0006 | 3.23 | 0.0077 | 0.0001 | 1.28 | 0.0738 |
| 4-cis-Hydroxycinnamate | 0.0001 | 0.0001 | 1.18 | 0.0595 | 0.0004 | 1.42 | 0.0234 |
| Caffeate | 0.0023 | 0.0028 | 1.26 | 0.0744 | nc | nc | nc |
| Citrate | 0.4600 | 0.5606 | 1.24 | 0.0165 | nc | nc | nc |
| Cystein | 0.0338 | 0.0450 | 1.61 | 0.0371 | nc | nc | nc |
| Dopamine | 0.0001 | 0.0002 | 1.38 | 0.0477 | nc | nc | nc |
| Trans-Ferulate | 0.0001 | 0.0002 | 1.74 | 0.0345 | 0.0003 | 2.39 | 0.0342 |
| Leucine | 0.0061 | 0.0101 | 1.84 | 0.0407 | nc | nc | nc |
| Phytol-2 | 0.0005 | 0.0006 | 1.36 | 0.0148 | nc | nc | nc |
| Quinate | 0.0322 | 0.0586 | 1.96 | 0.0012 | 0.0452 | 1.51 | 0.0146 |
| Shikimate | 0.0285 | 0.0510 | 1.89 | 0.0010 | 0.0391 | 1.45 | 0.0224 |
| Tyrosine | 0.0189 | 0.0223 | 1.21 | 0.0707 | nc | nc | nc |
| Plant Height (cm) | Shoot DW (g) | KY (g) | KN | TKW (g) | |
|---|---|---|---|---|---|
| Year 2010 | |||||
| WT | 181 ± 1.6 | 51.4 ± 2.6 | 29.9 ± 1.6 | 189 ± 10 | 162 ± 14.3 |
| gdh2 (m) | 181 ± 3.5 | 73 ± 5.9 (42) a | 40 ±5.4 (34) a | 247 ± 15 (30) a | 158 ± 13 |
| gdh2 (mm) | 180± 4.8 | 63 ± 5.8 (23) | 26.6 ± 0.8 | 195 ± 3 | 138 ± 5 |
| Year 2013 | |||||
| WT | 137± 3.6 | 84 ± 7 | 36.6 ± 5.1 | 156 ± 21 | 237 ± 7.7 |
| gdh2 (m) | 141 ± 5.5 | 106 ± 7.4 (26) a | 50.4 ± 2.6 (38) a | 216± 12 (38) a | 235 ± 8.0 |
| gdh2 (mm) | 131 ± 5.4 | 73.0 ± 10 | 31.5 ± 4.2 | 133 ± 20 | 246 ± 33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tercé-Laforgue, T.; Lothier, J.; Limami, A.M.; Rouster, J.; Lea, P.J.; Hirel, B. The Key Role of Glutamate Dehydrogenase 2 (GDH2) in the Control of Kernel Production in Maize (Zea mays L.). Plants 2023, 12, 2612. https://doi.org/10.3390/plants12142612
Tercé-Laforgue T, Lothier J, Limami AM, Rouster J, Lea PJ, Hirel B. The Key Role of Glutamate Dehydrogenase 2 (GDH2) in the Control of Kernel Production in Maize (Zea mays L.). Plants. 2023; 12(14):2612. https://doi.org/10.3390/plants12142612
Chicago/Turabian StyleTercé-Laforgue, Thérèse, Jérémy Lothier, Anis M. Limami, Jacques Rouster, Peter J. Lea, and Bertrand Hirel. 2023. "The Key Role of Glutamate Dehydrogenase 2 (GDH2) in the Control of Kernel Production in Maize (Zea mays L.)" Plants 12, no. 14: 2612. https://doi.org/10.3390/plants12142612
APA StyleTercé-Laforgue, T., Lothier, J., Limami, A. M., Rouster, J., Lea, P. J., & Hirel, B. (2023). The Key Role of Glutamate Dehydrogenase 2 (GDH2) in the Control of Kernel Production in Maize (Zea mays L.). Plants, 12(14), 2612. https://doi.org/10.3390/plants12142612








