Genetic Mapping and Analysis of a Compact Plant Architecture and Precocious Mutant in Upland Cotton
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification of Fruit Branch Types
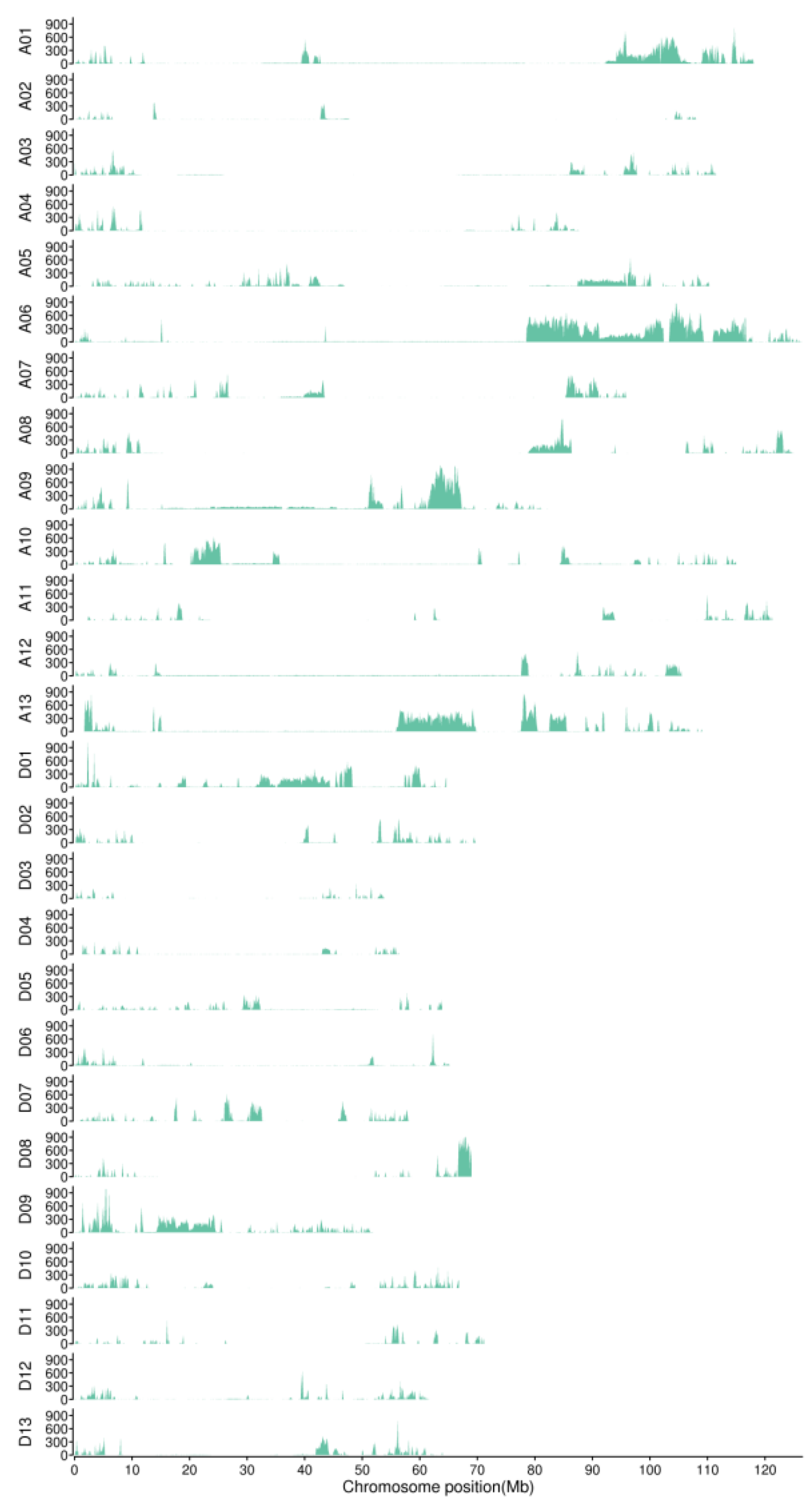
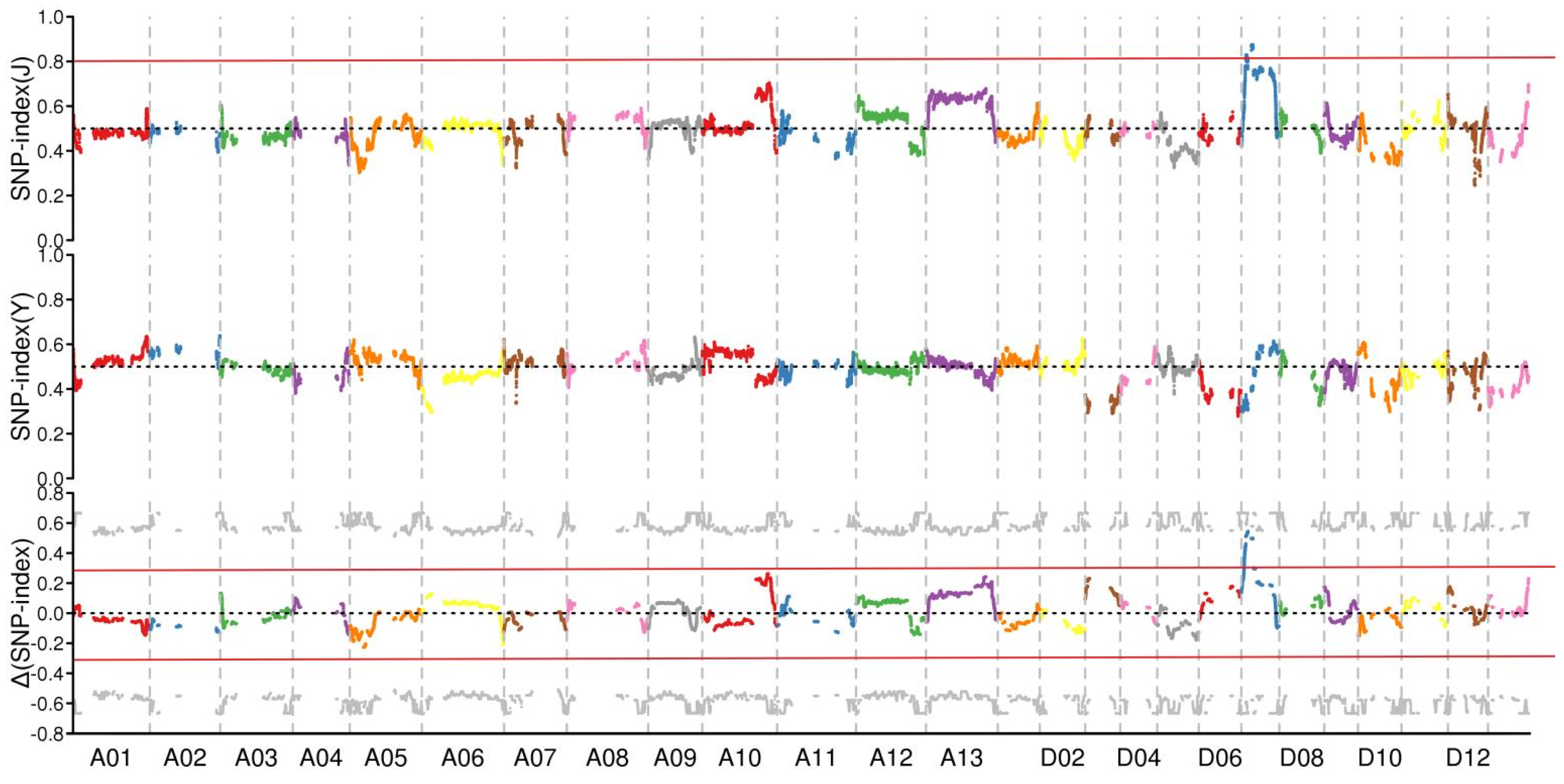
2.3. BSA Sequence Analysis
2.4. Candidate Gene Sequencing and Allele Test
2.5. Identification of Fruit Branch Types and Investigation of Flowering Period
2.6. Association Analysis of Candidate Genes
3. Results
3.1. Phenotype Analysis of 04N-11 Nulliplex-Branch Mutant
3.2. Nulliplex-Branch Traits Were Controlled by a Single Recessive Gene
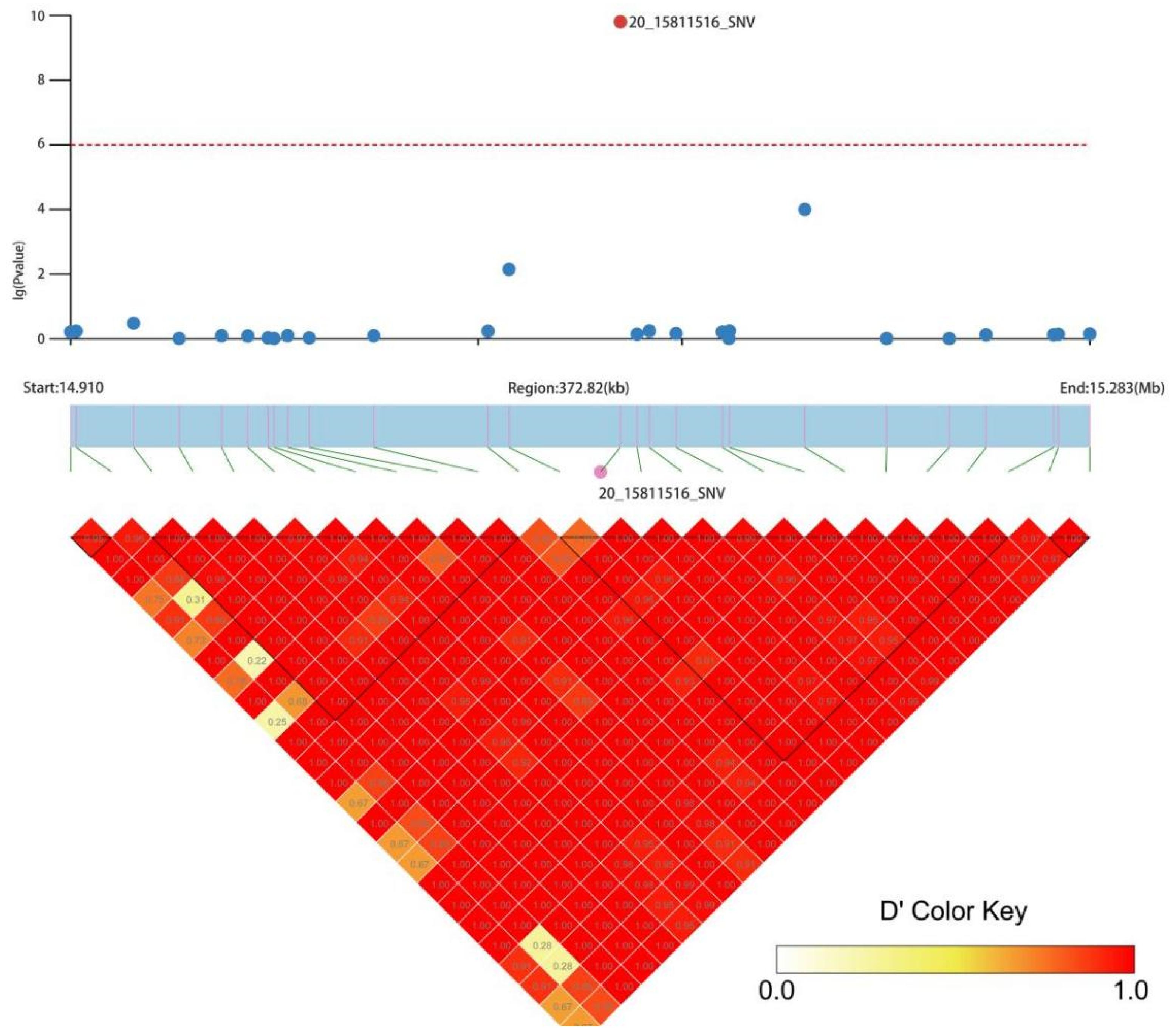
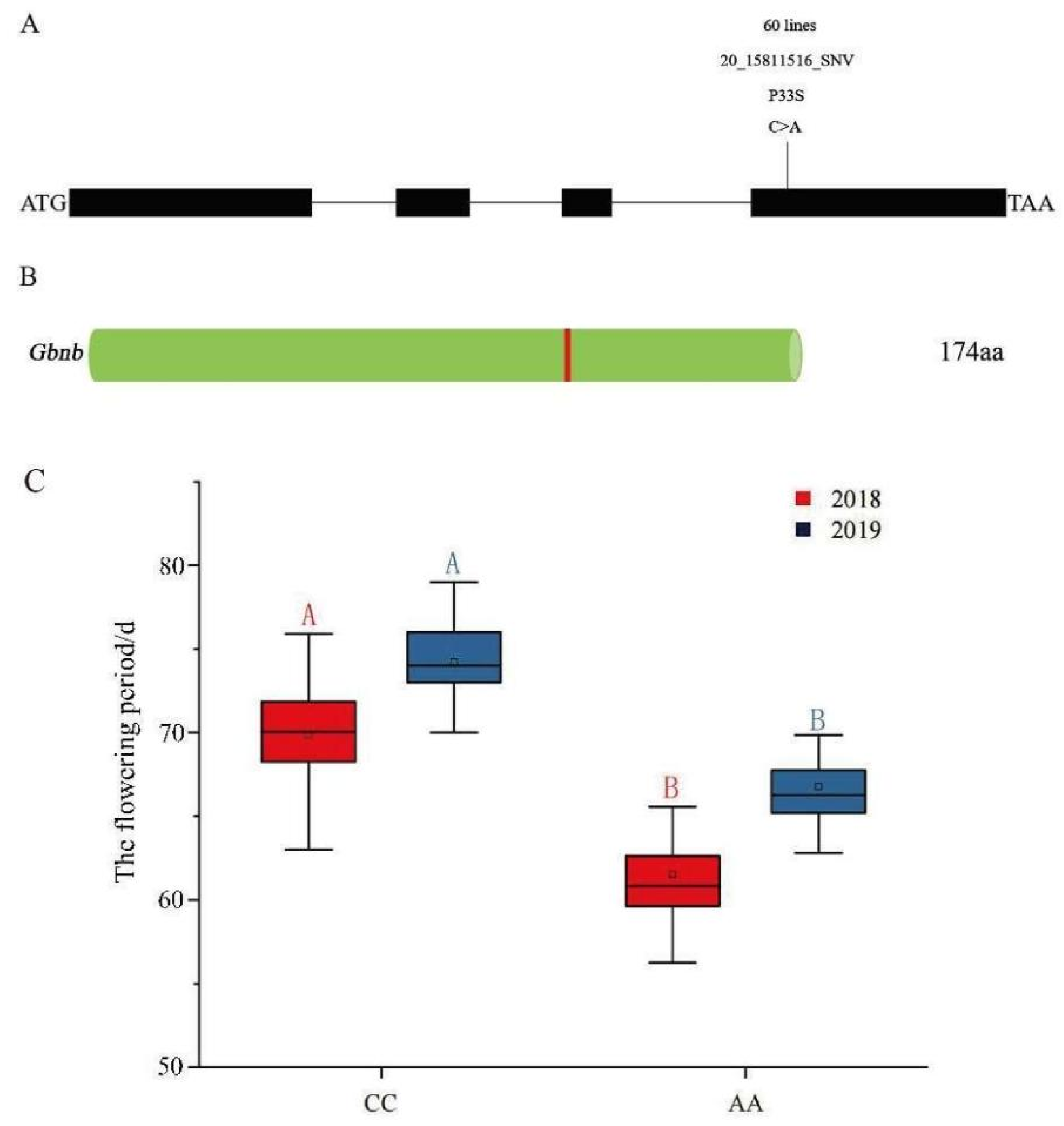
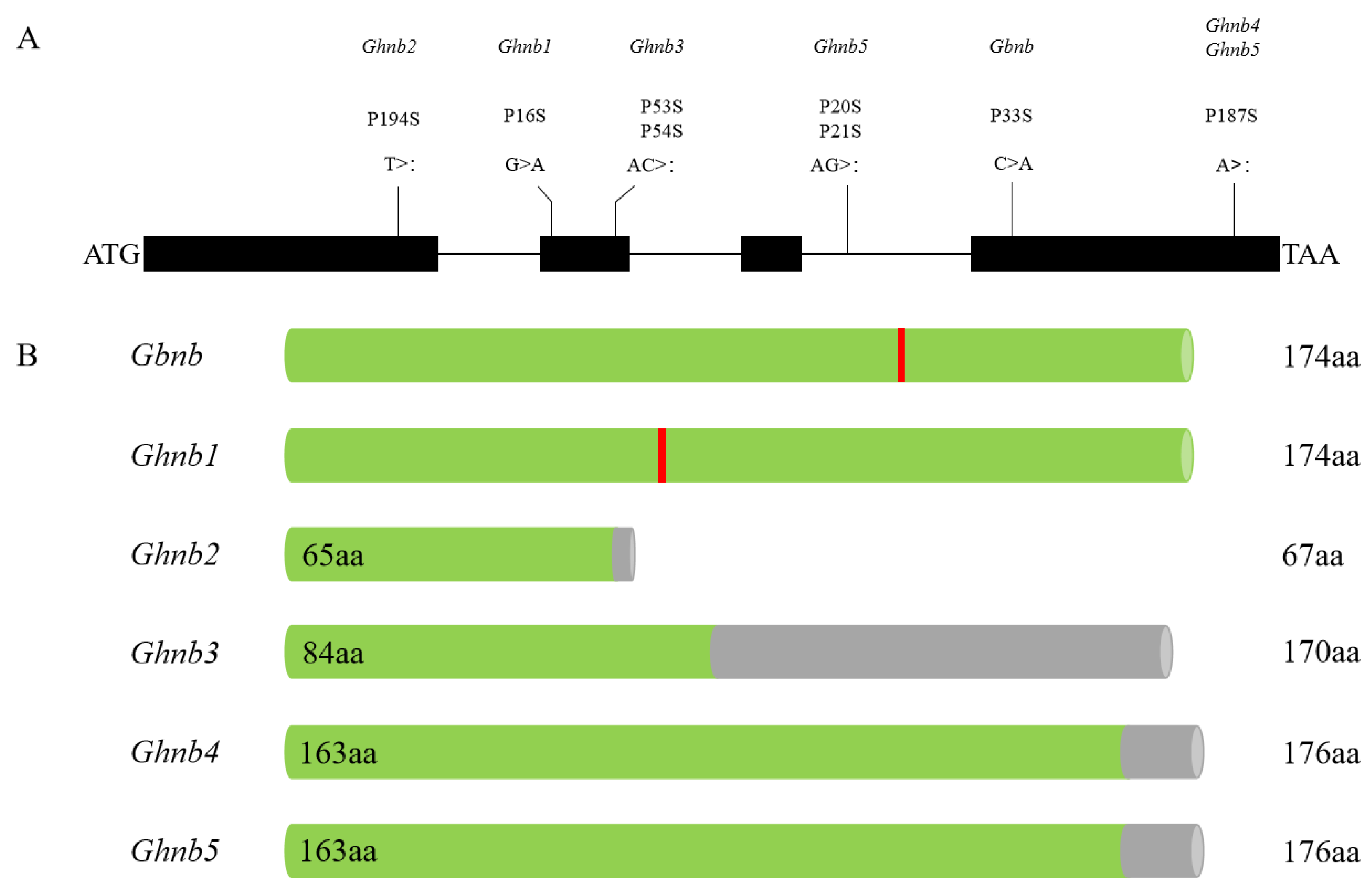
3.3. Cloning and Allelic Test of 04N-11 Nulliplex-Branch Candidate Gene
3.4. Association Analysis of Candidate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiao, T. National Bureau of Statistics: 2020 national cotton output flat slightly increase. China Fiber Insp. 2021, 97. [Google Scholar] [CrossRef]
- Mao, S.; Ma, X.; Cheng, S.; Wang, W.; Zhang, Y.; Huang, Q.; Wu, D. Analysis and strategy for the production and demand of high quality cotton in China. China Cotton 2020, 47, 1–5+20. [Google Scholar] [CrossRef]
- Li, X.; Wang, J.; Zhen, J.; Liang, Y.; Gong, Z.; Ai, X.; Zhang, Z.; Moming, M.; Guo, J. Explore to establish the cotton industry development pattern that the whole industry chain adds value in Xinxiang. Cotton Sci. 2020, 42, 20–25+29. [Google Scholar] [CrossRef]
- Yang, G.; Luo, X.; Nie, Y.; Zhang, X. Effects of Plant Density on Yield and Canopy Micro Environment in Hybrid Cotton. J. Integr. Agric. 2014, 13, 2154–2163. [Google Scholar] [CrossRef]
- Chen, W.; Yao, J.; Li, Y.; Zhao, L.; Liu, J.; Guo, Y.; Wang, J.; Yuan, L.; Liu, Z.; Lu, Y.; et al. Nulliplex-branch, a TERMINAL FLOWER 1 ortholog, controls plant growth habit in cotton. Theor. Appl. Genet. 2019, 132, 97–112. [Google Scholar] [CrossRef]
- Wen, T.; Liu, C.; Wang, T.; Wang, M.; Tang, F.; He, L. Genomic mapping and identification of candidate genes encoding nulliplex-branch trait in sea-island cotton (Gossypium barbadense L.) by multi-omics analysis. Mol. Breed. 2021, 41, 34. [Google Scholar] [CrossRef]
- Mao, L.; Zhang, L.; Evers, J.B.; Werf, W.; Liu, S.; Zhang, S.; Wang, B.; Li, Z. Yield components and quality of intercropped cotton in response to mepiquat chloride and plant density. Field Crops Res. 2015, 179, 63–71. [Google Scholar] [CrossRef]
- Meng, L.; Zhang, L.-Z.; Qi, H.-K.; DU, M.-W.; Zuo, Y.-L.; Zhang, M.-C.; Tian, X.-L.; Li, Z.-H. Optimizing the application of a novel harvest aid to improve the quality of mechanically harvested cotton in the North China Plain. J. Integr. Agric. 2021, 20, 2892–2899. [Google Scholar] [CrossRef]
- Zhang, A. Identification and Functional Analysis of Flowering Key Genes in Cotton. Master’s Thesis, Gansu Agricultural University, Lanzhou, China, 2021. [Google Scholar]
- Zou, C.; Wang, P.; Xu, Y. Bulked sample analysis in genetics, genomics and crop improvement. Plant Biotechnol. J. 2016, 14, 1941–1955. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Rehman, A.; Li, H.G.; Zhao, Z.B.; Sun, G.; Du, X.M. Mapping of dwarfing QTL of Ari1327, a semi-dwarf mutant of upland cotton. BMC Plant Biol. 2022, 22, 5. [Google Scholar] [CrossRef]
- Li, C.; Ling, F.; Su, G.; Sun, W.; Liu, H.; Su, Y.; Qi, X. Location and mapping of the NCLB resistance genes in maize by bulked segregant analysis (BSA) using whole genome re-sequencing. Mol. Breed. 2020, 40, 92. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.; Lin, J.; Wang, Y.; Fang, X. Identification of high resistance locus of rice stripe blight by BSA method. Acta Agric. Boreali-Sin. 2014, 29, 85–88. [Google Scholar] [CrossRef]
- Njogu, M.K.; Yang, F.; Li, J.; Wang, X.; Ogweno, J.O.; Chen, J. A novel mutation inTFL1homolog sustaining determinate growth in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2020, 133, 3323–3332. [Google Scholar] [CrossRef]
- Huang, Y. Functional Study on the Rice Dof Family Genes and Grain Size Gene WG7. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2019. [Google Scholar]
- Li, Q.; Yang, X.; Bai, G.; Warburton, M.L.; Mahuku, G.; Gore, M.; Dai, J.; Li, J.; Yan, J. Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor. Appl. Genet. 2010, 120, 753–763. [Google Scholar] [CrossRef]
- Nie, X.; Wen, T.; Shao, P.; Tang, B.; Nuriman-guli, A.; Yu, Y.; Du, X.; You, C.; Lin, Z. High-density genetic variation maps reveal the correlation between asymmetric interspecific introgressions and improvement of agronomic traits in Upland and Pima cotton varieties developed in Xinjiang, China. Plant J. 2020, 103, 677–689. [Google Scholar] [CrossRef]
- Chen, W.; Yao, J.; Chu, L.; Yuan, Z.; Li, Y.; Zhang, Y. Genetic mapping of the nulliplex-branch gene (gb_nb1) in cotton using next-generation sequencing. Theor. Appl. Genet. 2015, 128, 539–547. [Google Scholar] [CrossRef]
- Helin, D.; Zhao, X.; Li, P.; Zheng, C.; Sun, M.; Zhzng, S.; Zhzng, L.; Liu, S. Key technology of high quality and high yield cotton in southern Xinjiang. China Cotton 2016, 43, 39–40. [Google Scholar] [CrossRef]
- Si, Z.; Liu, H.; Zhu, J.; Chen, J.; Wang, Q.; Fang, L.; Gao, F.; Tian, Y.; Chen, Y.; Chang, L.; et al. Mutation of SELF-PRUNING homologs in cotton promotes short-branching plant architecture. J. Exp. Bot. 2018, 69, 2543–2553. [Google Scholar] [CrossRef]
- Guo, D.; Li, C.; Dong, R.; Li, X.; Xiao, X.; Huang, X. Molecular cloning and functional analysis of the FLOWERING LOCUS T (FT) homolog GhFT1 from Gossypium hirsutum. J. Integr. Plant Biol. 2015, 57, 522–533. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, M.; Zhang, X.; Guo, L.; Qi, T.; Wang, H.; Tang, H.; Zhang, J.; Xing, C. Development of InDel markers for the restorer gene and Rf1 assessment of their utility for marker-assisted selection in cotton. Euphytica 2017, 213, 251. [Google Scholar] [CrossRef]
- Jung, J.-K.; Park, S.-W.; Liu, W.Y.; Kang, B.-C. Discovery of single nucleotide polymorphism in Capsicum and SNP markers for cultivar identification. Euphytica 2010, 175, 91–107. [Google Scholar] [CrossRef]



| Year | Normal Branch | Nulliplex Branch | χ2 |
|---|---|---|---|
| 2019 | 278 | 110 | 1.98 |
| 2020 | 1455 | 440 | 3.27 |
| Normal Branch | Nulliplex Branch | |
|---|---|---|
| CC | 173 | 3 |
| AA | 3 | 57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chao, L.; Pan, Z.; Wang, J.; Wu, Y.; Shui, G.; Aini, N.; Tang, B.; Guo, C.; Han, P.; Shao, P.; et al. Genetic Mapping and Analysis of a Compact Plant Architecture and Precocious Mutant in Upland Cotton. Plants 2022, 11, 1483. https://doi.org/10.3390/plants11111483
Chao L, Pan Z, Wang J, Wu Y, Shui G, Aini N, Tang B, Guo C, Han P, Shao P, et al. Genetic Mapping and Analysis of a Compact Plant Architecture and Precocious Mutant in Upland Cotton. Plants. 2022; 11(11):1483. https://doi.org/10.3390/plants11111483
Chicago/Turabian StyleChao, Lei, Zhenyuan Pan, Jing Wang, Yuanlong Wu, Guangling Shui, Nurimanguli Aini, Binghui Tang, Chunping Guo, Peng Han, Panxia Shao, and et al. 2022. "Genetic Mapping and Analysis of a Compact Plant Architecture and Precocious Mutant in Upland Cotton" Plants 11, no. 11: 1483. https://doi.org/10.3390/plants11111483
APA StyleChao, L., Pan, Z., Wang, J., Wu, Y., Shui, G., Aini, N., Tang, B., Guo, C., Han, P., Shao, P., Tian, X., Chang, X., An, Q., Ma, C., You, C., Zhu, L., & Nie, X. (2022). Genetic Mapping and Analysis of a Compact Plant Architecture and Precocious Mutant in Upland Cotton. Plants, 11(11), 1483. https://doi.org/10.3390/plants11111483






