Abstract
As an important cash crop in China, apple has a good flavor and is rich in nutrients. Fungal attacks have become a major obstacle in apple cultivation. Colletotrichum gloeosporioides is one of the most devastating fungal pathogens in apple. Thus, discovering resistance genes in response to C. gloeosporioides may aid in designing safer control strategies and facilitate the development of apple resistance breeding. A previous study reported that ‘Hanfu’ autotetraploid apple displayed higher C. gloeosporioides resistance than ‘Hanfu’ apple, and the expression level of mdm-MIR390b was significantly upregulated in autotetraploid plants compared to that in ‘Hanfu’ plants, as demonstrated by digital gene expression (DGE) analysis. It is still unclear, however, whether mdm-MIR390b regulates apple anthracnose resistance. Apple MIR390b was transformed into apple ‘GL-3′ plants to identify the functions of mdm-MIR390b in anthracnose resistance. C. gloeosporioides treatment analysis indicated that the overexpression of mdm-MIR390b reduced fungal damage to apple leaves and fruit. Physiology analysis showed that mdm-MIR390b increased C. gloeosporioides resistance by improving superoxide dismutase (SOD) and peroxidase (POD) activity to alleviate the damage caused by O2− and H2O2. Our results demonstrate that mdm-MIR390b can improve apple plants’ anthracnose resistance.
1. Introduction
Apple (Malus × domestica), a delicious fresh and processed fruit, is cultivated in many countries worldwide [1]. The spread of fungal diseases is one of the most important factors that restricts the development of apple production [2]. Colletotrichum gloeosporioides is a destructive fungus that causes early severe defoliation and severely weakens tree vigor. Attenuating the effects of C. gloeosporioides would be significant for apple production. The use of chemical fungicides is effective. However, it causes serious damage to the environment [3]. In order to reduce the pollution of the environment, improving the defensive ability of plants has become a good choice. Therefore, finding plant disease resistance genes for molecular breeding is an effective means to prevent fungal diseases.
MicroRNAs (miRNAs) are small, noncoding, endogenous RNA molecules that are 20-24 nucleotides (nt) in length [4,5]. They have been proven as new regulators, controlling post-transcriptional mRNA stability, providing epigenetic modifications to characteristic regions of the genome, or allowing translation by complementarily binding to target nucleic acids [4,6,7]. In plants, miRNA biogenesis and the modes of action have been specifically described. Most MIR genes are transcribed as independent units by DNA-dependent RNA Polymerase II (Pol II), while some miRNA precursors are produced from introns of the genes encoding proteins [8]. Primary transcripts of MIRs (pri-miRNAs) from typical MIR genes are modified similarly to mRNA and folded into hairpin structures [5]. Then, pri-miRNAs are recognized and processed by the dicing complex containing DICER-LIKE RNase III endonucleases (DCLs), the RNA-binding protein HYPONASTIC LEAVES 1 (HYL1), and the zinc-finger protein SERRATE (SE) to produce miRNA:miRNA* duplexes [9,10,11]. After the 2′-OH position in the duplex is methylated by the small RNA methyltransferase HUA Enhancer 1 (HEN1), the duplex is exported to the cytoplasm by HASTY (HST) [12]. The mature miRNA strand in the duplex binds to different ARGONAUTE (AGO) proteins to form the RNA-induced silencing complex (RISC) for targets, and the miRNA* strand is damaged by SMALL RNA DEGRADING NUCLEASE (SDN) [5]. Eventually, mature miRNAs downregulate or repress their targets at the post-transcriptional or translational level and are therefore involved in plant growth and development and stress responses.
Plant miR390, an ancient and highly conserved miRNA, is the only one present in many species and at high abundances [13]. Two miR390s (ath-miR390a and ath-miR390b) were first discovered in Arabidopsis thaliana in the Arabidopsis Small RNA Project (ASRP) [14]. Later, miR390s were found in a variety of plants. There was one osa-miR390 in rice [15], three nta-miR390s in tobacco [16], two sly-miR390s in tomato [17], and two zma-miR390s in maize [18]. Many studies have shown that miR390 acts on target genes in a different regulatory manner compared to other miRNAs. It can produce a 21-nt-long trans-acting small interfering RNA (tasiRNA) by acting on a trans-acting siRNA gene (TAS), and then targets auxin response factor (ARF) [19]. The transcript transcribed by Pol II from the TAS3 gene is first cleaved by miR390 and the AGO1/7 complex to generate single-stranded RNA in a typical “two-hit” mode [20,21,22]. The single-stranded RNA is converted into double-stranded RNA by the plant-RNA-dependent RNA polymerase 6-centred (RDR6) complex [23,24]. Then, the double-stranded RNA bound to dsRNA-binding protein 4 (DRB4) is cleaved by DCL4 into short RNA fragments linked end-to-end, namely tasiRNA3 [19,25,26]. Finally, tasiRNA3 negatively regulates the expression of auxin response factor 2/3/4 (ARF2/3/4) [27,28,29]. In Arabidopsis, miR390 is specifically expressed at the sites of lateral root (LR) initiation to regulate the timing of LR growth through the miR390-tasiRNA3-ARF2/3/4 pathway [26]. The miR390-tasiRNA-ARF4 regulatory module contributes to auxin responses in LR developmental processes [30]. In flowering development, the miR390-tasiRNA3-ARF4 pathway delays woodland strawberry florescence by targeting two genes related to flowering (FveFUL and FveAP1) [31]. In poplar, the miR390/TAS3/ARFs module acts as a key regulator of increased salt tolerance via modulating the auxin signaling [32]. In plants, miR390 plays a significant role in diverse processes, including root growth, floral development, and abiotic stress responses. However, research on the fungal responses of miR390 is still rare.
In our earlier study, ‘Hanfu’ apple trees displayed lower C. gloeosporioides resistance than autotetraploid apple trees [33]. DGE analysis showed that MIR390b was differentially expressed in ‘Hanfu’ and autotetraploid apple trees [34]. In this study, bioinformatics analysis of miR390 was performed and showed that miR390 sequences were highly conservative in plants. When ‘Hanfu’ plants were attacked by C. gloeosporioides, we found that mdm-MIR390b could be dramatically induced. Then, a transgenic method was used to investigate the role of mdm-MIR390b in C. gloeosporioides infection responses in apple. Overexpression of mdm-MIR390b significantly enhanced apple anthracnose resistance under the C. gloeosporioides stress condition compared with ‘GL-3′ control plants. Under C. gloeosporioides infection, overexpressing-MIR390b lines showed improved SOD and POD activity against pathogen-induced reactive oxygen species (ROS) accumulation. The expression levels of disease resistance genes (PRs) were upregulated in plants overexpressing mdm-MIR390b. These results provide new insights into the connection between MIR390 and fungal stress in apple.
2. Results
2.1. Bioinformatics Analysis of miR390
In the miRbase database, six mdm-MIR390s were found and each mdm-MIR390 can produce a mature miRNA (Table 1). Analysis of the phylogenetic tree found that mdm-MIR390b, mdm-MIR390e, and mdm-MIR390f showed strong homology, and mdm-MIR390a, mdm-MIR390c, and mdm-MIR390d showed a closer relationship. Six mdm-MIR390s were clustered in the same branch. They showed a more distant homology to osa-MIR390, zma-MIR390s, and sbi-MIR390 (Figure 1a). Furthermore, MIR390s in dicot plants displayed higher sequence identity with mdm-MIR390s than those in monocot plants.

Table 1.
Apple MIR390 and miR390 information.

Figure 1.
Identification and sequence analysis of MIR390 family. (a) The phylogenetic analysis of MIR390s in plants. Bootstrap values were derived from 1000 replicates. The pink branches represent mdm-MIR390s. The cluster with the light blue background contains all mdm-MIR390s. The cosmic latte apple represents mdm-MIR390b. (b) MIR390s secondary structure analysis in apple.
Stem-loop structures of six precursor sequences were predicted by minimum free energy using RNAfold. As shown in Figure 1b, mature mdm-miR390s (21 nt) were generated from the 5’ arm of the stem-loop structure and there is one mismatched base pair in the miRNA:miRNA*duplex. The end of the miRNA:miRNA* duplex has a 2 nt overhang structure, which resulted from Dicer cleavage [35]. The results revealed that mdm-MIR390s have universal features of pre-miRNAs.
In fifteen species, the predicted miR390s contained two typical sequences produced from the 5′ arm of the MIR390 stem-loop. Most of them have the same sequence (5′-AAGCUCAGGAGGGAUAGCGCC-3′). Six mdm-miR390s also show this conserved sequence, and only three miR390s (nta-miR390a, sly-miR390a-5p, and stu-miR390) are different sequences (Figure 2a). The analysis of miR390 mature sequences’ alignment was visualized by WebLogo. This result showed that mature sequences had high identity, containing 20 bases that were completely conserved (Figure 2b).

Figure 2.
Mature sequence analysis of miR390s. (a) Sequence alignment of mdm-miR390s with other homologous sequences. (b) Conserved bases of miR390 mature sequences in fifteen species.
2.2. Expression Patterns of mdm-MIR390b in Apple
Previously, we found that the ‘Hanfu’ autotetraploid has stronger anthracnose resistance than ‘Hanfu’ [33]. DGE analysis of ‘Hanfu’ and autotetraploid apple trees revealed that mdm-MIR390b has a higher expression level in autotetraploid apple plants [34]. Thus, in ‘Hanfu’ and autotetraploid apple leaves, quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis was performed to examine the expression of mdm-MIR390b. Compared with ‘Hanfu’ apple leaves, the expression level of mdm-MIR390b was statistically increased, with an over three-fold higher relative expression level in autotetraploid apple leaves (Figure 3a). Meanwhile, we observed the expression of mdm-MIR390a-f in the ‘Hanfu’ leaf and fruit. In the leaf, the relative expression level of mdm-MIR390b was higher than that of five mdm-MIR390s. In the fruit, mdm-MIR390a and mdm-MIR390b displayed higher expression levels (Figure S1). To examine fungus-dependent changes in the expression of mdm-MIR390b, qRT-PCR was used to analyze its expression after C. gloeosporioides treatment in ‘Hanfu’ apple plants. Apple MIR390b expression was upregulated after C. gloeosporioides infection and peaked with an approximately six-fold higher relative expression level at 2 d. The relative expression levels of mdm-miR390 and tasiRNA3 were also examined. They showed a similar trend as the expression level of mdm-MIR390b under C. gloeosporioides treatment (Figure 3b).
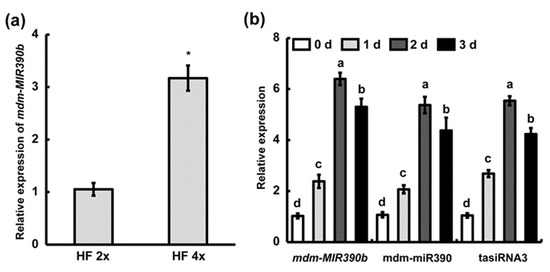
Figure 3.
The analysis of the mdm-MIR390b gene, mdm-miR390, and tasiRNA3by qRT-PCR. (a) mdm-MIR390b expression was examined in ‘Hanfu’ and autotetraploid apple. HF 2x represents ‘Hanfu’. HF 4x represents ‘Hanfu’ autotetraploid. The vertical bars represent SDs (n = 3). ‘*’ represents p < 0.05 (Student’s t-test). (b) Expression level of mdm-MIR390b, mdm-miR390, and tasiRNA3 under C. gloeosporioides infection in ‘Hanfu’. The vertical bars represent SDs (n = 3). The different letters represent significant differences at p < 0.05 level.
2.3. Overexpressing MIR390b in Apple ‘GL-3′
To investigate the function of mdm-MIR390b, we overexpressed mdm-MIR390b in apple ‘GL-3′ (Figure 4a). Three MIR390b-overexpressing lines under the control of the CaMV 35S promoter (MIR390b-line1, MIR390b-line2, and MIR390b-line3) were generated. The relative expression level of mdm-MIR390b was analyzed by qRT-PCR in apple ‘GL-3′ plants and MIR390b-overexpressing lines. As shown in Figure 4b, in the three mdm-MIR390b overexpression lines, the relative expression of mdm-MIR390b was significantly higher than that of ‘GL-3′. The expression level was, respectively, around 5.5, 4.0, and 5.0 times higher in MIR390b-line1, MIR390b-line2, and MIR390b-line3 than in ‘GL-3′. The expression level of mdm-miR390 in MIR390b-overexpressing plants was also significantly higher than that in ‘GL-3′ plants. Because the expression levels of mdm-MIR390b and mdm-miR390 in MIR390b-line1 and MIR390b-line3 plants were higher than those in MIR390b-line2 plants, the MIR390b-line1 and MIR390b-line3 plants were subjected to further investigation. Leaf regeneration experiments were performed using ‘GL-3′ and transgenic apple lines. On the right side of the glass dishes, the leaves of mdm-MIR390b-overexpressing lines again grew green resistant buds. However, on the left side of the dishes, the control leaves became white and lost their vitality (Figure 4c). These results indicated that the pRI101-MIR390b vector was transformed into apple ‘GL-3′ and three mdm-MIR390b-overexpressing lines were successfully obtained.
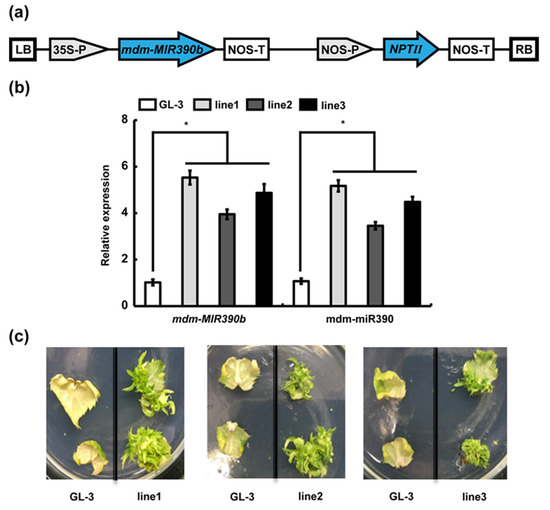
Figure 4.
Identification and analysis of transgenic plants. (a) Structural diagram of pRI101-MIR390b. (b) Expression analysis of mdm-MIR390b gene and mdm-miR390 in ‘GL-3′ and MIR390b-overexpressing plants. The vertical bars represent SDs (n = 3). ‘*’ represents p < 0.05 (Student’s t-test). (c) Leaf regeneration observation of ‘GL-3′ and MIR390b-overexpressing plants.
2.4. Overexpression of mdm-MIR390b Gene Elevates Apple Anthracnose Resistance
To clarify the function of mdm-MIR390b in C. gloeosporioides resistance, we examined the growth changes in mdm-MIR390b-overexpressing and ‘GL-3′ apple plants under C. gloeosporioides-stressed conditions. After apple plants were sprayed with fungal suspension, discrete small lesions were observed between mdm-MIR390b-overexpressing and ‘GL-3′ apple plants on the third day. Apple MIR390b-line1 and MIR390b-line3 plants displayed weaker disease situations than ‘GL-3′ plants. On the fifth day, larger brown spots were found in the leaves of ‘GL-3′, MIR390b-line1, and MIR390b-line3 plants. The control plants showed more severe disease conditions than overexpressing transgenic apple plants (Figure 5a). Quantitative analysis confirmed that the disease indexes of MIR390b-overexpressing plants were significantly lower than those of ‘GL-3′ plants after C. gloeosporioides treatment (Figure 5b). In apple fruit, the lesion diameter of the control fruit was greater than that of MIR390b-overexpressing plants. The overexpression of mdm-MIR390b inhibited the brown lesions that developed rapidly and the abundant conidia that were produced around the injection site. In contrast, the control apples exhibited more severe disease symptoms (Figure 5c,d). When the fruit were cut through the centers of lesions, V-shaped necrotic tissue areas were also observed. The lesions of control apples were 1.68-fold deeper than those in mdm-MIR390b-overexpressing apples (Figure 5e,f). These results indicate that the overexpression of mdm-MIR390b can enhance the anthracnose resistance of apple leaves and fruit.
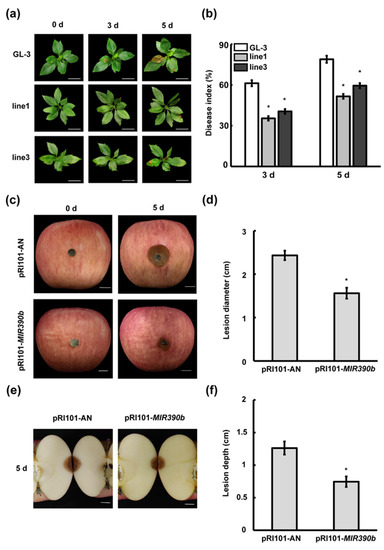
Figure 5.
Apple MIR390b enhances resistance to C. gloeosporioides in leaves and fruit. (a) Leaf phenotype of ‘GL-3′ and MIR390b-overexpressing plants under C. gloeosporioides infection, white bars = 2 cm. (b) Disease index of ‘GL-3′ and MIR390b-overexpressing plants under C. gloeosporioides infection. The vertical bars represent SDs (n = 3). ‘*’ represents p < 0.05 (Student’s t-test). (c) Phenotype of apple fruit transiently expressing pRI101-MIR390b and empty vector (control) under C. gloeosporioides infection, white bars = 1 cm. (d) Lesion diameter of apple fruit after infection with C. gloeosporioides for 5 d. The vertical bars represent SDs (n = 3). ‘*’ represents p < 0.05 (Student’s t-test). (e) V-shaped necrotic tissue caused by inoculation of C. gloeosporioides in wound, white bars = 1 cm. (f) Fruit lesion depth caused by wound inoculation of C. gloeosporioides. The vertical bars represent SDs (n = 3). ‘*’ represents p < 0.05 (Student’s t-test).
2.5. Physiological Changes in Apple Leaves after C. gloeosporioides Infection
ROS have important roles in pathogen–plant interaction and are involved in hypersensitive responses and signal transduction. To investigate the actions of leaves against C. gloeosporioides infection, O2− was detected in ‘GL-3′ and MIR390b-overexpressing plants by a chemical tissue staining method. Via nitro blue tetrazolium chloride (NBT) staining, more blue precipitations in ‘GL-3′ leaves were observed, indicating a greater level of O2− accumulation than in MIR390b-overexpressing leaves exposed to C. gloeosporioides stress (Figure 6a). SOD catalyzes the conversion of O2− into H2O2 and POD uses H2O2 as an electron acceptor to directly oxidize phenols or amines. They are the first line of defense against oxidative damage and remove excess free radicals in plants and improve the stress resistance of plants. Accordingly, SOD and POD activity was measured in ‘GL-3′ and MIR390b-overexpressing plants. Under normal conditions, there was no significant difference in SOD and POD activity between ‘GL-3′ and MIR390b-overexpressing plants. Under C. gloeosporioides treatment, the antioxidant enzyme activity of all apple plants increased, and compared with ‘GL-3′, the SOD and POD activity of MIR390b-line1 and MIR390b-line3 plants increased at 3 d (Figure 6b,c). In agreement with the results of NBT staining, the O2− content was much higher in the control plants than in MIR390b-overexpressing plants under C. gloeosporioides treatment. These results indicated that MIR390b-overexpressing plants probably achieved enhanced C. gloeosporioides resistance by increasing the activity of antioxidant enzymes in vivo in response to C. gloeosporioides stress.
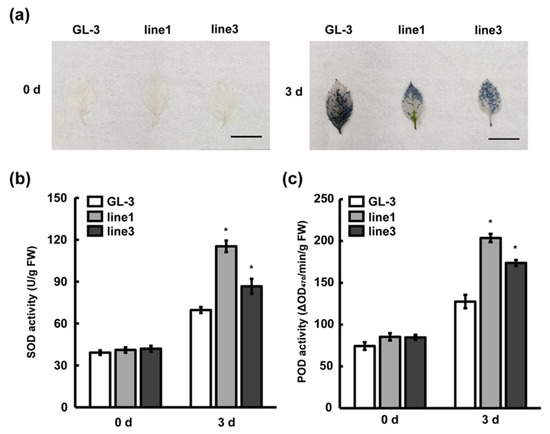
Figure 6.
Changes in ROS content and biochemical characteristics during C. gloeosporioides infection. (a) NBT staining of ‘GL-3′ and MIR390b-overexpressing plants’ leaves under C. gloeosporioides infection, black bars = 1 cm. (b) The activity of SOD in ‘GL-3′ and MIR390b-overexpressing plants during C. gloeosporioides infection. The vertical bars represent SDs (n = 3). ‘*’ represents p < 0.05 (Student’s t-test). (c) The activity of POD in ‘GL-3′ and MIR390b-overexpressing plants during C. gloeosporioides infection. The vertical bars represent SDs (n = 3). ‘*’ represents p < 0.05 (Student’s t-test).
2.6. Changes in Disease-Resistant Genes in MIR390b-Overexpressing Plants
Pathogenesis-related (PR) genes play unique roles in pathogen defense, and their expression is associated with enhanced resistance to pathogens [36,37]. To obtain clues regarding the molecular mechanism of mdm-MIR390b-mediated anthracnose resistance, the expression of four previously known C. gloeosporioides response genes in MIR390b-line1 and MIR390b-line3 plants was examined by qRT-PCR analysis. The qRT-PCR results demonstrated that the transcription levels of the selected four marker genes (PR2, PR3-1, PR10-1, and PR10-2) were remarkably upregulated in MIR390b-overexpressing plants compared with the controls (Figure 7).
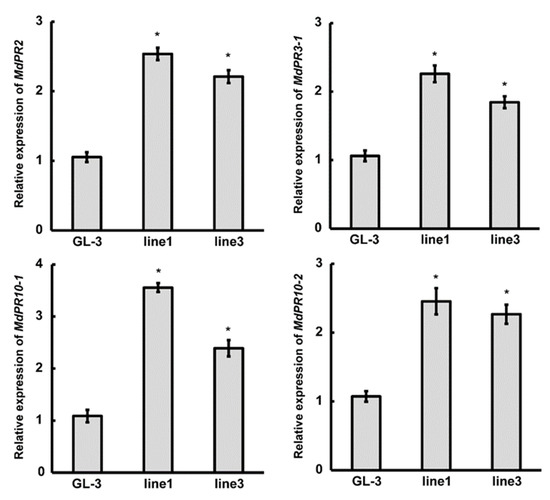
Figure 7.
Expression analysis of disease-resistant marker genes in ‘GL-3′ and MIR390b-overexpressing plants. Leaves of 4-week-old ‘GL-3′, MIR390b-line1, and MIR390b-line3 plants were harvested. Expression of PR2, PR3-1, PR10-1, and PR10-2 was measured in control and transgenic apple plants by qRT-PCR. The vertical bars represent SDs (n = 3). ‘*’ represents p < 0.05 (Student’s t-test).
3. Discussion
Plant miRNAs ubiquitously exist in various organisms and play imperative roles in multiple life processes [38,39]. Defining the role of miRNAs in the plant stress response is a hotspot of research. Recently, the mechanism of miR156 regulating salt tolerance in apple has been clearly studied [40]. In wheat, tae-miR171a was validated to have an obvious response to drought memory by regulating the expression of target genes [41]. The molecular mechanism of miR482b expression in tomato infected by Botrytis cinerea was revealed [42]. In plants, miR390 is an ancient family of miRNAs and is conserved among species [13]. Our analysis showed that mature miR390 sequences were conserved and six mdm-miR390 sequences were consistent (Figure 2). In terms of LR growth, miR390 promotes LR growth and the cleavage of TAS3 by AGO7 results in the production of tasiRNAs, which target mRNAs encoding ARFs [29,32]. In Physcomitrella patens, the overexpression of miR390 elevated miR390-triggered tasiRNA accumulation, decreased the level of tasiRNA targets, and caused the slower formation of gametophores [43]. It was reported that the overexpression of cgo-miR390s affected the development of reproductive organs in Cymbidium goeringii [44]. The effects of miR390 in plant growth and development have been investigated relatively clearly. However, studies on MIR390 in the biotic stress response remain largely lacking. In addition, apple miRNAs’ regulatory networks were analyzed by deep small RNA-seq and the emergence, evolution, and diversification of miR390 in land plants were elaborated by a series of bioinformatics analyses [19,27]. Thus, the functions of miR390 should be further verified by the transgenic strategy in apple. ‘GL-3′ apple plants that overexpressed mdm-MIR390b were used to analyze the specific role of mdm-MIR390b in response to C. gloeosporioides infection (Figure 4). Our research clearly demonstrated that the overexpression of mdm-MIR390b in apple improved its resistance to C. gloeosporioides infection (Figure 5). In a recent study, researchers found that phased small RNA involved systemic signaling in plants. Two tasiRNA3 derived from TAS3a and synthesized within several hours of Pseudomonas syringae DC 3000 infection were the early mobile signal; they then cleaved ARF2/3/4 to induce systemic acquired resistance (SAR) [45]. In MIR390b-overexpressing plants, the expression level of tasiRNA3 was higher compared with ‘GL-3′ plants. By contrast, qRT-PCR analysis of MdARF2/3/4 showed that they had a lower expression level compared with ‘GL-3′ plants (Figure S2). We speculate that apple anthracnose resistance was elevated by the miR390-tasiRNA3-ARFs pathway. The role of this pathway during C. gloeosporioides infection needs to be elucidated.
ROS, as important signaling molecules, have been studied for their roles in plant immunity [46]. A transient increase in ROS induces the expression of resistance proteins in plants, but the continuous high accumulation of ROS can lead to negative effects in plant cells [47]. ROS were observed to accumulate in many plants after the perception of pathogens and are considered to be closely related to the defense response of the plant [48,49,50]. One approach to evaluating the abilities of plant defense is to observe the accumulation of ROS in plants. NBT staining is convenient and efficient, used in many studies used to detect ROS. After C. gloeosporioides treatment, NBT staining showed the higher accumulation of O2− in ‘GL-3′ plants with respect to MIR390b-overexpressing plants (Figure 6a). Thus, the low level of ROS in the MIR390b-overexpressing plants probably enhanced the apple anthracnose resistance. A chloroplast-derived ROS burst destroyed the antioxidant enzyme system and caused cell death and leaf necrosis [51]. In research on postharvest longan, Lasiodiplodia theobromae-induced pericarp browning and disease development was due to the reduction of ROS scavenging ability and an increase in ROS production [52]. Excess ROS caused by pathogens severely harms plants, and plants have evolved using multiple methods to scavenge ROS [48,53]. One of the adaptive mechanisms is that the plants reduce the ROS content in vivo through antioxidant enzymes [48]. High SOD and POD activity is conducive to maintaining the physiological redox homeostasis to protect the plants from ROS-mediated damage. In the immune species Chrysanthemum makinoi var. wakasaense, SOD and POD activity increased significantly with Puccnia horiana inoculation [54]. The activity of SOD and POD increased at the sugarcane jointing stage, which was beneficial to deal with excessive ROS. The removal of superfluous ROS in sugarcane positively regulated resistance to smut [55]. High activity of ROS-scavenging enzymes (SOD) and high content of ascorbic acid and glutathione can inhibit the growth of Peronophythora litchii in harvested litchis, thereby enhancing disease resistance [56]. In agreement with these results, SOD and POD activity in MIR390b-overexpressing plants was higher than that in ‘GL-3′ plants (Figure 6b,c). In addition, the MIR390b-overexpressing fruit showed a small lesion diameter and depth, while the control fruit displayed serious necrotic tissue zones (Figure 5e,f). Hence, we speculated that the fruit might improve the anthracnose resistance by boosting SOD and POD activity. Active oxygen metabolism effectively prevented pathogens from stimulating membrane lipid peroxidation, causing the loss of cellular compartmentalization and the inhibition of photosynthetic electron transport [51,52]. PR genes were induced by pathogen infection and PR proteins may act as antimicrobial components in the plant defense response [36,57]. We found a remarkable upregulation of PR genes in MIR390b-overexpressing plants compared with ‘GL-3′ plants (Figure 7). The finding indicated that mdm-MIR390b strengthened apple’s resistance to C. gloeosporioides by improving the expression level of the PR gene. Thus, it is reasonable to conclude that mdm-MIR390b enhances resistance against pathogens by changing the activity of antioxidant enzymes and the expression levels of PR genes.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Apple tissue culture plants including ‘GL-3′, ‘Hanfu’, and a ‘Hanfu’ autotetraploid were cultured in Murashige and Skoog culture medium supplemented with 0.1 mg/L gibberellic acid 3, 0.2 mg/L 3-indoleacetic acid, and 0.3 mg/L 6-benzylaminopurine (apple subculture medium). They were placed at (24 ± 1) °C under long-day conditions (16 h:8 h, light:dark). ‘Hanfu’ apple fruit were collected at a commercially mature stage from an orchard at Shenyang Agriculture University. ‘Hanfu’ was used in qRT-PCR and fungal inoculation experiments. The ‘Hanfu’ autotetraploid was used in qRT-PCR. ‘GL-3′ was used for Agrobacterium-mediated leaf transformation, histochemical staining, determination of antioxidant enzymes, and fungal inoculation experiments. The fruit without physical injuries and with uniform shape and size were selected for fungal inoculation experiments.
4.2. Pathogen Culture and Inoculation Method
The phytopathogenic fungus Colletotrichum gloeosporioides was cultured for 10 days on potato dextrose agar medium at 28 °C in the dark. Conidia were then collected in sterile water, and the suspension population was adjusted to 107 spores/mL using a bacterial counting chamber. For fungal inoculation treatments, the plants of ‘Hanfu’ and ‘GL-3′ were sprayed with the above suspension. The inoculated plants were subsequently placed in the dark box at 25 °C for disease development. After 1, 2, 3, 4, and 5 d, plant leaves were used for photography or further experiments. At least three plants were used per treatment. The disease index was calculated according to a previous report [58].
4.3. Phylogenetic and Sequence Analysis
The precursor and mature sequences of the MIR390 family were downloaded from a public miRNA database, miRbase (https://www.mirbase.org/, accessed on 20 November 2022) [59]. Apple miR390* sequences were obtained from a comprehensive functional plant miRNA database, PmiREN (https://www.pmiren.com/, accessed on 20 November 2022) [60]. MIR390 precursors were used for phylogenetic analysis. The phylogenetic tree was constructed using MEGA X software with the Maximum Likelihood (ML) method under the Kimura 2-parameter model [61]. The primary tree was optimized by a web server, Evolview v3 [62].
Secondary structure prediction was performed with the web server RNAfold (http://rna.tbi.univie.ac.at/, accessed on 20 November 2022). Fold algorithms and basic options were set as follows: avoid isolated base pairs and minimum free energy (MFE) and partition function. Multiple sequence alignment of mature miR390 was executed by MEGA X and the result was processed using GeneDoc software. In fifteen plants, miR390 sequences were shown using the WebLogo online tool (http://weblogo.berkeley.edu/logo.cgi, accessed on 20 November 2022). The parameters were set to default.
ID numbers of apple MIR390 homologous sequences were as follows: ath-MIR390a (MI0001000), ath-MIR390b (MI0001001), osa-MIR390 (MI0001690), bna-MIR390a (MI0006447), bna-MIR390b (MI0006448), bna-MIR390c (MI0006449), vvi-MIR390 (MI0006552), ghr-MIR390a (MI0005647), ghr-MIR390b (MI0005648), ghr-MIR390c (MI0005649), mtr-MIR390 (MI0005586), sbi-MIR390 (MI0010887), zma-MIR390a (MI0013209), zma-MIR390b (MI0013244), ppe-MIR390 (MI0026099), nta-MIR390a (MI0021391), nta-MIR390b (MI0021392), nta-MIR390c (MI0021393), cme-MIR390a (MI0023238), cme-MIR390b (MI0018164), cme-MIR390c (MI0023239), sly-MIR390a (MI0029117), sly-MIR390b (MI0029123), stu-MIR390 (MI0025988), fve-MIR390a (MI0036404), fve-MIR390b (MI0036405).
4.4. Nucleic Acid Extraction and qRT-PCR Analysis
Plants frozen in liquid nitrogen were grounded into powder and we extracted DNA and RNA using the cetyltrimethylammonium bromide (CTAB) extraction method [58]. The nucleic acid concentration, nucleic acid quality, and cDNA synthesis were performed according to established protocols [58].
qRT-PCR was performed on ABI QuantStudio™ 6 Flex real-time PCR instrument. Each reaction solution was a total volume of 10 μL containing 5μL 2 × UltraSYBR Mixture (CWBIO, CW2601M), 0.5 μL of cDNA, 0.5 μL of 0.2 μM mdm-MIR390b-QF and 0.2 μM mdm-MIR390b-QR primers (Table S1), and 3.5 μL ddH2O. The reactions were initially polymerase activation at 95 °C for 5 min, followed by 40 cycles at 95 °C for 15s and 60 °C for 1 min. MdEF-1α (NCBI-ID: DQ341381) was an internal reference gene. For detecting the relative expression of mdm-miR390 and tasiRNA3, SuperReal PreMix (TIANGEN BIOTECH, FP206-02) was used. The previously reported methods were performed [31]. Apple 5SrRNA was selected as an internal reference. There were three biological and technical replicates. Relative expression levels of all examined genes and small RNA were calculated using the 2−ΔΔCt method [63].
4.5. Vector Construction and Transformation into Apple
mdm-MIR390b (103 bp) was amplified from the genomic DNA of ‘Hanfu’ with the mdm-MIR390b-F and mdm-MIR390b-R primers listed in Table S1 by PCR. The product was ligated into a pRI101-AN binary plant transformation vector at double restriction sites (Xba Ⅰ and Sac Ⅰ) to construct pRI101-MIR390b. The PCR mixture and cycle conditions were obtained from the instruction manual (KOD, TOYOBO, Osaka, Japan).
pRI101-MIR390b was transformed into apple ‘GL-3′ by the Agrobacterium tumefaciens-mediated (EHA105) method [64]. Resistant buds from separate apple leaf explants were chosen as independent transgenic events for further identification.
4.6. Confirmation of Transgenic Plants
Healthy ‘GL-3′ and putative transgenic plants were selected. Their leaves were cut into two parts along the direction of the vertical vein, and we then placed leaf fragments on apple subculture medium supplemented with 25 mg/L kanamycin, 250 mg/L cefotaxim sodium salt, and 250 mg/L timentin. After 15 days in the dark, they were moved to a light culture.
4.7. Agroinfiltration in Apple Fruit
A. tumefaciens (EHA105) containing pRI101-AN and pRI101-MIR390b were used for the infection of fruit. The preparation of the infection suspension was performed as previously described [65]. The fruit were wounded approximately 5 mm deep with a 1 mL syringe, and then 200 μL A. tumefaciens suspension was injected into apple fruit using a needleless syringe. The wounded fruit were treated in darkness at 25 °C. After 5 d, each injected site was treated with a 6-mm-diameter mycelial disc of C. gloeosporioides. Next, all apple fruit were placed in damp plastic bags and stored at 25 °C in the dark. After 5 d of incubation, the lesion diameter and depth were measured. Ten fruit were infiltrated with each construct and five different fruit were evaluated for apple disease conditions.
4.8. Histochemical Staining
Four-week-old healthy apple plants were subjected to the inoculum suspension for 3 d. Then, the leaves of three independent plants were stained by NBT. The previously reported methods were performed [66,67]. The experiments were conducted three times.
4.9. Determination of Antioxidant Enzymes
The apple leaves were isolated from three independent apple plants treated with pathogen C. gloeosporioides for 3 d. SOD and POD activity was measured according to previously reported methods [68]. The experiments were conducted three times.
4.10. Statistical Analysis
The analysis of differences between the data was performed with the IBM SPSS 26 software (Chicago, IL, USA). All values are presented as mean ± SD (Standard Deviation) as indicated. The single asterisk (Student’s t-test) and different letters (one-way ANOVA Duncan’s test) indicate significant differences at the p < 0.05 level.
5. Conclusions
In conclusion, this study demonstrated that mdm-MIR390b plays critical roles during C. gloeosporioides infection in apple. The apple MIR390b gene showed inductive expression during C. gloeosporioides infection. mdm-MIR390b-overexpressing plants were created and had higher anthracnose resistance by ROS deoxidation and higher-level expression of PR than control plants. Moreover, mdm-MIR390b provides a reference for resistance breeding in apple.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11233299/s1, Figure S1: Expression analysis of mdm-MIR390a, mdm-MIR390b, mdm-MIR390c, mdm-MIR390d and mdm-MIR390e/f in ‘Hanfu’ leaf and fruit; Figure S2: Expression analysis of tasiRNA3, MdARF2, MdARF3 and MdARF4 in ‘GL-3’ and MIR390b-overexpressing plants; Table S1: Primer sequences used in this study.
Author Contributions
Conceptualization: Y.M. and F.W., Funding acquisition: Y.M. and F.W., Formal analysis: J.S. and Q.J., Project administration: Y.M., Resources: Y.M. and F.W., Supervision: Y.M. and F.W., Validation: Q.J., F.W., J.S., S.Z. and X.D., Visualization: Q.J. and J.S., Supervision: Y.M. and F.W., Writing—original draft: J.S., Writing—review and editing: Y.M. and J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32272655) and the Project of the Education Department of Liaoning Province (No. LJKZ0647).
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank the editor and the anonymous reviewers for their efforts to improve the manuscript.
Conflicts of Interest
The authors declare that they have no conflicts of interest to report.
References
- Wang, N.; Wolf, J.; Zhang, F. Towards Sustainable Intensification of Apple Production in China—Yield Gaps and Nutrient Use Efficiency in Apple Farming Systems. J. Integr. Agric. 2016, 15, 716–725. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, R.; Gleason, M.L.; Sun, G. Sustainable Apple Disease Management in China: Challenges and Future Directions for a Transforming Industry. Plant Dis. 2022, 106, 786–799. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Lan, C.; Capriotti, L.; Ah-Fong, A.; Nino Sanchez, J.; Hamby, R.; Heller, J.; Zhao, H.; Glass, N.L.; Judelson, H.S.; et al. Spray-Induced Gene Silencing for Disease Control Is Dependent on the Efficiency of Pathogen RNA Uptake. Plant Biotechnol. J. 2021, 19, 1756–1768. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, P.; Voinnet, O. Revisiting the Principles of MicroRNA Target Recognition and Mode of Action. Nat. Rev. Mol. Cell Biol. 2009, 10, 141–148. [Google Scholar] [CrossRef]
- Rogers, K.; Chen, X. Biogenesis, Turnover, and Mode of Action of Plant MicroRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef]
- Iwakawa, H.; Tomari, Y. The Functions of MicroRNAs: MRNA Decay and Translational Repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef]
- Yu, Y.; Jia, T.; Chen, X. The ‘How’ and ‘Where’ of Plant MicroRNAs. New Phytol. 2017, 216, 1002–1017. [Google Scholar] [CrossRef]
- Wang, J.; Mei, J.; Ren, G. Plant MicroRNAs: Biogenesis, Homeostasis, and Degradation. Front. Plant Sci. 2019, 10, 360. [Google Scholar] [CrossRef]
- Park, W.; Li, J.; Song, R.; Messing, J.; Chen, X. CARPEL FACTORY, a Dicer Homolog, and HEN1, a Novel Protein, Act in MicroRNA Metabolism in Arabidopsis Thaliana. Curr. Biol. 2002, 12, 1484–1495. [Google Scholar] [CrossRef]
- Dong, Z.; Han, M.-H.; Fedoroff, N. The RNA-Binding Proteins HYL1 and SE Promote Accurate in Vitro Processing of Pri-MiRNA by DCL1. Proc. Natl. Acad. Sci. USA 2008, 105, 9970–9975. [Google Scholar] [CrossRef]
- Cuperus, J.T.; Montgomery, T.A.; Fahlgren, N.; Burke, R.T.; Townsend, T.; Sullivan, C.M.; Carrington, J.C. Identification of MIR390a Precursor Processing-Defective Mutants in Arabidopsis by Direct Genome Sequencing. Proc. Natl. Acad. Sci. USA 2010, 107, 466–471. [Google Scholar] [CrossRef]
- Cambiagno, D.A.; Giudicatti, A.J.; Arce, A.L.; Gagliardi, D.; Li, L.; Yuan, W.; Lundberg, D.S.; Weigel, D.; Manavella, P.A. HASTY Modulates MiRNA Biogenesis by Linking Pri-MiRNA Transcription and Processing. Mol. Plant 2021, 14, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Chávez Montes, R.A.; De Rosas-Cárdenas, F.F.; De Paoli, E.; Accerbi, M.; Rymarquis, L.A.; Mahalingam, G.; Marsch-Martínez, N.; Meyers, B.C.; Green, P.J.; de Folter, S. Sample Sequencing of Vascular Plants Demonstrates Widespread Conservation and Divergence of MicroRNAs. Nat. Commun. 2014, 5, 3722. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, A.M.; Allen, E.; Givan, S.; Smith, D.; Carrington, J.C.; Kasschau, K.D. ASRP: The Arabidopsis Small RNA Project Database. Nucleic Acids Res. 2005, 33, D637–D640. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Girke, T.; Jain, P.K.; Zhu, J.-K. Cloning and Characterization of MicroRNAs from Rice. Plant Cell 2005, 17, 1397–1411. [Google Scholar] [CrossRef]
- Tang, S.; Wang, Y.; Li, Z.; Gui, Y.; Xiao, B.; Xie, J.; Zhu, Q.-H.; Fan, L. Identification of Wounding and Topping Responsive Small RNAs in Tobacco (Nicotiana tabacum). BMC Plant Biol. 2012, 12, 28. [Google Scholar] [CrossRef]
- Kravchik, M.; Sunkar, R.; Damodharan, S.; Stav, R.; Zohar, M.; Isaacson, T.; Arazi, T. Global and Local Perturbation of the Tomato MicroRNA Pathway by a Trans-Activated DICER-LIKE 1 Mutant. J. Exp. Bot. 2014, 65, 725–739. [Google Scholar] [CrossRef]
- Zhang, L.; Chia, J.-M.; Kumari, S.; Stein, J.C.; Liu, Z.; Narechania, A.; Maher, C.A.; Guill, K.; McMullen, M.D.; Ware, D. A Genome-Wide Characterization of MicroRNA Genes in Maize. PLoS Genet. 2009, 5, e1000716. [Google Scholar] [CrossRef]
- Xia, R.; Zhu, H.; An, Y.; Beers, E.P.; Liu, Z. Apple MiRNAs and TasiRNAs with Novel Regulatory Networks. Genome Biol. 2012, 13, R47. [Google Scholar] [CrossRef]
- Axtell, M.J.; Jan, C.; Rajagopalan, R.; Bartel, D.P. A Two-Hit Trigger for SiRNA Biogenesis in Plants. Cell 2006, 127, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7-MiR390 Interaction and Dual Functionality in TAS3 Trans-Acting SiRNA Formation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef]
- Jouannet, V.; Moreno, A.B.; Elmayan, T.; Vaucheret, H.; Crespi, M.D.; Maizel, A. Cytoplasmic Arabidopsis AGO7 Accumulates in Membrane-Associated SiRNA Bodies and Is Required for Ta-SiRNA Biogenesis. EMBO J. 2012, 31, 1704–1713. [Google Scholar] [CrossRef] [PubMed]
- Rajeswaran, R.; Pooggin, M.M. RDR6-Mediated Synthesis of Complementary RNA Is Terminated by MiRNA Stably Bound to Template RNA. Nucleic Acids Res. 2012, 40, 594–599. [Google Scholar] [CrossRef]
- Sakurai, Y.; Baeg, K.; Lam, A.Y.W.; Shoji, K.; Tomari, Y.; Iwakawa, H. Cell-Free Reconstitution Reveals the Molecular Mechanisms for the Initiation of Secondary SiRNA Biogenesis in Plants. Proc. Natl. Acad. Sci. USA 2021, 118, e2102889118. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. MicroRNA-Directed Phasing during Trans-Acting SiRNA Biogenesis in Plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. MiR390, Arabidopsis TAS3 TasiRNAs, and Their AUXIN RESPONSE FACTOR Targets Define an Autoregulatory Network Quantitatively Regulating Lateral Root Growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef]
- Xia, R.; Xu, J.; Meyers, B.C. The Emergence, Evolution, and Diversification of the MiR390-TAS3-ARF Pathway in Land Plants. Plant Cell 2017, 29, 1232–1247. [Google Scholar] [CrossRef]
- Ma, J.; Li, C.; Gao, P.; Qiu, Y.; Zong, M.; Zhang, H.; Wang, J. Melon Shoot Organization 1, Encoding an AGRONAUTE7 Protein, Plays a Crucial Role in Plant Development. Theor. Appl. Genet. 2022, 135, 2875–2890. [Google Scholar] [CrossRef]
- Hobecker, K.V.; Reynoso, M.A.; Bustos-Sanmamed, P.; Wen, J.; Mysore, K.S.; Crespi, M.; Blanco, F.A.; Zanetti, M.E. The MicroRNA390/TAS3 Pathway Mediates Symbiotic Nodulation and Lateral Root Growth. Plant Physiol. 2017, 174, 2469–2486. [Google Scholar] [CrossRef]
- Yoon, E.K.; Yang, J.H.; Lim, J.; Kim, S.H.; Kim, S.-K.; Lee, W.S. Auxin Regulation of the MicroRNA390-Dependent Transacting Small Interfering RNA Pathway in Arabidopsis Lateral Root Development. Nucleic Acids Res. 2010, 38, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Guan, Y.; Zhang, Z.; Li, H. MiR390-TasiRNA3-ARF4 Pathway Is Involved in Regulating Flowering Time in Woodland Strawberry. Plant Cell Rep. 2022, 41, 921–934. [Google Scholar] [CrossRef]
- He, F.; Xu, C.; Fu, X.; Shen, Y.; Guo, L.; Leng, M.; Luo, K. The MicroRNA390/TRANS-ACTING SHORT INTERFERING RNA3 Module Mediates Lateral Root Growth under Salt Stress via the Auxin Pathway. Plant Physiol. 2018, 177, 775–791. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, F.; Zhang, Z.; Fu, J.; Ma, Y. Characterization of Fungi Resistance in Two Autotetraploid Apple Cultivars. Sci. Hortic. 2017, 220, 27–35. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, L.; Zhang, F.; Ou, C.; Wang, F.; Zhang, Z. Involvement of Auxin and Brassinosteroid in Dwarfism of Autotetraploid Apple (Malus × domestica). Sci. Rep. 2016, 6, 26719. [Google Scholar] [CrossRef]
- Blaszczyk, J.; Tropea, J.E.; Bubunenko, M.; Routzahn, K.M.; Waugh, D.S.; Court, D.L.; Ji, X. Crystallographic and Modeling Studies of RNase III Suggest a Mechanism for Double-Stranded RNA Cleavage. Structure 2001, 9, 1225–1236. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xu, C.; Wei, H.; Fan, W.; Li, T. Two Pathogenesis-Related Proteins Interact with Leucine-Rich Repeat Proteins to Promote Alternaria Leaf Spot Resistance in Apple. Hortic. Res. 2021, 8, 219. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, F.; Yang, S.; Zhang, Y.; Xue, H.; Wang, Y.; Yan, S.; Wang, Y.; Zhang, Z.; Ma, Y. MdWRKY100 Encodes a Group I WRKY Transcription Factor in Malus Domestica That Positively Regulates Resistance to Colletotrichum Gloeosporioides Infection. Plant Sci. 2019, 286, 68–77. [Google Scholar] [CrossRef]
- Li, S.; Castillo-González, C.; Yu, B.; Zhang, X. The Functions of Plant Small RNAs in Development and in Stress Responses. Plant J. 2017, 90, 654–670. [Google Scholar] [CrossRef]
- Axtell, M.J.; Bartel, D.P. Antiquity of MicroRNAs and Their Targets in Land Plants. Plant Cell 2005, 17, 1658–1673. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, F.; Jiang, Q.; Yang, S.; Yue, P.; Wang, F.; Zhang, Y.; Li, L.; He, P.; et al. The MiR156/SPL Module Regulates Apple Salt Stress Tolerance by Activating MdWRKY100 Expression. Plant Biotechnol. J. 2021, 19, 311–323. [Google Scholar] [CrossRef]
- Yue, H.; Zhang, H.; Su, N.; Sun, X.; Zhao, Q.; Weining, S.; Nie, X.; Yue, W. Integrate Small RNA and Degradome Sequencing to Reveal Drought Memory Response in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2022, 23, 5917. [Google Scholar] [CrossRef]
- Wu, F.; Xu, J.; Gao, T.; Huang, D.; Jin, W. Molecular Mechanism of Modulating MiR482b Level in Tomato with Botrytis cinerea Infection. BMC Plant Biol. 2021, 21, 496. [Google Scholar] [CrossRef]
- Cho, S.H.; Coruh, C.; Axtell, M.J. MiR156 and MiR390 Regulate TasiRNA Accumulation and Developmental Timing in Physcomitrella Patens. Plant Cell 2012, 24, 4837–4849. [Google Scholar] [CrossRef]
- Xu, Z.; Liu, Q.; Chen, Y.; He, Y.; Hu, F. MiR390 Family of Cymbidium goeringii Is Involved in the Development of Reproductive Organs in Transgenic Arabidopsis. BMC Plant Biol. 2022, 22, 149. [Google Scholar] [CrossRef] [PubMed]
- Shine, M.B.; Zhang, K.; Liu, H.; Lim, G.; Xia, F.; Yu, K.; Hunt, A.G.; Kachroo, A.; Kachroo, P. Phased Small RNA–Mediated Systemic Signaling in Plants. Sci. Adv. 2022, 8, eabm8791. [Google Scholar] [CrossRef]
- Wang, N.; Fan, X.; He, M.; Hu, Z.; Tang, C.; Zhang, S.; Lin, D.; Gan, P.; Wang, J.; Huang, X.; et al. Transcriptional Repression of TaNOX10 by TaWRKY19 Compromises ROS Generation and Enhances Wheat Susceptibility to Stripe Rust. Plant Cell 2022, 34, 1784–1803. [Google Scholar] [CrossRef]
- Kandhol, N.; Singh, V.P.; Wang, Y.; Chen, Z.-H.; Tripathi, D.K. Ca2+ Sensor-Mediated ROS Homeostasis: Defense without Yield Penalty. Trends Plant Sci. 2022, 27, 834–836. [Google Scholar] [CrossRef]
- Lehmann, S.; Serrano, M.; L’Haridon, F.; Tjamos, S.E.; Metraux, J.-P. Reactive Oxygen Species and Plant Resistance to Fungal Pathogens. Phytochemistry 2015, 112, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Chae, H.B.; Kim, M.G.; Kang, C.H.; Park, J.H.; Lee, E.S.; Lee, S.-U.; Chi, Y.H.; Paeng, S.K.; Bae, S.B.; Wi, S.D.; et al. Redox Sensor QSOX1 Regulates Plant Immunity by Targeting GSNOR to Modulate ROS Generation. Mol. Plant 2021, 14, 1312–1327. [Google Scholar] [CrossRef]
- Hu, B.; Zhou, Y.; Zhou, Z.; Sun, B.; Zhou, F.; Yin, C.; Ma, W.; Chen, H.; Lin, Y. Repressed OsMESL Expression Triggers Reactive Oxygen Species-Mediated Broad-Spectrum Disease Resistance in Rice. Plant Biotechnol. J. 2021, 19, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yin, C.; Strasser, R.J.; Govindjee; Yang, C.; Qiang, S. Reactive Oxygen Species from Chloroplasts Contribute to 3-Acetyl-5-Isopropyltetramic Acid-Induced Leaf Necrosis of Arabidopsis Thaliana. Plant Physiol. Biochem. 2012, 52, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Lin, H.; Zhang, S.; Lin, Y.; Wang, H.; Lin, M.; Hung, Y.-C.; Chen, Y. The Roles of ROS Production-Scavenging System in Lasiodiplodia Theobromae (Pat.) Griff. & Maubl.-Induced Pericarp Browning and Disease Development of Harvested Longan Fruit. Food Chem. 2018, 247, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yang, J.; Wang, Y.; Du, J.; Zhang, J.; Luisi, B.F.; Liang, W. Broad-Spectrum Chemicals Block ROS Detoxification to Prevent Plant Fungal Invasion. Curr. Biol. 2022, 32, 3886–3897.e6. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, J.; Xia, X.; Xu, Y.; Sun, J.; Gu, J.; Sun, H.; Lei, H.; Chen, F.; Jiang, J.; et al. Comparative Analysis of Leaf Trichomes, Epidermal Wax And Defense Enzymes Activities in Response to Puccinia Horiana in Chrysanthemum and Ajania Species. Hortic. Plant J. 2020, 6, 191–198. [Google Scholar] [CrossRef]
- Deng, Q.; Wu, J.; Chen, J.; Shen, W. Physiological Mechanisms of Improved Smut Resistance in Sugarcane Through Application of Silicon. Front. Plant Sci. 2020, 11, 568130. [Google Scholar] [CrossRef]
- Jiang, X.; Lin, H.; Lin, M.; Chen, Y.; Wang, H.; Lin, Y.; Shi, J.; Lin, Y. A Novel Chitosan Formulation Treatment Induces Disease Resistance of Harvested Litchi Fruit to Peronophythora Litchii in Association with ROS Metabolism. Food Chem. 2018, 266, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Ma, S.; Chen, X.; Shen, X.; Yin, C.; Mao, Z. Transcriptome Changes Associated with Apple (Malus domestica) Root Defense Response after Fusarium proliferatum f. Sp. malus domestica Infection. BMC Genomics 2022, 23, 484. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Yang, S.; Wang, Y.; Zhang, B.; Zhang, F.; Xue, H.; Jiang, Q.; Ma, Y. Overexpression of Chitinase Gene Enhances Resistance to Colletotrichum gloeosporioides and Alternaria alternata in Apple (Malus × domestica). Sci. Hortic. 2021, 277, 109779. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Guo, Z.; Kuang, Z.; Zhao, Y.; Deng, Y.; He, H.; Wan, M.; Tao, Y.; Wang, D.; Wei, J.; Li, L.; et al. PmiREN2.0: From Data Annotation to Functional Exploration of Plant MicroRNAs. Nucleic Acids Res. 2022, 50, D1475–D1482. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A Webserver for Visualization, Annotation, and Management of Phylogenetic Trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Chen, K.; Tang, X.; Song, M.; Guo, Y.; Liu, L.; Xue, H.; Dai, H.; Zhang, Z.; Chen, K.; Tang, X.; et al. Functional Identification of MdMYB5 Involved in Secondary Cell Wall Formation in Apple. Fruit Res. 2021, 1, 6. [Google Scholar] [CrossRef]
- Dai, H.; Li, W.; Han, G.; Yang, Y.; Ma, Y.; Li, H.; Zhang, Z. Development of a Seedling Clone with High Regeneration Capacity and Susceptibility to Agrobacterium in Apple. Sci. Hortic. 2013, 164, 202–208. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Mao, K.; Zhao, C.; Zhao, X.-Y.; Zhang, H.-L.; Shu, H.-R.; Hao, Y.-J. MdCOP1 Ubiquitin E3 Ligases Interact with MdMYB1 to Regulate Light-Induced Anthocyanin Biosynthesis and Red Fruit Coloration in Apple. Plant Physiol. 2012, 160, 1011–1022. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, F.; Wang, Y.; Xue, H.; Jiang, Q.; Shi, J.; Dai, H.; Zhang, Z.; Li, L.; He, P.; et al. MdHAL3, a 4′-Phosphopantothenoylcysteine Decarboxylase, Is Involved in the Salt Tolerance of Autotetraploid Apple. Plant Cell Rep. 2020, 39, 1479–1491. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Guo, Y.; Song, M.; Liu, L.; Xue, H.; Dai, H.; Zhang, Z. Dual Role of MdSND1 in the Biosynthesis of Lignin and in Signal Transduction in Response to Salt and Osmotic Stress in Apple. Hortic. Res. 2020, 7, 204. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Li, Z.; Yang, J.; Zu, Y. Responses of Antioxidant Enzymes and Key Resistant Substances in Perennial Ryegrass (Lolium perenne L.) to Cadmium and Arsenic Stresses. BMC Plant Biol. 2022, 22, 145. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).