Genome-Wide Association Study of Healthful Flavonoids among Diverse Mandarin Accessions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mapping Population
2.2. Chemical Reagents and Standard Compounds
2.3. Sample Preparation and Flavonoid Extraction
2.4. Analysis of Flavonoids with LC-MS/MS and Peak Detection and Quantification
2.5. GWAS
3. Results
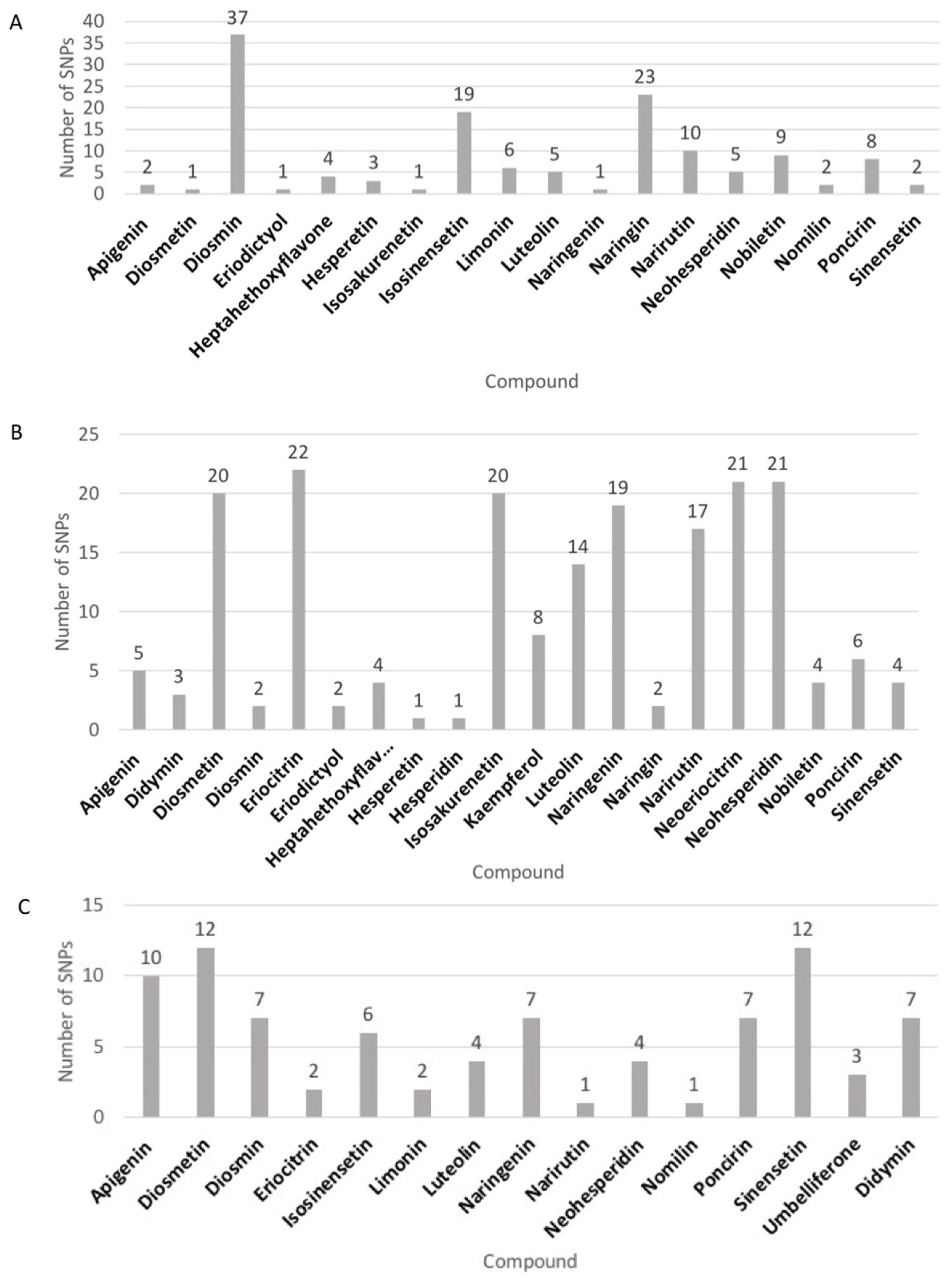
3.1. Analysis of 28 Target Flavonoids among Diverse Mandarin Accessions
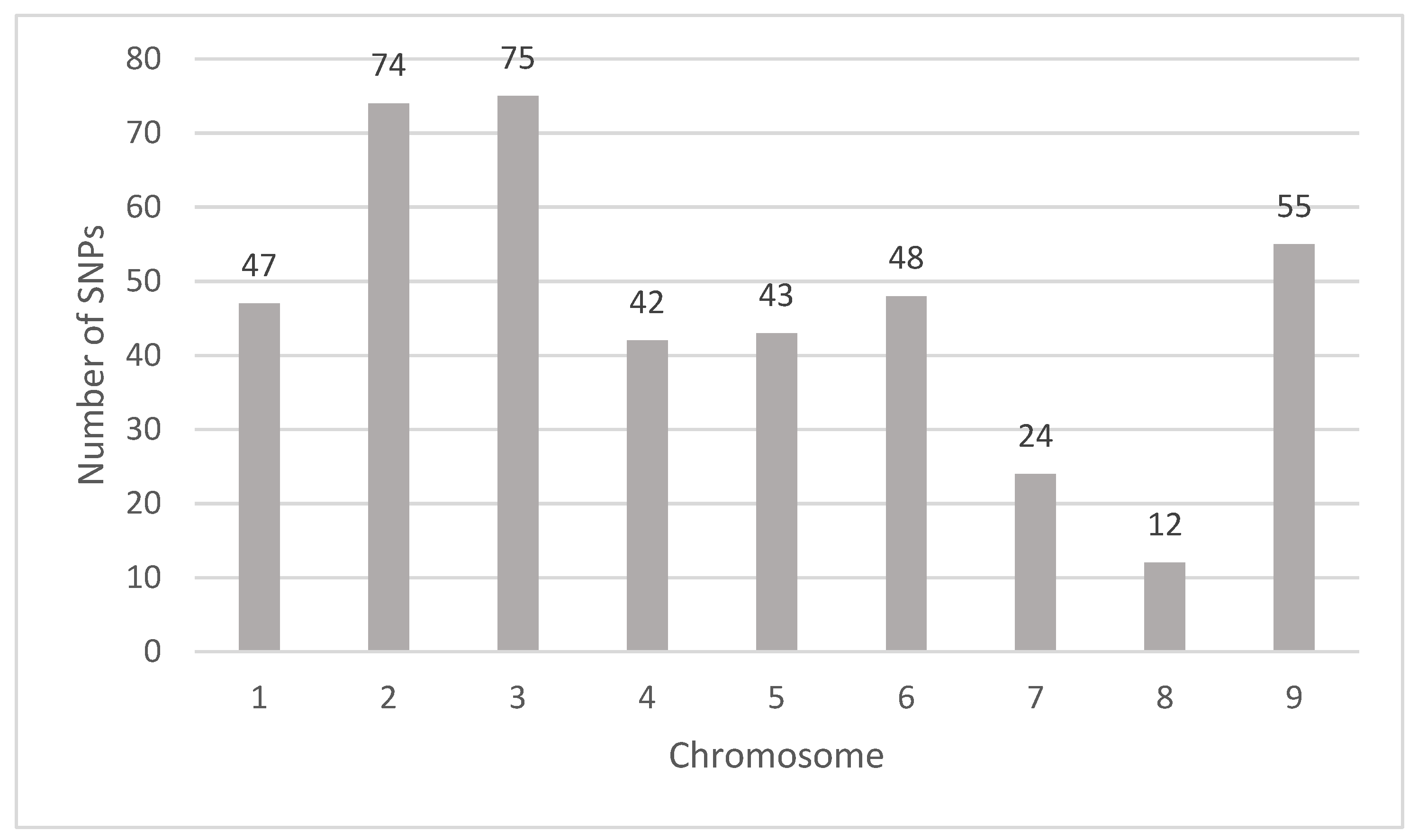
3.2. GWAS
3.3. Candidate Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mollet, B.; Rowland, I. Functional foods: At the frontier between food and pharma. Curr. Opin. Biotechnol. 2002, 13, 483–485. [Google Scholar] [CrossRef]
- Park, T.; Kim, Y. Phytochemicals as potential agents for prevention and treatment of obesity and metabolic diseases. In Anti-Obesity Drug Discovery and Development; Rahman, A., Choudhary, M.I., Eds.; Bentham: Dubai, United Arab Emirates, 2011; Volume 1, pp. 150–185. [Google Scholar]
- Xi, W.; Zhang, Y.; Sun, Y.; Shen, Y.; Ye, X.; Zhou, Z. Phenolic composition of Chinese wild mandarin (Citrus reticulata Blanco.) pulps and their antioxidant properties. Ind. Crops Prod. 2014, 52, 466–474. [Google Scholar] [CrossRef]
- Goulas, V.; Manganaris, G.A. Exploring the phytochemical content and the antioxidant potential of Citrus fruits grown in Cyprus. Food Chem. 2012, 131, 39–47. [Google Scholar] [CrossRef]
- Wang, S.; Yang, C.; Tu, H.; Zhou, J.; Liu, X.; Cheng, Y.; Luo, J.; Deng, X.; Zhang, H.; Xu, J. Characterization and metabolic diversity of flavonoids in citrus species. Sci. Rep. 2017, 7, 10549. [Google Scholar] [CrossRef] [PubMed]
- Grosser, J.W.; Gmitter, F.G., Jr.; Dutt, M.; Calovic, M.; Ling, P.; Castle, B. Highlights of the University of Florida, Citrus Research and Education Center’s comprehensive citrus breeding and genetics program. Acta Hortic. 2015, 1065, 405–413. [Google Scholar] [CrossRef]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genome 2008, 1, 15–20. [Google Scholar] [CrossRef]
- Khan, M.A.; Korban, S.S. Association mapping in forest trees and fruit crops. J. Exp. Bot. 2012, 63, 4045–4060. [Google Scholar] [CrossRef] [Green Version]
- Dilla-Ermita, C.J.; Tandayu, E.; Lozada, D.N.; Nora, L.; Bartolome, V.; Vera Cruz, C.; Ardales, E.Y.; Diaz, M.G.; Mendioro, M.S.; Thomson, M.J. Genome-wide association analysis of bacterial blight resistance in diverse rice germplasm. Philipp. J. Crop Sci. 2014, 39, 40. [Google Scholar]
- Riedelsheimer, C.; Lisec, J.; Czedik-Eysenberg, A.; Sulpice, R.; Flis, A.; Grieder, C.; Altmann, T.; Stitt, M.; Willmitzer, L.; Melchinger, A.E. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc. Natl. Acad. Sci. USA 2012, 109, 8872–8877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, Z.; Tan, R.; Yuan, J.; Bales, C.; Du, W.; Zhang, S.; Chilvers, M.I.; Schmidt, C.; Song, Q.; Cregan, P.B.; et al. Genome-wide association mapping of quantitative resistance to sudden death syndrome in soybean. BMC Genom. 2014, 15, 809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwata, H.; Minamikawa, M.F.; Kajiya-Kanegae, H.; Ishimori, M.; Hayashi, T. Genomics-assisted breeding in fruit trees. Breed Sci. 2016, 66, 100–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, K.; Wang, L.; Zhu, G.; Fang, W.; Chen, C.; Luo, J. Genetic diversity, linkage disequilibrium, and association mapping analyses of peach (Prunus persica) landraces in China. Tree Genet. Genomes 2012, 8, 975–990. [Google Scholar] [CrossRef]
- Kumar, S.; Garrick, D.J.; Bink, M.C.; Whitworth, C.; Chagné, D.; Volz, R.K. Novel genomic approaches unravel genetic architecture of complex traits in apple. BMC Genom. 2013, 14, 393. [Google Scholar] [CrossRef] [Green Version]
- Myles, S.; Boyko, A.R.; Owens, C.L.; Brown, P.J.; Grassi, F.; Aradhya, M.K.; Prins, B.; Reynolds, A.; Chia, J.-M.; Ware, D.; et al. Genetic structure and domestication history of the grape. Proc. Natl. Acad. Sci. USA 2011, 108, 3530–3535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minamikawa, M.F.; Nonaka, K.; Kaminuma, E.; Kajiya-Kanegae, H.; Onogi, A.; Goto, S.; Yoshioka, T.; Imai, A.; Hamada, H.; Hayashi, T.; et al. Genome-wide association study and genomic prediction in citrus: Potential of genomics-assisted breeding for fruit quality traits. Sci. Rep. 2017, 7, 4721. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.E.; Sands, D.C. The breeder’s dilemma—Yield or nutrition? Nat. Biotechnol. 2006, 24, 1078. [Google Scholar] [CrossRef]
- Brachi, B.; Morris, G.P.; Borevitz, J.O. Genome-wide association studies in plants: The missing heritability is in the field. Genome Biol. 2011, 12, 232. [Google Scholar] [CrossRef] [Green Version]
- Treutter, D. Significance of flavonoids in plant resistance: A review. Environ. Chem. Lett. 2006, 4, 147. [Google Scholar] [CrossRef]
- De Vos, R.C.; Moco, S.; Lommen, A.; Keurentjes, J.J.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Bressolle, F.; Bromet-Petit, M.; Audran, M. Validation of liquid chromatographic and gas chromatographic methods Applications to pharmacokinetics. J. Chromatogr. B Biomed. Appl. 1996, 686, 3–10. [Google Scholar] [CrossRef]
- Hiraoka, Y.; Ferrante, S.; Moragues, M.; Federici, C.; Close, T.; Roose, M. LD Analysis and Phylogeny Inference in Citrus with High-density SNP Array data. In Proceedings of the International Plant & Animal Genome XXV, San Diego, CA, USA, 13–17 January 2018. [Google Scholar]
- Anderson, C.A.; Pettersson, F.H.; Clarke, G.M.; Cardon, L.R.; Morris, A.P.; Zondervan, K.T. Data quality control in genetic case-control association studies. Nat. Protoc. 2010, 5, 1564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.-y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X. Multiple testing corrections for imputed SNPs. Genet. Epidemiol. 2011, 35, 154–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikitin, A.; Egorov, S.; Daraselia, N.; Mazo, I. Pathway studio—The analysis and navigation of molecular networks. Bioinformatics 2003, 19, 2155–2157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, H.; Wu, N.; Chang, Y.; Li, X.; Xiao, J.; Xiong, L. Carotenoid deficiency impairs ABA and IAA biosynthesis and differentially affects drought and cold tolerance in rice. Plant Mol. Biol. 2013, 83, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.M.; Zeef, L.A.; Ellis, J.; Goodacre, R.; Turner, S.R. Identification of Novel Genes in Arabidopsis Involved in Secondary Cell Wall Formation Using Expression Profiling and Reverse Genetics. Plant Cell 2005, 17, 2281–2295. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Teng, Q.; Zhong, R.; Ye, Z.H. The four Arabidopsis reduced wall acetylation genes are expressed in secondary wall-containing cells and required for the acetylation of xylan. Plant Cell Physiol. 2011, 52, 1289–1301. [Google Scholar] [CrossRef]
- Luo, J.; Nishiyama, Y.; Fuell, C.; Taguchi, G.; Elliott, K.; Hill, L.; Tanaka, Y.; Kitayama, M.; Yamazaki, M.; Bailey, P.; et al. Convergent evolution in the BAHD family of acyl transferases: Identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 2007, 50, 678–695. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, C.; Gmitter, F.G. QTL mapping of mandarin (Citrus reticulata) fruit characters using high-throughput SNP markers. Tree Genet. Genomes 2016, 12, 77. [Google Scholar] [CrossRef]
- Rafalski, A. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 2002, 5, 94–100. [Google Scholar] [CrossRef]
- Chadchawan, S.; Chokwiwatkul, R.; Tantipirom, N.; Khunpolwatatna, N.; Imyim, A.; Suriya-aroonroj, D.; Buaboocha, T.; Pongpanich, M.; Comai, L. Identification of genes involving in salt tolerance using GWAS data based on Na+ content in local Thai rice leaves. Genom. Genet. 2017, 10, 27–37. [Google Scholar] [CrossRef]
- Suwarno, W.B.; Pixley, K.V.; Palacios-Rojas, N.; Kaeppler, S.M.; Babu, R. Genome-wide association analysis reveals new targets for carotenoid biofortification in maize. Theor. Appl. Genet. 2015, 128, 851–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visioni, A.; Tondelli, A.; Francia, E.; Pswarayi, A.; Malosetti, M.; Russell, J.; Thomas, W.; Waugh, R.; Pecchioni, N.; Romagosa, I.; et al. Genome-wide association mapping of frost tolerance in barley (Hordeum vulgare L.). BMC Genom. 2013, 14, 424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Z.; Zhang, J.; Cai, S.; Chen, X.; Quan, X.; Zhang, G. Association mapping for total polyphenol content, total flavonoid content and antioxidant activity in barley. BMC Genom. 2018, 19, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhodes, D.H.; Hoffmann, L., Jr.; Rooney, W.L.; Ramu, P.; Morris, G.P.; Kresovich, S. Genome-wide association study of grain polyphenol concentrations in global sorghum [Sorghum bicolor (L.) Moench] germplasm. J. Agric. Food Chem. 2014, 62, 10916–10927. [Google Scholar] [CrossRef]
- Shao, Y.; Jin, L.; Zhang, G.; Lu, Y.; Shen, Y.; Bao, J. Association mapping of grain color, phenolic content, flavonoid content and antioxidant capacity in dehulled rice. Theor. Appl. Genet. 2011, 122, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.J.; Dwyer, J.T.; Beecher, G.R.; Bhagwat, S.A.; Gebhardt, S.E.; Haytowitz, D.B.; Holden, J.M. Flavanones in oranges, tangerines (mandarins), tangors, and tangelos: A compilation and review of the data from the analytical literature. J. Food Compos. Anal. 2006, 19, S66–S73. [Google Scholar] [CrossRef]
- Caballero, A.; Tenesa, A.; Keightley, P.D. The nature of genetic variation for complex traits revealed by GWAS and regional heritability mapping analyses. Genetics 2015, 201, 1601–1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hindu, V.; Palacios-Rojas, N.; Babu, R.; Suwarno, W.B.; Rashid, Z.; Usha, R.; Saykhedkar, G.R.; Nair, S.K. Identification and validation of genomic regions influencing kernel zinc and iron in maize. Theor. Appl. Genet. 2018, 131, 1443–1457. [Google Scholar] [CrossRef] [Green Version]
- Tripoli, E.; La Guardia, M.; Giammanco, S.; Di Majo, D.; Giammanco, M. Citrus flavonoids: Molecular structure, biological activity and nutritional properties: A review. Food Chem. 2007, 104, 466–479. [Google Scholar] [CrossRef]
- Lin, C.F.; Leu, Y.L.; Al-Suwayeh, S.A.; Ku, M.C.; Hwang, T.L.; Fang, J.Y. Anti-inflammatory activity and percutaneous absorption of quercetin and its polymethoxylated compound and glycosides: The relationships to chemical structures. Eur. J. Pharm. Sci. 2012, 47, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, H.; Guo, L.; Zhao, H.; Ho, C.T. Chemistry and bioactivity of nobiletin and its metabolites. J. Funct. Foods 2014, 6, 2–10. [Google Scholar] [CrossRef]
- Kubasek, W.L.; Shirley, B.W.; McKillop, A.; Goodman, H.M.; Briggs, W.; Ausubel, F.M. Regulation of Flavonoid Biosynthetic Genes in Germinating Arabidopsis Seedlings. Plant Cell 1992, 4, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Arcas, M.C.; Botía, J.M.; Ortuño, A.M.; Del Río, J.A. UV irradiation Alters the Levels of Flavonoids Involved in the Defence Mechanism of Citrus aurantium Fruits against Penicillium digitatum. Eur. J. Plant Pathol. 2000, 106, 617–622. [Google Scholar] [CrossRef]
- Liu, C.; Long, J.; Zhu, K.; Liu, L.; Yang, W.; Zhang, H.; Li, L.; Xu, Q.; Deng, X. Characterization of a Citrus R2R3-MYB Transcription Factor that Regulates the Flavonol and Hydroxycinnamic Acid Biosynthesis. Sci. Rep. 2016, 6, 25352. [Google Scholar] [CrossRef]
- Koca, U.; Berhow, M.A.; Febres, V.J.; Champ, K.I.; Carrillo-Mendoza, O.; Moore, G.A. Decreasing unpalatable flavonoid components in Citrus: The effect of transformation construct. Physiol. Plant. 2009, 137, 101–114. [Google Scholar] [CrossRef]
- Moriguchi, T.; Kita, M.; Tomono, Y.; Endo-Inagaki, T.; Omura, M. Gene expression in flavonoid biosynthesis: Correlation with flavonoid accumulation in developing citrus fruit. Physiol. Plant. 2001, 111, 66–74. [Google Scholar] [CrossRef]
- Killiny, N.; Jones, S.E.; Nehela, Y.; Hijaz, F.; Dutt, M.; Gmitter, F.G.; Grosser, J.W. All roads lead to Rome: Towards understanding different avenues of tolerance to huanglongbing in citrus cultivars. Plant Physiol. Biochem. 2018, 129, 1–10. [Google Scholar] [CrossRef]
- Massenti, R.; Lo Bianco, R.; Sandhu, A.K.; Gu, L.; Sims, C. Huanglongbing modifies quality components and flavonoid content of ‘Valencia’ oranges. J. Sci. Food Agric. 2016, 96, 73–78. [Google Scholar] [CrossRef]

| # | Compound | Peel | Peel | Peel | Pulp | Pulp | Pulp | Seed | Seed | Seed |
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Max. | Min. | Mean | Max. | Min. | Mean | Max. | Min. | ||
| 1 | Apigenin | 0.01 z | 1.88 | 0 | 0.0002 | 0.03 | 0 | 0.0004 | 0.07 | 0 |
| 2 | Coumarin | 0.006 | 0.1 | 0 | 0.0007 | 0.16 | 0 | 0 | 0 | 0 |
| 3 | Didymin | 3.71 | 118.59 | 0 | 5.19 | 133.51 | 0.005 | 0.12 | 2.93 | 0 |
| 4 | Diosmetin | 0.56 | 58.99 | 0 | 0.008 | 0.88 | 0 | 0.008 | 0.35 | 0 |
| 5 | Diosmin | 0.003 | 0.02 | 0 | 0.0006 | 0.06 | 0 | 0.02 | 0.46 | 0 |
| 6 | Eriocitrin | 0.32 | 9.16 | 0 | 0.35 | 22.55 | 0 | 0.03 | 2.03 | 0 |
| 7 | Eriodictyol | 0.007 | 0.04 | 0 | 0.05 | 0.05 | 0 | 0.0001 | 0.01 | 0 |
| 8 | Heptamethoxyflavone | 50.7 | 1767.33 | 0 | 3.17 | 148.92 | 0.03 | 2.37 | 54.59 | 0.16 |
| 9 | Hesperetin | 0.05 | 11.28 | 0 | 0.003 | 0.18 | 0 | 0.02 | 0.94 | 0 |
| 10 | Hesperidin | 14.5 | 575.78 | 0.05 | 14.07 | 266.89 | 0.02 | 0.55 | 17.3 | 0.002 |
| 11 | Isosakurenetin | 0.005 | 0.07 | 0 | 0.0004 | 0.1 | 0 | 0.0005 | 0.05 | 0 |
| 12 | Isosinensetin | 82.99 | 1372.64 | 0 | 3.47 | 76.36 | 0 | 0.32 | 10.95 | 0 |
| 13 | Kaempferol | 0 | 0 | 0 | 0 | 0 | 0 | 0.0006 | 0.08 | 0 |
| 14 | Limonin | 9.2 | 231.94 | 0 | 13.65 | 410.42 | 0.03 | 148.17 | 718.93 | 5.24 |
| 15 | Luteolin | 0.002 | 0.04 | 0 | 0.0006 | 0.07 | 0 | 0.001 | 0.04 | 0 |
| 16 | Naringenin | 0.03 | 1.43 | 0 | 0.004 | 0.45 | 0 | 0.0006 | 0.07 | 0 |
| 17 | Naringin | 0.25 | 14.05 | 0 | 0.4 | 20.77 | 0 | 0.02 | 2.35 | 0 |
| 18 | Narirutin | 4.91 | 125.13 | 0 | 6.69 | 243.81 | 0 | 0.31 | 6.21 | 0 |
| 19 | Neodiosmin | 12.41 | 321.22 | 0 | 4.22 | 108.55 | 0 | 0.54 | 3.46 | 0.03 |
| 20 | Neoeriocitrin | 0.01 | 1.49 | 0 | 0.02 | 1.84 | 0 | 0.01 | 0.86 | 0 |
| 21 | Neohesperidin | 1.95 | 58.19 | 0 | 2.59 | 86.45 | 0 | 0.08 | 4.67 | 0 |
| 22 | Nobiletin | 471.84 | 0.02 | 0.03 | 28.72 | 597.4 | 0.32 | 18.73 | 204.64 | 2.08 |
| 23 | Nomilin | 0.88 | 39.72 | 0 | 1.86 | 43.07 | 0 | 24.56 | 134.1 | 0.24 |
| 24 | Poncirin | 0.16 | 14.65 | 0 | 0.19 | 11.32 | 0 | 0.11 | 20.42 | 0 |
| 25 | Rutin | 0.16 | 3.72 | 0 | 0.04 | 0.62 | 0 | 0.05 | 0.38 | 0 |
| 26 | Sinensetin | 42.43 | 1302.52 | 0 | 2.1 | 83.56 | 0 | 0.167 | 13.62 | 0 |
| 27 | Tangeretin | 444.28 | 0.98 | 4.86 | 152.36 | 1989.3 | 0.38 | 175.13 | 1218.65 | 10.36 |
| 28 | Umbelliferone | 0.006 | 0.13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Compound | Sample Type | Marker | Chromosome | Position | Gene | Proportion of Variance Explained | p-Value |
|---|---|---|---|---|---|---|---|
| Apigenin | peel | AX-160026423 | 1 | 28455844 | Ciclev10007429m.g | 0.065 | 2.72 × 10−7 |
| Apigenin | pulp | AX-161017449 | 3 | 8373806 | Ciclev10019809m.g | 0.066 | 1.41 × 10−7 |
| Apigenin | seed | AX-160120038 | 6 | 18733554 | Ciclev10013344m.g | 0.070 | 1.57 × 10−7 |
| Didymin | seed | AX-160548920 | 6 | 500457 | Ciclev10011912m.g | 0.080 | 1.69 × 10−8 |
| Didymin | peel | AX-160548920 | 6 | 500457 | Ciclev10011912m.g | 0.070 | 4.94 × 10−8 |
| Diosmetin | seed | AX-160428826 | 5 | 34679192 | Ciclev10003655m.g | 0.195 | 9.47 × 10−20 |
| Diosmetin | peel | AX-159831918 | 1 | 27881393 | Ciclev10009180m.g | 0.048 | 1.13 × 10−8 |
| Diosmetin | pulp | AX-160808091 | 6 | 23828780 | Ciclev10011357m.g | 0.052 | 2.99 × 10−6 |
| Diosmin | pulp | AX-160953115 | 1 | 4957396 | Ciclev10007412m.g | 0.103 | 2.98 × 10−11 |
| Diosmin | peel | AX-160796087 | 8 | 23855395 | Ciclev10029047m.g | 0.059 | 5.87 × 10−7 |
| Diosmin | seed | AX-160362613 | 2 | 35355786 | Ciclev10014319m.g | 0.060 | 1.13 × 10−6 |
| Eriocitrin | seed | AX-160981611 | 3 | 5235434 | Ciclev10021722m.g | 0.095 | 7.55 × 10−10 |
| Eriocitrin | peel | AX-160487850 | 3 | 35789790 | Ciclev10020864m.g | 0.068 | 8.05 × 10−8 |
| Eriodictyol | seed | AX-160014487 | 7 | 1494553 | Ciclev10025311m.g | 0.094 | 8.81 × 10−10 |
| Eriodictyol | pulp | AX-160808091 | 6 | 23828780 | Ciclev10011357m.g | 0.052 | 2.99 × 10−6 |
| Heptamethoxyflavone | pulp | AX-160478010 | 4 | 23838038 | Ciclev10032432m.g | 0.073 | 2.9 × 10−8 |
| Heptamethoxyflavone | seed | AX-159875671 | 1 | 23588919 | Ciclev10008059m.g | 0.067 | 2.91 × 10−7 |
| Hesperetin | pulp | AX-159872199 | 6 | 24383951 | Ciclev10012224m.g | 0.059 | 5.63 × 10−7 |
| Hesperetin | seed | AX-160906983 | 5 | 13059166 | Ciclev10002443m.g | 0.058 | 2.05 × 10−6 |
| Hesperidin | seed | AX-160261315 | 5 | 34690939 | Ciclev10002520m.g | 0.070 | 1.39 × 10−7 |
| Hesperidin | Pulp | AX-159823569 | 7 | 92972 | Ciclev10025023m.g | 0.050 | 4.85 × 10−6 |
| Isosakurenetin | seed | AX-159926254 | 2 | 12136093 | Ciclev10014778m.g | 0.066 | 3.27 × 10−7 |
| Isosakurenetin | pulp | AX-160865092 | 3 | 40172143 | Ciclev10024487m.g | 0.054 | 1.96 × 10−6 |
| Isosinensetin | pulp | AX-160385265 | 5 | 34255060 | Ciclev10001494m.g | 0.077 | 1.82 × 10−8 |
| Isosinensetin | peel | AX-160729031 | 3 | 27830592 | Ciclev10019217m.g | 0.053 | 2.39 × 10−6 |
| Kaempferol | seed | AX-159946295 | 4 | 18414143 | Ciclev10032158m.g | 0.208 | 4.51 × 10−21 |
| Limonin | peel | AX-160548920 | 6 | 500457 | Ciclev10011912m.g | 0.076 | 1.24 × 10−8 |
| Limonin | pulp | AX-160121385 | 4 | 21318491 | Ciclev10033394m.g | 0.054 | 8.40 × 10−6 |
| Luteolin | seed | AX-159825934 | 2 | 3781683 | Ciclev10016037m.g | 0.294 | 1.07 × 10−30 |
| Luteolin | pulp | AX-160742943 | 6 | 20232219 | Ciclev10012009m.g | 0.067 | 1.11 × 10−7 |
| Luteolin | peel | AX-160742943 | 6 | 20232219 | Ciclev10012009m.g | 0.065 | 1.73 × 10−7 |
| Naringenin | peel | AX-160808091 | 6 | 23828780 | Ciclev10011357m.g | 0.070 | 7.57 × 10−8 |
| Naringenin | seed | AX-159951409 | 5 | 24061380 | Ciclev10001582m.g | 0.068 | 2.38 × 10−7 |
| Naringenin | pulp | AX-160808091 | 6 | 23828780 | Ciclev10011357m.g | 0.063 | 2.49 × 10−7 |
| Naringin | pulp | AX-160259155 | 3 | 42212133 | Ciclev10022115m.g | 0.127 | 1.02 × 10−13 |
| Naringin | seed | AX-159969067 | 6 | 25181051 | Ciclev10011245m.g | 0.060 | 1.30 × 10−6 |
| Narirutin | seed | AX-160529374 | 5 | 24023209 | Ciclev10001022m.g | 0.099 | 3.28 × 10−10 |
| Narirutin | pulp | AX-160871858 | 2 | 10250942 | Ciclev10016536m.g | 0.057 | 1.79 × 10−6 |
| Neoeriocitrin | seed | AX-160981611 | 3 | 5235434 | Ciclev10021722m.g | 0.090 | 1.89 × 10−9 |
| Neohesperidin | pulp | AX-160292039 | 4 | 7374156 | Ciclev10033003m.g | 0.119 | 6.81 × 10−13 |
| Neohesperidin | peel | AX-159868580 | 5 | 21083453 | Ciclev10001079m.g | 0.100 | 5.47 × 10−11 |
| Neohesperidin | seed | AX-160395823 | 9 | 21603308 | Ciclev10005135m.g | 0.090 | 2.29 × 10−9 |
| Nobiletin | pulp | AX-160014948 | 5 | 33018623 | Ciclev10000065m.g | 0.060 | 5.61 × 10−7 |
| Nobiletin | seed | AX-160432194 | 2 | 27643973 | Ciclev10014906m.g | 0.058 | 2.27 × 10−6 |
| Nomilin | peel | AX-160423747 | 6 | 17478449 | Ciclev10011307m.g | 0.066 | 1.19 × 10−7 |
| Nomilin | pulp | AX-160298860 | 3 | 3171806 | Ciclev10022532m.g | 0.060 | 5.22 × 10−7 |
| Poncirin | seed | AX-159969067 | 6 | 25181051 | Ciclev10011245m.g | 0.098 | 3.39 × 10−10 |
| Poncirin | peel | AX-160261315 | 5 | 34690939 | Ciclev10002520m.g | 0.063 | 2.68 × 10−7 |
| Poncirin | pulp | AX-160395823 | 9 | 21603308 | Ciclev10005135m.g | 0.060 | 4.68 × 10−7 |
| Sinensetin | peel | AX-160947519 | 9 | 29959571 | Ciclev10006155m.g | 0.106 | 1.25 × 10−11 |
| Sinensetin | seed | AX-160284313 | 3 | 40137702 | Ciclev10020460m.g | 0.077 | 3.48 × 10−8 |
| Sinensetin | pulp | AX-160947519 | 9 | 29959571 | Ciclev10006155m.g | 0.070 | 5.58 × 10−8 |
| Associated Compound/Samples | “Given Name” | GWAS Identified SNP | Chr. | Physical Position | Annotation | Arabidopsis Orthologue | Citrus Gene | Citrus LOC | Predicted Citrus Annotation | Study References |
|---|---|---|---|---|---|---|---|---|---|---|
| Didymin/peel | MYB96 | AX-160475114 | 3 | 7201172 | Myb transcription factor | AT5G62470 | Ciclev10020967m.g | LOC18048626 | Citrus clementina myb-related protein 306 | [27] |
| Naringenin/seed | MPL12.14 | AX-161026143 | 2 | 34869109 | protein REDUCED WALL ACETYLATION 1 | AT5G46340 | Ciclev10014824m.g | LOC18053255 | REDUCED WALL ACETYLATION 3 | [28,29] |
| Heptamethoxyflavone/seed | AT3G29635 | AX-159875671 | 1 | 23596472 | HXXXD-type acyl-transferase-like protein | AT3G29635 | Ciclev10008059m.g | LOC18055186 | malonyl-CoA:anthocyanidin 5-O-glucoside-6″-O-malonyltransferase | [30] |
| Naringenin/peel | CRTISO | AX-160782343 | 6 | 19581218 | carotenoid isomerase | AT1G06820 | Ciclev10011230m.g | LOC18038313 | prolycopene isomerase, chloroplastic | [31,32] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mattia, M.R.; Du, D.; Yu, Q.; Kahn, T.; Roose, M.; Hiraoka, Y.; Wang, Y.; Munoz, P.; Gmitter, F.G., Jr. Genome-Wide Association Study of Healthful Flavonoids among Diverse Mandarin Accessions. Plants 2022, 11, 317. https://doi.org/10.3390/plants11030317
Mattia MR, Du D, Yu Q, Kahn T, Roose M, Hiraoka Y, Wang Y, Munoz P, Gmitter FG Jr. Genome-Wide Association Study of Healthful Flavonoids among Diverse Mandarin Accessions. Plants. 2022; 11(3):317. https://doi.org/10.3390/plants11030317
Chicago/Turabian StyleMattia, Matthew R., Dongliang Du, Qibin Yu, Tracy Kahn, Mikeal Roose, Yoko Hiraoka, Yu Wang, Patricio Munoz, and Fred G. Gmitter, Jr. 2022. "Genome-Wide Association Study of Healthful Flavonoids among Diverse Mandarin Accessions" Plants 11, no. 3: 317. https://doi.org/10.3390/plants11030317
APA StyleMattia, M. R., Du, D., Yu, Q., Kahn, T., Roose, M., Hiraoka, Y., Wang, Y., Munoz, P., & Gmitter, F. G., Jr. (2022). Genome-Wide Association Study of Healthful Flavonoids among Diverse Mandarin Accessions. Plants, 11(3), 317. https://doi.org/10.3390/plants11030317








