Conservation and Divergence of Phosphoenolpyruvate Carboxylase Gene Family in Cotton
Abstract
:1. Introduction
2. Methods and Materials
2.1. Collection of Genome Sequences
2.2. Genetic Identification, Multiple Sequence Alignment and Phylogenetic Analysis
2.3. Conserved Motif, Exon-Intron Structure and Promoter Analysis
2.4. Chromosomal Distribution and Subcellular Localization
2.5. Gene Duplication, Collinearity Analysis and Selection Pressure Analysis
2.6. Plant Materials and Stress Treatments
2.7. Gene Expression Patterns of the PEPC Gene Family
3. Results
3.1. Genome-Wide Identification and Biophysical Characteristics of the PEPCs
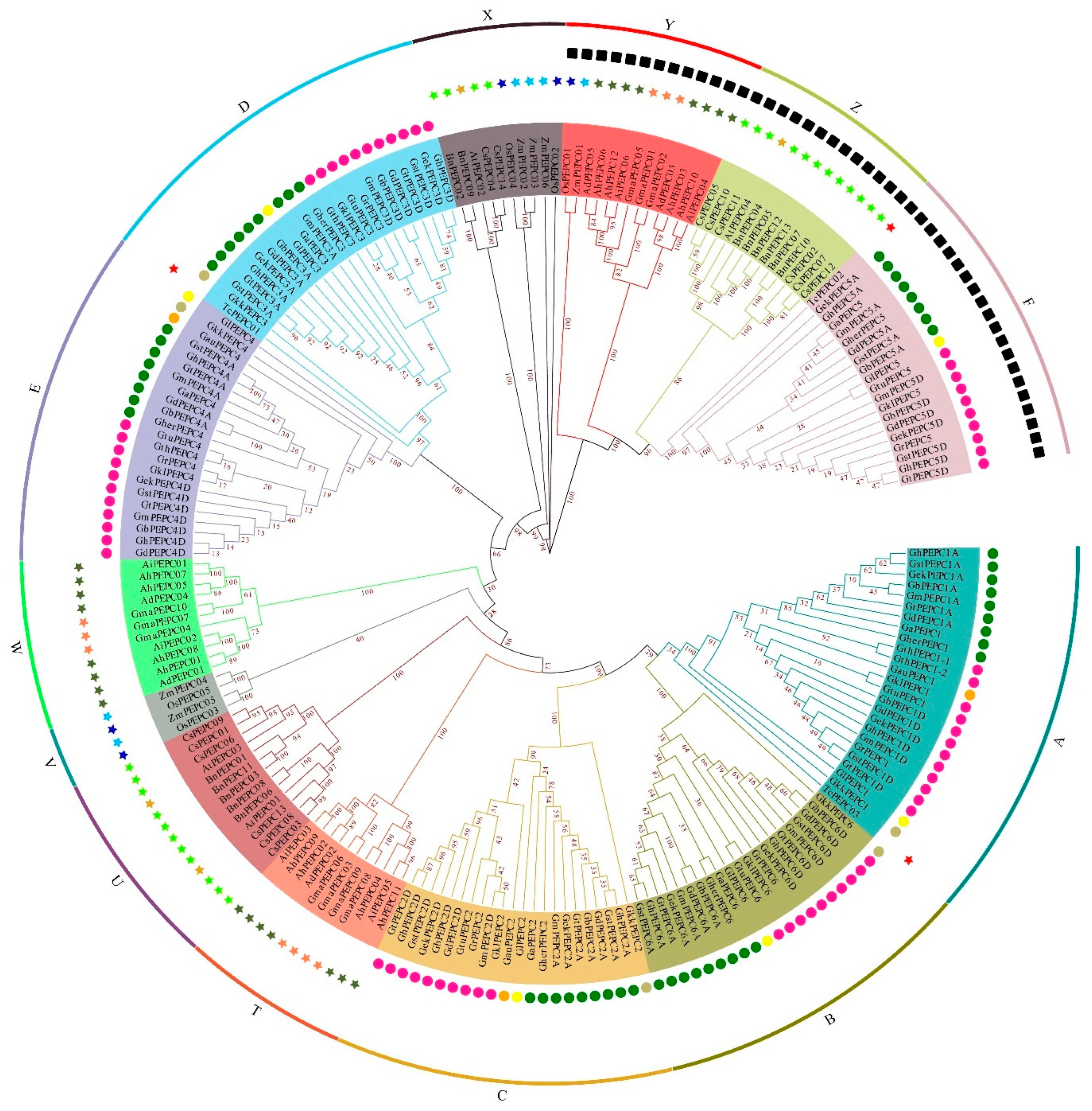
3.2. Phylogenetic Analysis of the PEPC Gene Family
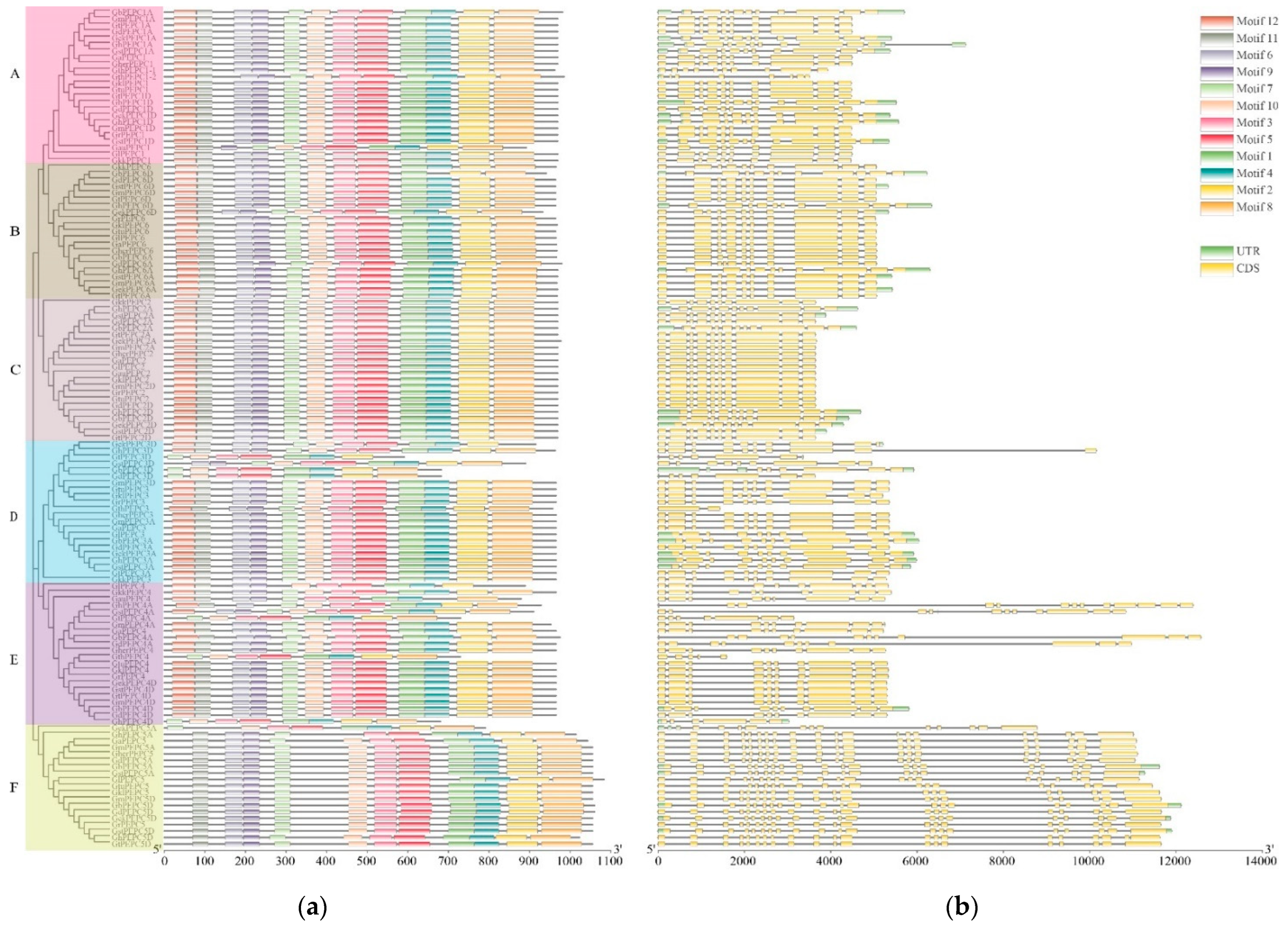
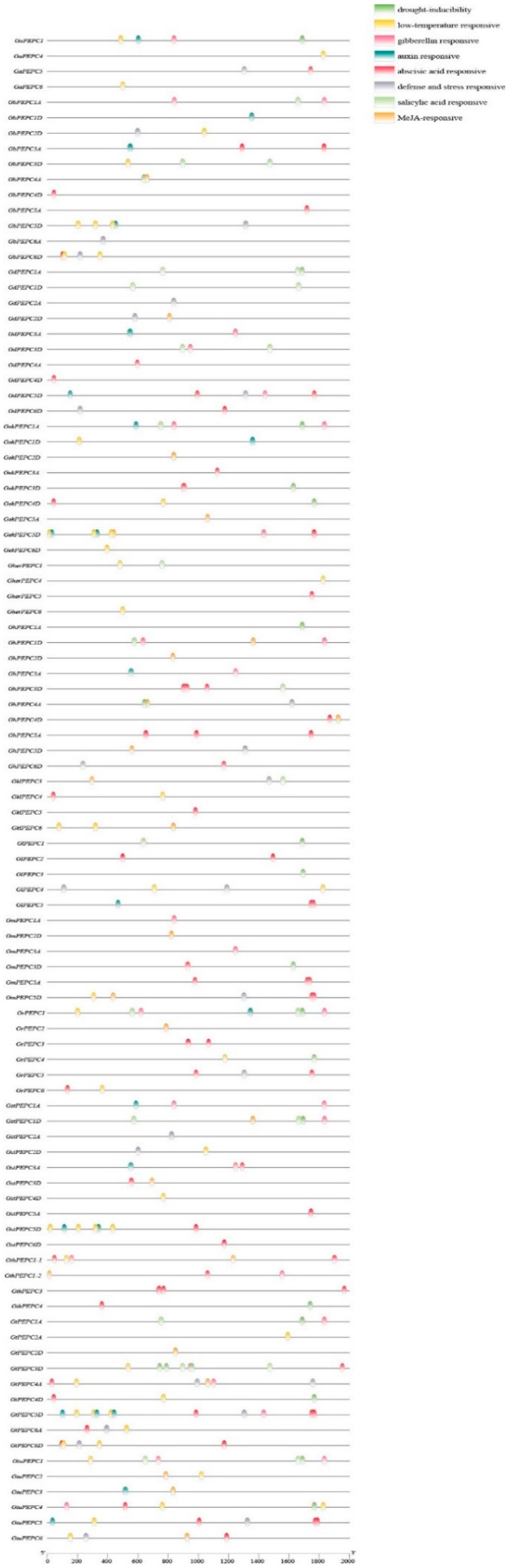
3.3. Conserved Motif, Gene Structure and Promoter Analysis
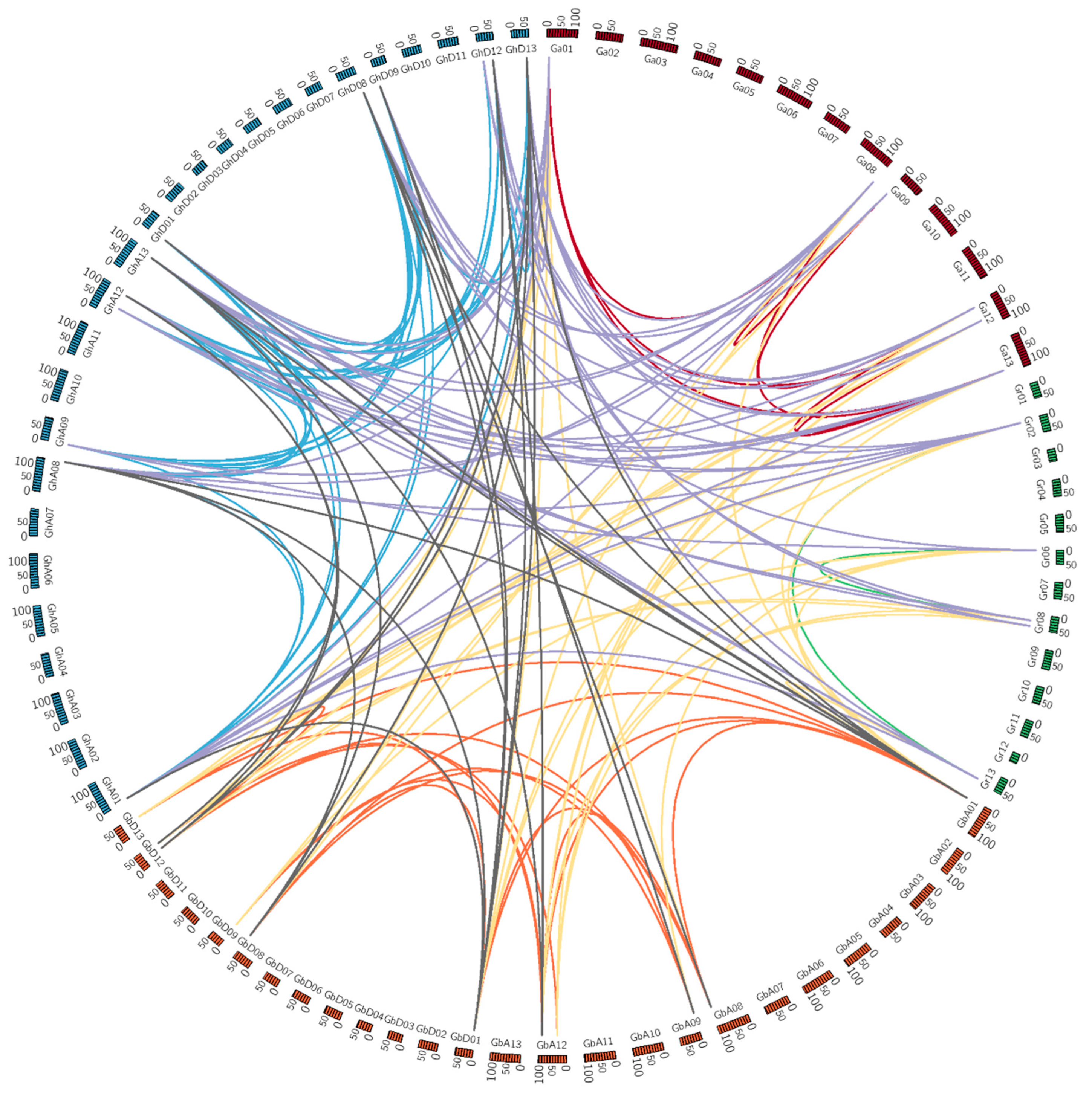
3.4. Chromosomal Distribution and Gene Duplications of Cotton PEPC Family
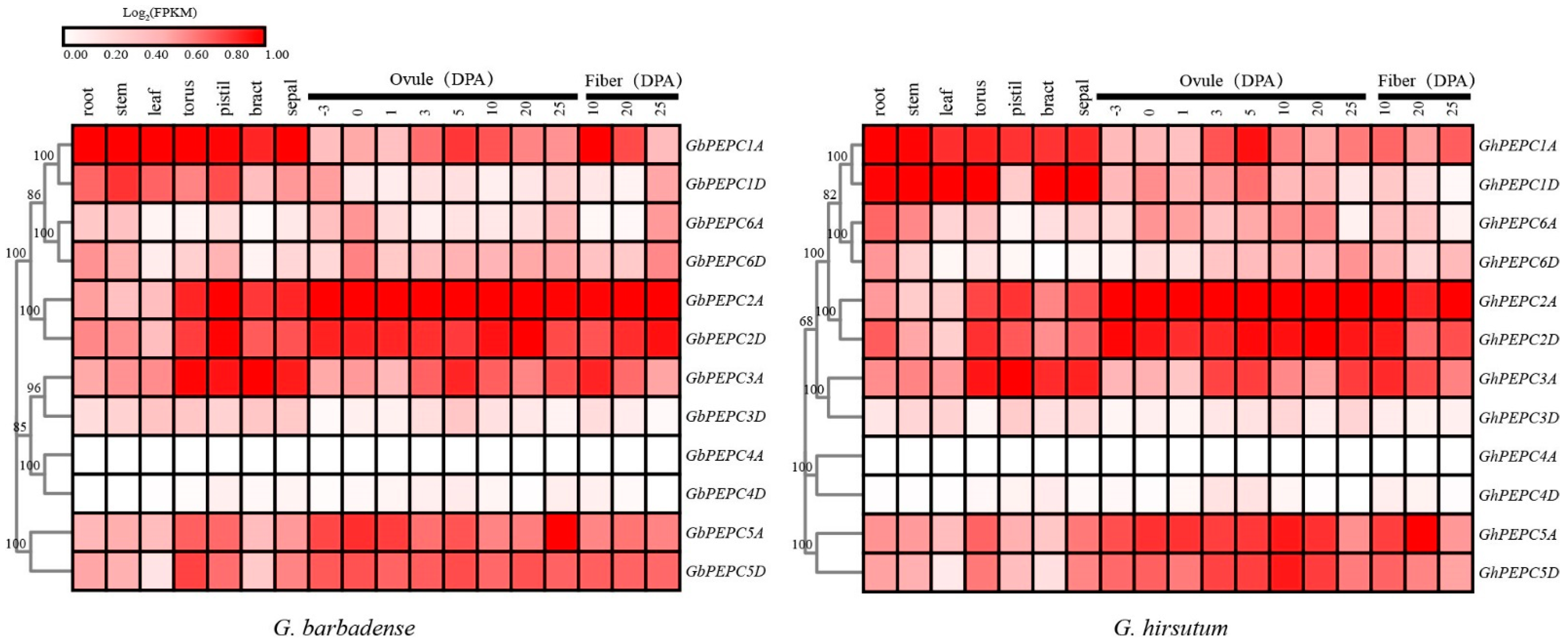
3.5. Temporal and Spatial Expression of PEPC Genes in Cotton
3.6. Abiotic Stress-Induced Aberrant Expression of PEPC Genes
4. Discussion
4.1. Conservation and Divergence of PEPC Genes in Cotton
4.2. Temporal and Spatial Expression of PEPC Genes Indicates Their Specific Function in Different Biological Processes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chollet, R.; Vidal, J.; Oleary, M.H. Phosphoenolpyruvate carboxylase: A ubiquitous, highly regulated enzyme in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 273–298. [Google Scholar] [CrossRef] [Green Version]
- Izui, K.; Matsumura, H.; Furumoto, T.; Kai, Y. Phosphoenolpyruvate carboxylase: A New Era of Structural Biology. Annu. Rev. Plant Biol. 2004, 55, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Kai, Y.; Matsumura, H.; Izui, K. Phosphoenolpyruvate carboxylase: Three-dimensional structure and molecular mechanisms. Arch. Biochem. Biophys. 2003, 414, 170–179. [Google Scholar] [CrossRef]
- O’Leary, B.; Fedosejevs, E.T.; Hill, A.T.; Bettridge, J.; Park, J.; Rao, S.K.; Leach, C.A.; Plaxton, W.C. Tissue-specific expression and post-translational modifications of plant- and bacterial-type phosphoenolpyruvate carboxylase isozymes of the castor oil plant, Ricinus communis L. J. Exp. Bot. 2011, 62, 5485–5495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Leary, B.; Park, J.; Plaxton, W.C. The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): Recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem. J. 2011, 436, 15–34. [Google Scholar] [CrossRef] [Green Version]
- Cousins, A.B.; Baroli, I.; Badger, M.R.; Ivakov, A.; Lea, P.J.; Leegood, R.C.; von Caemmerer, S. The Role of Phosphoenolpyruvate Carboxylase during C4 Photosynthetic Isotope Exchange and Stomatal Conductance. Plant Physiol. 2007, 145, 1006–1017. [Google Scholar] [CrossRef] [Green Version]
- Asai, N.; Nakajima, N.; Tamaoki, M.; Kamada, H.; Kondo, N. Role of malate synthesis mediated by phosphoenolpyruvate carboxylase in guard cells in the regulation of stomatal movement. Plant Cell Physiol. 2000, 41, 10–15. [Google Scholar] [CrossRef]
- González, M.-C.; Sánchez, R.; Cejudo, F.J. Abiotic stresses affecting water balance induce phosphoenolpyruvate carboxylase expression in roots of wheat seedlings. Planta 2003, 216, 985–992. [Google Scholar] [CrossRef]
- Cheng, G.; Wang, L.; Lan, H. Cloning of PEPC-1 from a C4 halophyte Suaeda aralocaspica without Kranz anatomy and its recombinant enzymatic activity in responses to abiotic stresses. Enzym. Microb. Technol. 2016, 83, 57–67. [Google Scholar] [CrossRef]
- Liu, D.; Hu, R.; Zhang, J.; Guo, H.-B.; Cheng, H.; Li, L.; Borland, A.; Qin, H.; Chen, J.-G.; Muchero, W.; et al. Overexpression of an Agave Phosphoenolpyruvate Carboxylase Improves Plant Growth and Stress Tolerance. Cells 2021, 10, 582. [Google Scholar] [CrossRef]
- Giuliani, R.; Karki, S.; Covshoff, S.; Lin, H.C.; Coe, R.A.; Koteyeva, N.K.; Evans, M.A.; Quick, W.P.; von Caemmerer, S.; Furbank, R.T.; et al. Transgenic maize phosphoenolpyruvate carboxylase alters leaf-atmosphere CO2 and 13CO2 exchanges in Oryza sativa. Photosynth. Res. 2019, 142, 153–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Li, X.; Dai, C.; Zhou, J.; Yan, T.; Zhang, J. Improved short-term drought response of transgenic rice over-expressing maize C4 phosphoenolpyruvate carboxylase via calcium signal cascade. J. Plant Physiol. 2017, 218, 206–221. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, X.; Lu, W.; Wei, X.; Zhang, Q.; Lv, C.; Song, N. Enhanced photorespiration in transgenic rice over-expressing maize C4 phosphoenolpyruvate carboxylase gene contributes to alleviating low nitrogen stress. Plant Physiol. Biochem. 2018, 130, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Tang, Y.; Zhang, J.; Yang, M.; Xu, Y. RNA Interference-based Suppression of Phosphoenolpyruvate Carboxylase Results in Susceptibility of Rapeseed to Osmotic Stress. J. Integr. Plant Biol. 2010, 52, 585–592. [Google Scholar] [CrossRef]
- Deng, X.; Cai, J.; Li, Y.; Fei, X. Expression and knockdown of the PEPC1 gene affect carbon flux in the biosynthesis of triacylglycerols by the green alga Chlamydomonas reinhardtii. Biotechnol. Lett. 2014, 36, 2199–2208. [Google Scholar] [CrossRef]
- Osorio, S.; Vallarino, J.G.; Szecowka, M.; Ufaz, S.; Tzin, V.; Angelovici, R.; Galili, G.; Fernie, A.R. Alteration of the Interconversion of Pyruvate and Malate in the Plastid or Cytosol of Ripening Tomato Fruit Invokes Diverse Consequences on Sugar But Similar Effects on Cellular Organic Acid, Metabolism, and Transitory Starch Accumulation. Plant Physiol. 2012, 161, 628–643. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, R.; Cejudo, F.J. Identification and Expression Analysis of a Gene Encoding a Bacterial-Type Phosphoenolpyruvate Carboxylase from Arabidopsis and Rice. Plant Physiol. 2003, 132, 949–957. [Google Scholar] [CrossRef] [Green Version]
- Besnard, G.; Pinçon, G.; D’Hont, A.; Hoarau, J.-Y.; Cadet, F.; Offmann, B. Characterisation of the phosphoenolpyruvate carboxylase gene family in sugarcane (Saccharum spp.). Theor. Appl. Genet. 2003, 107, 470–478. [Google Scholar] [CrossRef]
- Wang, N.; Zhong, X.; Cong, Y.; Wang, T.; Yang, S.; Li, Y.; Gai, J. Genome-wide Analysis of Phosphoenolpyruvate Carboxylase Gene Family and Their Response to Abiotic Stresses in Soybean. Sci. Rep. 2016, 6, 38448. [Google Scholar] [CrossRef]
- Waseem, M.; Ahmad, F. The phosphoenolpyruvate carboxylase gene family identification and expression analysis under abiotic and phytohormone stresses in Solanum lycopersicum L. Gene 2018, 690, 11–20. [Google Scholar] [CrossRef]
- Crétin, C.; Santi, S.; Keryer, E.; Lepinie, L.; Tagu, D.; Vidai, J.; Gadal, P. The phosphoenolpyruvate carboxylase gene family of Sorghum: Promoter structures, amino acid sequences and expression of genes. Gene 1991, 99, 87–94. [Google Scholar] [CrossRef]
- Li, X.-R.; Wang, L.; Ruan, Y.-L. Developmental and molecular physiological evidence for the role of phosphoenolpyruvate carboxylase in rapid cotton fibre elongation. J. Exp. Bot. 2009, 61, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Li, J.; Guo, X.; Jin, S.; Zhang, X. Metabolic engineering of cottonseed oil biosynthesis pathway via RNA interference. Sci. Rep. 2016, 6, 33342. [Google Scholar] [CrossRef]
- Zhao, Y.; Huang, Y.; Wang, Y.; Cui, Y.; Liu, Z.; Hua, J. RNA interference of GhPEPC2 enhanced seed oil accumulation and salt tolerance in Upland cotton. Plant Sci. 2018, 271, 52–61. [Google Scholar] [CrossRef]
- Zhao, Y.; Guo, A.; Wang, Y.; Hua, J. Evolution of PEPC gene family in Gossypium reveals functional diversification and GhPEPC genes responding to abiotic stresses. Gene 2019, 698, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Wendel, J.F.; Gundlach, H.; Guo, H.; Jenkins, J.; Jin, D.; Llewellyn, D.; Showmaker, K.C.; Shu, S.; Udall, J.; et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 2012, 492, 423–427. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S.; et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef]
- Peng, R.; Jones, D.C.; Liu, F.; Zhang, B. From sequencing to genome editing for cotton improvement. Trends Biotechnol. 2020, 39, 221–224. [Google Scholar] [CrossRef]
- Udall, J.A.; Long, E.; Ramaraj, T.; Conover, J.L.; Yuan, D.; Grover, C.E.; Gong, L.; Arick, M.; Masonbrink, R.E.; Peterson, D.G.; et al. The Genome Sequence of Gossypioides kirkii Illustrates a Descending Dysploidy in Plants. Front. Plant Sci. 2019, 10, 1541. [Google Scholar] [CrossRef]
- Chen, Z.J.; Sreedasyam, A.; Ando, A.; Song, Q.; De Santiago, L.M.; Hulse-Kemp, A.M.; Ding, M.; Ye, W.; Kirkbride, R.C.; Jenkins, J.; et al. Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 2020, 52, 525–533. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2018, 51, 224–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, X.; Huang, G.; He, S.; Yang, Z.; Sun, G.; Ma, X.; Li, N.; Zhang, X.; Sun, J.; Liu, M.; et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 2018, 50, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Wu, Z.; Percy, R.G.; Bai, M.; Li, Y.; Frelichowski, J.E.; Hu, J.; Wang, K.; Yu, J.Z.; Zhu, Y. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat. Genet. 2020, 52, 516–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udall, J.A.; Long, E.; Hanson, C.; Yuan, D.; Ramaraj, T.; Conover, J.L.; Gong, L.; Arick, M.A.; Grover, C.E.; Peterson, D.G.; et al. De Novo Genome Sequence Assemblies of Gossypium raimondii and Gossypium turneri. G3 Genes Genomes Genet. 2019, 9, 3079–3085. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Cai, X.; Wang, Q.; Wang, P.; Zhang, Y.; Cai, C.; Xu, Y.; Wang, K.; Zhou, Z.; Wang, C.; et al. Genome sequencing of the Australian wild diploid species Gossypium australe highlights disease resistance and delayed gland morphogenesis. Plant Biotechnol. J. 2019, 18, 814–828. [Google Scholar] [CrossRef] [Green Version]
- Nowicki, M.; Bzhalava, D.; Bała, P. Massively Parallel Implementation of Sequence Alignment with Basic Local Alignment Search Tool Using Parallel Computing in Java Library. J. Comput. Biol. 2018, 25, 871–881. [Google Scholar] [CrossRef]
- Matsuda, F.; Tsugawa, H.; Fukusaki, E. Method for Assessing the Statistical Significance of Mass Spectral Similarities Using Basic Local Alignment Search Tool Statistics. Anal. Chem. 2013, 85, 8291–8297. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2014, 43, D257–D260. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 2, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [Green Version]
- Kohel, R.J.; Richmond, T.R.; Lewis, C.F. Texas Marker-1. Description of a Genetic Standard for Gossypium hirsutum L. 1. Crop Sci. 1970, 10, 670–671. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S.; Chan, C.-K.K. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods Mol. Bioinform. 2016, 1374, 339–361. [Google Scholar] [CrossRef]
- Pollier, J.; Rombauts, S.; Goossens, A. Analysis of RNA-Seq Data with TopHat and Cufflinks for Genome-Wide Expression Analysis of Jasmonate-Treated Plants and Plant Cultures. Jasmonate Signal. 2013, 1011, 305–315. [Google Scholar] [CrossRef]
- Cai, C.; Li, C.; Sun, R.; Zhang, B.; Nichols, R.L.; Hake, K.D.; Pan, X. Small RNA and degradome deep sequencing reveals important roles of microRNAs in cotton (Gossypium hirsutum L.) response to root-knot nematode Meloidogyne incognita infection. Genomics 2021, 113, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Nichols, R.L.; Qiu, L.; Sun, R.; Zhang, B.; Pan, X. Small RNA Sequencing Reveals Regulatory Roles of MicroRNAs in the Development of Meloidogyne incognita. Int. J. Mol. Sci. 2019, 20, 5466. [Google Scholar] [CrossRef] [Green Version]
- Westhoff, P.; Gowik, U. Evolution of C4 Phosphoenolpyruvate Carboxylase. Genes and Proteins: A Case Study with the Genus Flaveria. Ann. Bot. 2004, 93, 13–23. [Google Scholar] [CrossRef] [Green Version]
- Gennidakis, S.; Rao, S.; Greenham, K.; Uhrig, R.G.; O’Leary, B.; Snedden, W.A.; Lu, C.; Plaxton, W.C. Bacterial- and plant-type phosphoenolpyruvate carboxylase polypeptides interact in the hetero-oligomeric Class-2 PEPC complex of developing castor oil seeds. Plant J. 2007, 52, 839–849. [Google Scholar] [CrossRef]
- Cao, J.; Cheng, G.; Wang, L.; Maimaitijiang, T.; Lan, H. Genome-Wide Identification and Analysis of the Phosphoenolpyruvate Carboxylase Gene Family in Suaeda aralocaspica, an Annual Halophyte with Single-Cellular C4 Anatomy. Front. Plant Sci. 2021, 12, 665279. [Google Scholar] [CrossRef]
- Zhang, M.; Zeng, J.Y.; Long, H.; Xiao, Y.H.; Yan, X.Y.; Pei, Y. Auxin Regulates Cotton Fiber Initiation via GhPIN-Mediated Auxin Transport. Plant Cell Physiol. 2017, 58, 385–397. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, H.; Xi, J.; Zeng, J.; Huang, J.; Li, B.; Song, S.; Zhao, J.; Pei, Y. Auxin Directly Upregulates GhRAC13 Expression to Promote the Onset of Secondary Cell Wall Deposition in Cotton Fibers. Front. Plant Sci. 2020, 11, 581983. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon–intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bläsing, O.E.; Ernst, K.; Streubel, M.; Westhoff, P.; Svensson, P. The non-photosynthetic phosphoenolpyruvate carboxylases of the C4 dicot Flaveria trinervia-implications for the evolution of C4 photosynthesis. Planta 2002, 215, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Engelmann, S.; Bläsing, O.E.; Westhoff, P.; Svensson, P. Serine 774 and amino acids 296 to 437 comprise the major C4 determinants of the C4 phosphoenolpyruvate carboxylase of Flaveria trinervia. FEBS Lett. 2002, 524, 11–14. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J.; et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef] [Green Version]
- Bandyopadhyay, A.; Datta, K.; Zhang, J.; Yang, W.; Raychaudhuri, S.; Miyao, M.; Datta, S. Enhanced photosynthesis rate in genetically engineered indica rice expressing pepc gene cloned from maize. Plant Sci. 2007, 172, 1204–1209. [Google Scholar] [CrossRef]
- García-Mauriño, S.; Monreal, J.; Alvarez, R.; Vidal, J.; Echevarría, C. Characterization of salt stress-enhanced phosphoenolpyruvate carboxylase kinase activity in leaves of Sorghum vulgare: Independence from osmotic stress, involvement of ion toxicity and significance of dark phosphorylation. Planta 2003, 216, 648–655. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.; Li, Z.; Wedegaertner, T.C.; Jaconis, S.; Wan, S.; Zhao, Z.; Liu, Z.; Liu, Y.; Zheng, J.; Hake, K.D.; et al. Conservation and Divergence of Phosphoenolpyruvate Carboxylase Gene Family in Cotton. Plants 2022, 11, 1482. https://doi.org/10.3390/plants11111482
Wei Y, Li Z, Wedegaertner TC, Jaconis S, Wan S, Zhao Z, Liu Z, Liu Y, Zheng J, Hake KD, et al. Conservation and Divergence of Phosphoenolpyruvate Carboxylase Gene Family in Cotton. Plants. 2022; 11(11):1482. https://doi.org/10.3390/plants11111482
Chicago/Turabian StyleWei, Yangyang, Zhaoguo Li, Tom C. Wedegaertner, Susan Jaconis, Sumei Wan, Zilin Zhao, Zhen Liu, Yuling Liu, Juyun Zheng, Kater D. Hake, and et al. 2022. "Conservation and Divergence of Phosphoenolpyruvate Carboxylase Gene Family in Cotton" Plants 11, no. 11: 1482. https://doi.org/10.3390/plants11111482
APA StyleWei, Y., Li, Z., Wedegaertner, T. C., Jaconis, S., Wan, S., Zhao, Z., Liu, Z., Liu, Y., Zheng, J., Hake, K. D., Peng, R., & Zhang, B. (2022). Conservation and Divergence of Phosphoenolpyruvate Carboxylase Gene Family in Cotton. Plants, 11(11), 1482. https://doi.org/10.3390/plants11111482






