A Genome-Wide Association Study Identifying Single-Nucleotide Polymorphisms for Iron and Zinc Biofortification in a Worldwide Barley Collection
Abstract
:1. Introduction
2. Results
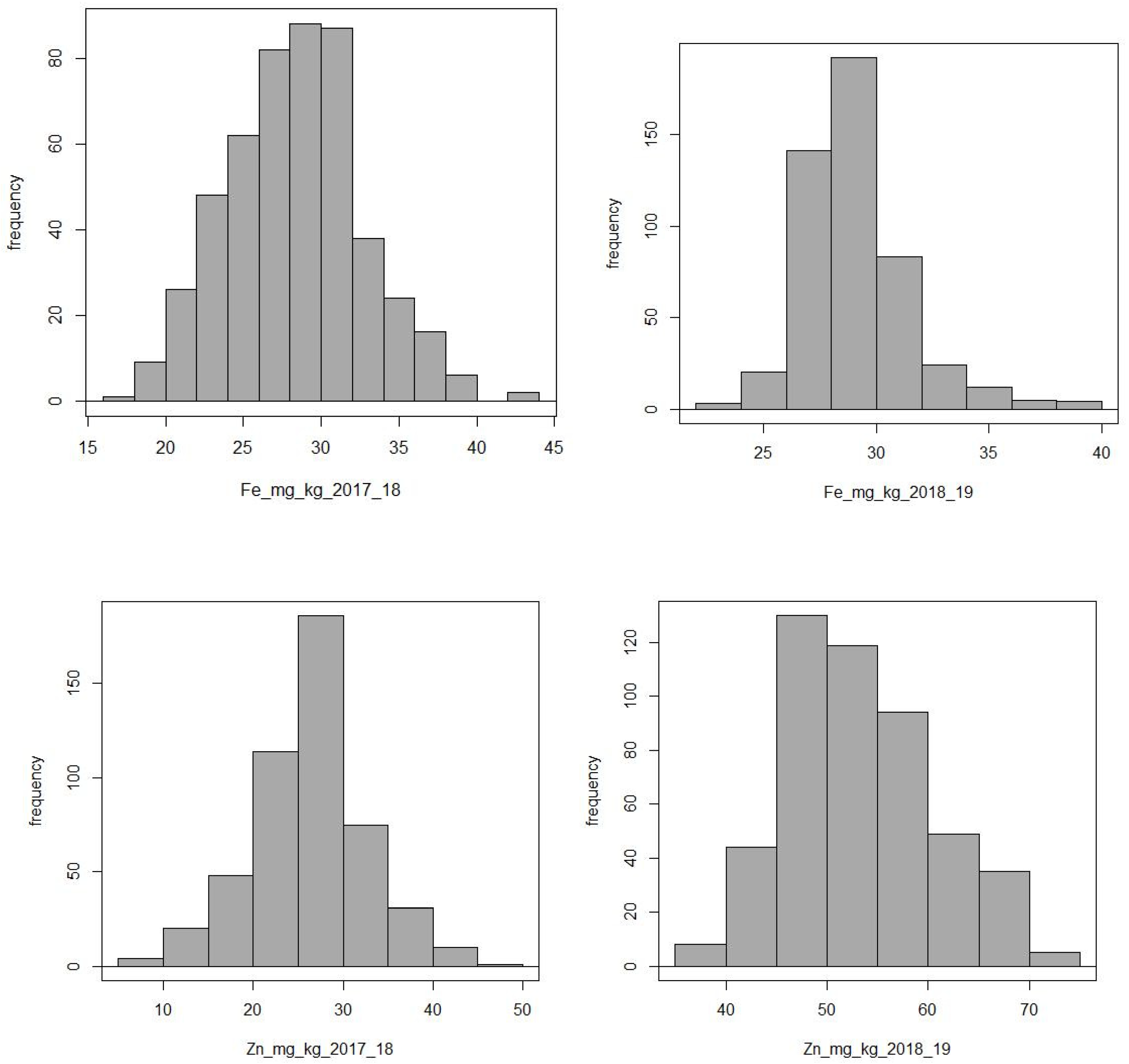
2.1. Phenotype Results
2.2. Population Structure and Linkage Disequilibrium
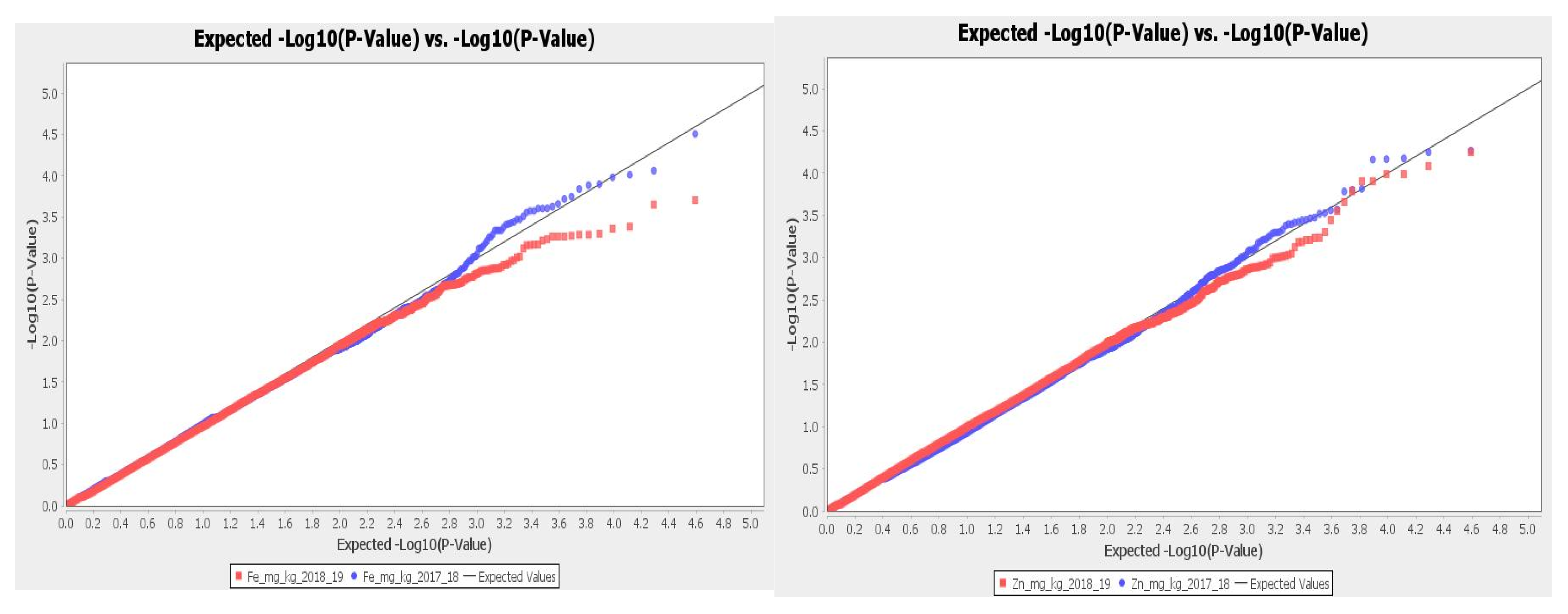
2.3. GWAS Results
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Experimental Design
4.2. Iron and Zinc Content
4.3. Genotyping
4.4. Population Structure, Kinship and Linkage Disequilibrium
4.5. LD Decay
4.6. Genome-Wide Association Analysis (GWAS)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- White, P.J.; Broadley, M.R. Biofortification of crops with seven mineral elements often lacking in human diets–iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol. 2009, 182, 49–84. [Google Scholar] [CrossRef] [PubMed]
- WHO. Malnutrution. Available online: https://www.who.int/news-room/fact-sheets/detail/malnutrition (accessed on 1 April 2022).
- Tam, E.; Keats, E.C.; Rind, F.; Das, J.K.; Bhutta, Z.A. Micronutrient supplementation and fortification interventions on health and development outcomes among children under-five in low-and middle-income countries: A systematic review and meta-analysis. Nutrients 2020, 12, 289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouis, H. Reducing mineral and vitamin deficiencies through biofortification: Progress under HarvestPlus. In Hidden Hunger: Strategies to Improve Nutrition Quality; Karger Publishers: Basel, Switzerland, 2018; Volume 118, pp. 112–122. [Google Scholar] [CrossRef] [Green Version]
- Grando, S.; Macpherson, H.G. Food Barley: Importance, Uses and Local Knowledge; ICARDA: Aleppo, Syria, 2005. [Google Scholar]
- Baik, B.-K.; Ullrich, S.E. Barley for food: Characteristics, improvement, and renewed interest. J. Cereal Sci. 2008, 48, 233–242. [Google Scholar] [CrossRef]
- Dawson, I.K.; Russell, J.; Powell, W.; Steffenson, B.; Thomas, W.T.; Waugh, R. Barley: A translational model for adaptation to climate change. New Phytol. 2015, 206, 913–931. [Google Scholar] [CrossRef] [PubMed]
- Sakellariou, M.; Mylona, P.V. New Uses for Traditional Crops: The Case of Barley Biofortification. Agronomy 2020, 10, 1964. [Google Scholar] [CrossRef]
- Zhou, Z.; Shi, X.; Zhao, G.; Qin, M.; Ibba, M.I.; Wang, Y.; Li, W.; Yang, P.; Wu, Z.; Lei, Z. Identification of novel genomic regions and superior alleles associated with Zn accumulation in wheat using a genome-wide association analysis method. Int. J. Mol. Sci. 2020, 21, 1928. [Google Scholar] [CrossRef] [Green Version]
- Fukao, Y.; Ferjani, A.; Tomioka, R.; Nagasaki, N.; Kurata, R.; Nishimori, Y.; Fujiwara, M.; Maeshima, M. iTRAQ analysis reveals mechanisms of growth defects due to excess zinc in Arabidopsis. Plant Physiol. 2011, 155, 1893–1907. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Zhang, S.; Yang, L.; Wang, Y.; Yu, B.; Zhang, F. Comparative proteomics illustrates the complexity of Fe, Mn and Zn deficiency-responsive mechanisms of potato (Solanum tuberosum L.) plants in vitro. Planta 2019, 250, 199–217. [Google Scholar] [CrossRef]
- Satyavathi, C.T.; Tomar, R.S.; Ambawat, S.; Kheni, J.; Padhiyar, S.M.; Desai, H.; Bhatt, S.; Shitap, M.; Meena, R.C.; Singhal, T. Stage specific comparative transcriptomic analysis to reveal gene networks regulating iron and zinc content in pearl millet [Pennisetum glaucum (L.) R. Br.]. Sci. Rep. 2022, 12, 276. [Google Scholar] [CrossRef]
- Kaur, G.; Shukla, V.; Kumar, A.; Kaur, M.; Goel, P.; Singh, P.; Shukla, A.; Meena, V.; Kaur, J.; Singh, J. Integrative analysis of hexaploid wheat roots identifies signature components during iron starvation. J. Exp. Bot. 2019, 70, 6141–6161. [Google Scholar] [CrossRef] [Green Version]
- Lucini, L.; Bernardo, L. Comparison of proteome response to saline and zinc stress in lettuce. Front. Plant Sci. 2015, 6, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruber, B.D.; von Wirén, N. Improvement of Mineral Nutrition: A Source and Sink for Candidate Genes. In Biotechnological Approaches to Barley Improvement; Springer: Berlin/Heidelberg, Germany, 2014; pp. 101–112. [Google Scholar]
- Gyawali, S.; Otte, M.L.; Chao, S.; Jilal, A.; Jacob, D.L.; Amezrou, R.; Verma, R.P.S. Genome wide association studies (GWAS) of element contents in grain with a special focus on zinc and iron in a world collection of barley (Hordeum vulgare L.). J. Cereal Sci. 2017, 77, 266–274. [Google Scholar] [CrossRef]
- Mamo, B.E.; Barber, B.L.; Steffenson, B.J. Genome-wide association mapping of zinc and iron concentration in barley landraces from Ethiopia and Eritrea. J. Cereal Sci. 2014, 60, 497–506. [Google Scholar] [CrossRef]
- El-Haramein, F.; Grando, S. Determination of iron and zinc content in food barley. In Proceedings of the 10th International Barley Genetics Symposium, Alexandria, Egypt, 5–10 April 2008; pp. 603–606. [Google Scholar]
- Yan, J.; Wang, F.; Yang, R.; Xiao, T.; Fahima, T.; Saranga, Y.; Korol, A.; Nevo, E.; Cheng, J. Natural variation in grain iron and zinc concentrations of wild barley, Hordeum spontaneum, populations from Israel. In Advance in Barley Sciences; Springer: Berlin/Heidelberg, Germany, 2013; pp. 169–183. [Google Scholar] [CrossRef]
- Wiegmann, M.; Thomas, W.T.; Bull, H.J.; Flavell, A.J.; Zeyner, A.; Peiter, E.; Pillen, K.; Maurer, A. Wild barley serves as a source for biofortification of barley grains. Plant Sci. 2019, 283, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Rathan, N.D.; Sehgal, D.; Thiyagarajan, K.; Singh, R.; Singh, A.-M.; Govindan, V. Identification of genetic loci and candidate genes related to grain zinc and iron concentration using a zinc-enriched wheat ‘Zinc-Shakti’. Front. Genet. 2021, 12, 652653. [Google Scholar] [CrossRef] [PubMed]
- Ouelhadj, A.; Kaminski, M.; Mittag, M.; Humbeck, K. Receptor-like protein kinase HvLysMR1 of barley (Hordeum vulgare L.) is induced during leaf senescence and heavy metal stress. J. Exp. Bot. 2007, 58, 1381–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricachenevsky, F.K.; Menguer, P.K.; Sperotto, R.A. kNACking on heaven’s door: How important are NAC transcription factors for leaf senescence and Fe/Zn remobilization to seeds? Front. Plant Sci. 2013, 4, 226. [Google Scholar] [CrossRef] [Green Version]
- Lonergan, P.F.; Pallotta, M.A.; Lorimer, M.; Paull, J.G.; Barker, S.J.; Graham, R.D. Multiple genetic loci for zinc uptake and distribution in barley (Hordeum vulgare). New Phytol. 2009, 184, 168–179. [Google Scholar] [CrossRef]
- Sadeghzadeh, B.; Rengel, Z.; Li, C. Quantitative trait loci (QTL) of seed Zn accumulation in barley population clipper X Sahara. J. Plant Nutr. 2015, 38, 1672–1684. [Google Scholar] [CrossRef] [Green Version]
- Reuscher, S.; Kolter, A.; Hoffmann, A.; Pillen, K.; Krämer, U. Quantitative trait loci and inter-organ partitioning for essential metal and toxic analogue accumulation in barley. PLoS ONE 2016, 11, e0153392. [Google Scholar] [CrossRef]
- Detterbeck, A.; Nagel, M.; Rensch, S.; Weber, M.; Börner, A.; Persson, D.P.; Schjoerring, J.K.; Christov, V.; Clemens, S. The search for candidate genes associated with natural variation of grain Zn accumulation in barley. Biochem. J. 2019, 476, 1889–1909. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.K.; Pandey, A.; Hamurcu, M.; Hakki, E.E.; Gezgin, S. Role of molecular approaches in improving genetic variability of micronutrients and their utilization in breeding programs. In Wheat and Barley Grain Biofortification; Elsevier: Amsterdam, The Netherlands, 2020; pp. 27–52. [Google Scholar] [CrossRef]
- Conte, S.S.; Walker, E.L. Genetic and biochemical approaches for studying the yellow stripe-like transporter family in plants. Curr. Top. Membr. 2012, 69, 295–322. [Google Scholar] [CrossRef] [PubMed]
- Herzig, P.; Backhaus, A.; Seiffert, U.; Von Wirén, N.; Pillen, K.; Maurer, A. Genetic dissection of grain elements predicted by hyperspectral imaging associated with yield-related traits in a wild barley NAM population. Plant Sci. 2019, 285, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Prasanna, R.; Sharma, V.; Rajawat, M.V.S.; Nishanth, S.; Saxena, A.K. Prospecting plant–microbe interactions for enhancing nutrient availability and grain biofortification. In Wheat and Barley Grain Biofortification; Elsevier: Amsterdam, The Netherlands, 2020; pp. 203–228. [Google Scholar] [CrossRef]
- Tiong, J.; McDonald, G.K.; Genc, Y.; Pedas, P.; Hayes, J.E.; Toubia, J.; Langridge, P.; Huang, C.Y. H v ZIP 7 mediates zinc accumulation in barley (H ordeum vulgare) at moderately high zinc supply. New Phytol. 2014, 201, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Ram, S.; Narwal, S.; Gupta, O.P.; Pandey, V.; Singh, G.P. Anti-nutritional factors and bioavailability: Approaches, challenges, and opportunities. In Wheat and Barley Grain Biofortification; Elsevier: Amsterdam, The Netherlands, 2020; pp. 101–128. [Google Scholar] [CrossRef]
- Holme, I.B.; Wendt, T.; Gil-Humanes, J.; Deleuran, L.C.; Starker, C.G.; Voytas, D.F.; Brinch-Pedersen, H. Evaluation of the mature grain phytase candidate HvPAPhy_a gene in barley (Hordeum vulgare L.) using CRISPR/Cas9 and TALENs. Plant Mol. Biol. 2017, 95, 111–121. [Google Scholar] [CrossRef]
- Takahashi, M.; Nakanishi, H.; Kawasaki, S.; Nishizawa, N.K.; Mori, S. Enhanced tolerance of rice to low iron availability in alkaline soils using barley nicotianamine aminotransferase genes. Nat. Biotechnol. 2001, 19, 466–469. [Google Scholar] [CrossRef]
- Pequerul, A.; Perez, C.; Madero, P.; Val, J.; Monge, E. A rapid wet digestion method for plant analysis. In Optimization of Plant Nutrition; Springer: Berlin/Heidelberg, Germany, 1993; pp. 3–6. Available online: https://link.springer.com/chapter/10.1007/978-94-017-2496-8_1 (accessed on 1 April 2022).
- Alcock, N.W. A hydrogen-peroxide digestion system for tissue trace-metal analysis. Biol. Trace Elem. Res. 1987, 13, 363–370. [Google Scholar] [CrossRef]
- Sen Gupta, D.; Thavarajah, D.; McGee, R.J.; Coyne, C.J.; Kumar, S.; Thavarajah, P. Genetic diversity among cultivated and wild lentils for iron, zinc, copper, calcium and magnesium concentrations. Aust. J. Crop Sci. 2016, 10, 1381–1387. [Google Scholar] [CrossRef]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef] [Green Version]
- Udupa, S.M.; Robertson, L.D.; Weigand, F.; Baum, M.; Kahl, G. Allelic variation at (TAA)n microsatellite loci in a world collection of chickpea (Cicer arietinum L.) germplasm. Mol. Gen. Genet. 1999, 261, 354–363. [Google Scholar] [CrossRef]
- Bayer, M.M.; Rapazote-Flores, P.; Ganal, M.; Hedley, P.E.; Macaulay, M.; Plieske, J.; Ramsay, L.; Russell, J.; Shaw, P.D.; Thomas, W. Development and evaluation of a barley 50k iSelect SNP array. Front. Plant Sci. 2017, 8, 1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheet, P.; Stephens, M. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006, 78, 629–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raj, A.; Stephens, M.; Pritchard, J.K. fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 2014, 197, 573–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, s13742-13015-10047-13748. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Remington, D.L.; Thornsberry, J.M.; Matsuoka, Y.; Wilson, L.M.; Whitt, S.R.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 2001, 98, 11479–11484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colmsee, C.; Beier, S.; Himmelbach, A.; Schmutzer, T.; Stein, N.; Scholz, U.; Mascher, M. BARLEX–the barley draft genome explorer. Mol. Plant 2015, 8, 964–966. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.; Chapman, B.; Wang, P.; Zhang, Q.; Zhou, G.; Zhang, X.-q.; Barrero, R.A.; Bellgard, M.I.; Li, C. BarleyVarDB: A database of barley genomic variation. Database 2020, 2020, baaa091. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Boudiar, R.; Casas, A.M.; Igartua, E.; Contreras-Moreira, B. BARLEYMAP: Physical and genetic mapping of nucleotide sequences and annotation of surrounding loci in barley. Mol. Breed. 2015, 35, 13. [Google Scholar] [CrossRef]

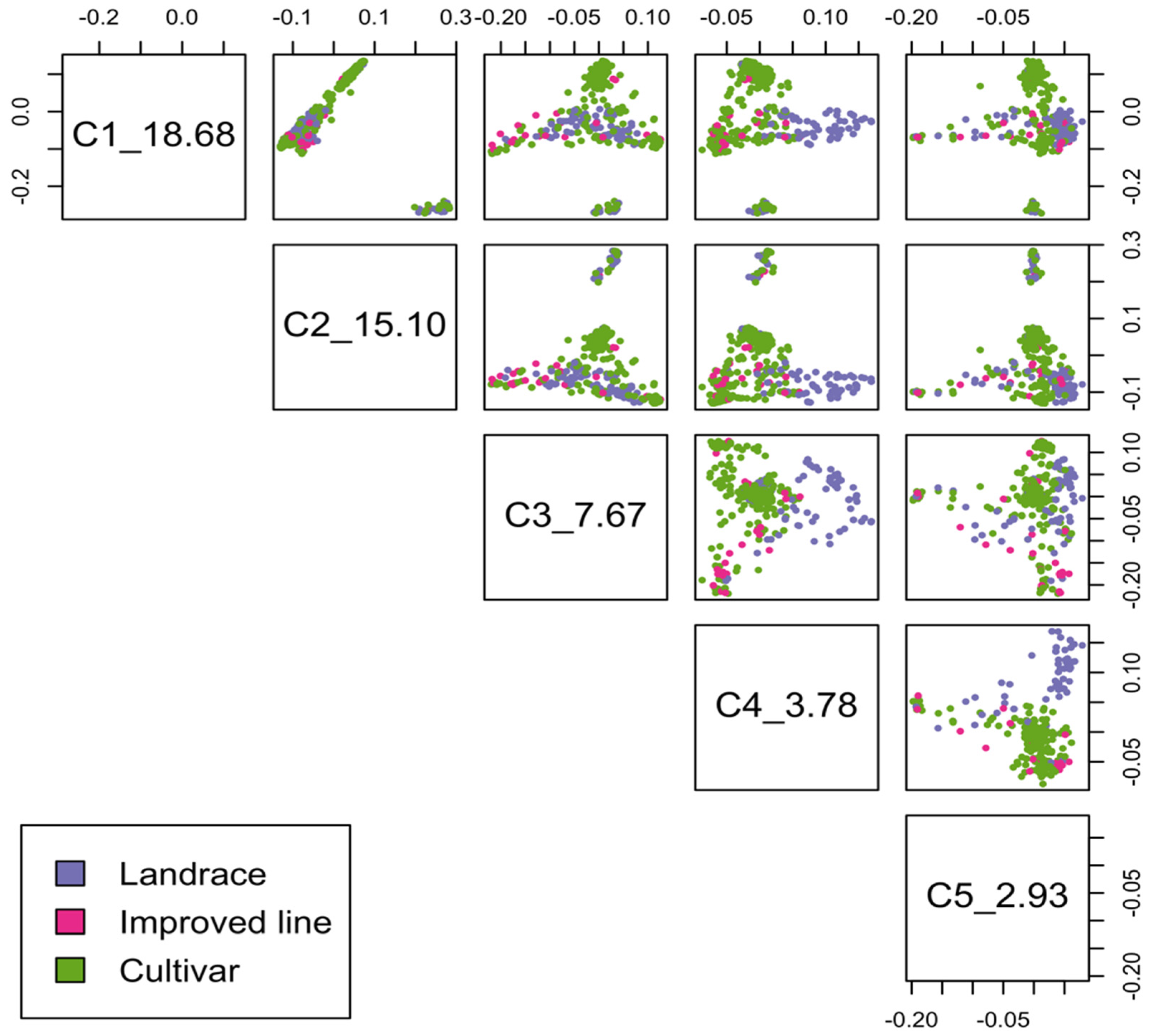

| Subpopulation | Cultivar | Improved Line | Landrace | Total |
|---|---|---|---|---|
| 1 | 14 | 3 | 74 | 91 |
| 2 | 4 | 16 | 35 | 55 |
| 3 | 6 | 3 | 230 | 238 |
| 4 | 19 | 2 | 16 | 37 |
| 5 | 52 | 4 | 18 | 74 |
| Total | 95 | 28 | 373 | 495 |
| Trait | SNP | Chr | Pos | p | Adjusted p | R2 | Gene ID | Candidate Gene |
|---|---|---|---|---|---|---|---|---|
| Zn_2017_18 | JHI-Hv50k-2016-198443 | 3H | 598684782 | 0.000054068 | 0.002378992 | 3.426 | HORVU3Hr1G082300 | Protein NRT1/PTR FAMILY 5.10 |
| Zn 2017_18 | JHI-Hv50k-2016-263938 | 4H | 615385811 | 0.00005644 | 0.00248336 | 3.409 | HORVU4Hr1G080420 | Tetratricopeptide repeat protein 7A |
| Zn_2017_18 | JHI-Hv50k-2016-184521 | 3H | 498961078 | 0.000066521 | 0.002926924 | 3.342 | HORVU3Hr1G065530 | Zinc finger BED domain-containing protein 1 |
| Zn_2017_18 | JHI-Hv50k-2016-184502 | 3H | 498958727 | 0.000068109 | 0.002996796 | 3.332 | HORVU3Hr1G065530 | Zinc finger BED domain-containing protein 1 |
| Zn_2017_18 | JHI-Hv50k-2016-198449 | 3H | 598790707 | 0.000068898 | 0.003031512 | 3.328 | HORVU3Hr1G082310 | 5′-3′ exoribonuclease 3 |
| Zn_2018_19 | JHI-Hv50k-2016-232541 | 4H | 26342827 | 0.000056102 | 0.001346448 | 3.419 | marker for oxidative stress response protein | |
| Zn_2018_19 | SCRI_RS_115755 | 3H | 664905936 | 0.000056102 | 0.001968216 | 3.265 | HORVU3Hr1G102520 | Nascent polypeptide-associated complex subunit alpha-like protein 3 |
| Zn_2018_19 | JHI-Hv50k-2016-44040 | 1H | 518159560 | 0.00010246 | 0.00245904 | 3.175 | HORVU1Hr1G077680 | Ethylene-responsive transcription factor 1B |
| Zn_2018_19 | JHI-Hv50k-2016-44085 | 1H | 518229092 | 0.00010246 | 0.00245904 | 3.175 | HORVU1Hr1G077710 | FAR1-related sequence 5 |
| Zn_2018_19 | JHI-Hv50k-2016-73691 | 2H | 29669343 | 0.00012464 | 0.00299136 | 3.096 | HORVU2Hr1G013690 | Undescribed |
| Zn_2018_19 | JHI-Hv50k-2016-73694 | 2H | 29669609 | 0.00012464 | 0.00299136 | 3.096 | HORVU2Hr1G013690 | Undescribed |
| Zn_2018_19 | JHI-Hv50k-2016-73663 | 2H | 29624393 | 0.0001632 | 0.0039168 | 2.988 | HORVU2Hr1G013680 | Elongation factor 1-alpha |
| Trait | SNP | Chr | Pos | p | Adjusted p | R2 | Gene ID | Candidate Gene |
|---|---|---|---|---|---|---|---|---|
| Fe 2018–2019 | JHI-Hv50k-2016-114559 | 2H | 690696206 | 0.000198 | 0.00396 | 2.925 | HORVU2Hr1G100330 | Nuclear pore complex protein-related |
| Fe 2018–2019 | JHI-Hv50k-2016-260401 | 4H | 598066009 | 0.000222 | 0.00444 | 2.88 | HORVU4Hr1G075360 | Alanine:glyoxylate aminotransferase 2 |
| Fe 2017–2018 | JHI-Hv50k-2016-197724 | 3H | 595054022 | 0.0000312 | 0.0013104 | 3.584 | HORVU3Hr1G081540 | Transcription initiation factor TFIID subunit 9 |
| Fe 2017–2018 | JHI-Hv50k-2016-197775 | 3H | 595221701 | 0.0000869 | 0.0036498 | 3.177 | HORVU3Hr1G081590 | 4;5-DOPA dioxygenase extradiol |
| Fe 2017–2018 | JHI-Hv50k-2016-457547 | 7H | 32324334 | 0.0000978 | 0.0041076 | 3.13 | HORVU3Hr1G081580 | 4;5-DOPA dioxygenase extradiol |
| Fe 2017–2018 | JHI-Hv50k-2016-488291 | 7H | 498765300 | 0.000105 | 0.00441 | 3.103 | HORVU7Hr1G082900 | 23S rRNA (uracil(1939)-C(5))-methyltransferase RlmD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nyiraguhirwa, S.; Grana, Z.; Ouabbou, H.; Iraqi, D.; Ibriz, M.; Mamidi, S.; Udupa, S.M. A Genome-Wide Association Study Identifying Single-Nucleotide Polymorphisms for Iron and Zinc Biofortification in a Worldwide Barley Collection. Plants 2022, 11, 1349. https://doi.org/10.3390/plants11101349
Nyiraguhirwa S, Grana Z, Ouabbou H, Iraqi D, Ibriz M, Mamidi S, Udupa SM. A Genome-Wide Association Study Identifying Single-Nucleotide Polymorphisms for Iron and Zinc Biofortification in a Worldwide Barley Collection. Plants. 2022; 11(10):1349. https://doi.org/10.3390/plants11101349
Chicago/Turabian StyleNyiraguhirwa, Solange, Zahra Grana, Hassan Ouabbou, Driss Iraqi, Mohammed Ibriz, Sujan Mamidi, and Sripada M. Udupa. 2022. "A Genome-Wide Association Study Identifying Single-Nucleotide Polymorphisms for Iron and Zinc Biofortification in a Worldwide Barley Collection" Plants 11, no. 10: 1349. https://doi.org/10.3390/plants11101349
APA StyleNyiraguhirwa, S., Grana, Z., Ouabbou, H., Iraqi, D., Ibriz, M., Mamidi, S., & Udupa, S. M. (2022). A Genome-Wide Association Study Identifying Single-Nucleotide Polymorphisms for Iron and Zinc Biofortification in a Worldwide Barley Collection. Plants, 11(10), 1349. https://doi.org/10.3390/plants11101349









