Challenges in Medicinal and Aromatic Plants DNA Barcoding—Lessons from the Lamiaceae
Abstract
:1. Introduction
1.1. Introducing the Historical Importance and Status of Medicinal Plants
1.2. Increasing TM Supply Demand Threatening “Wild Type” Stock
1.3. Herbal Medicines Quality Assurance Strategies
2. DNA Barcoding—Lessons from the Lamiaceae
2.1. A Carvone Focussed Market and Hybridisation: Mentha L.—Mentheae: Nepetioideae
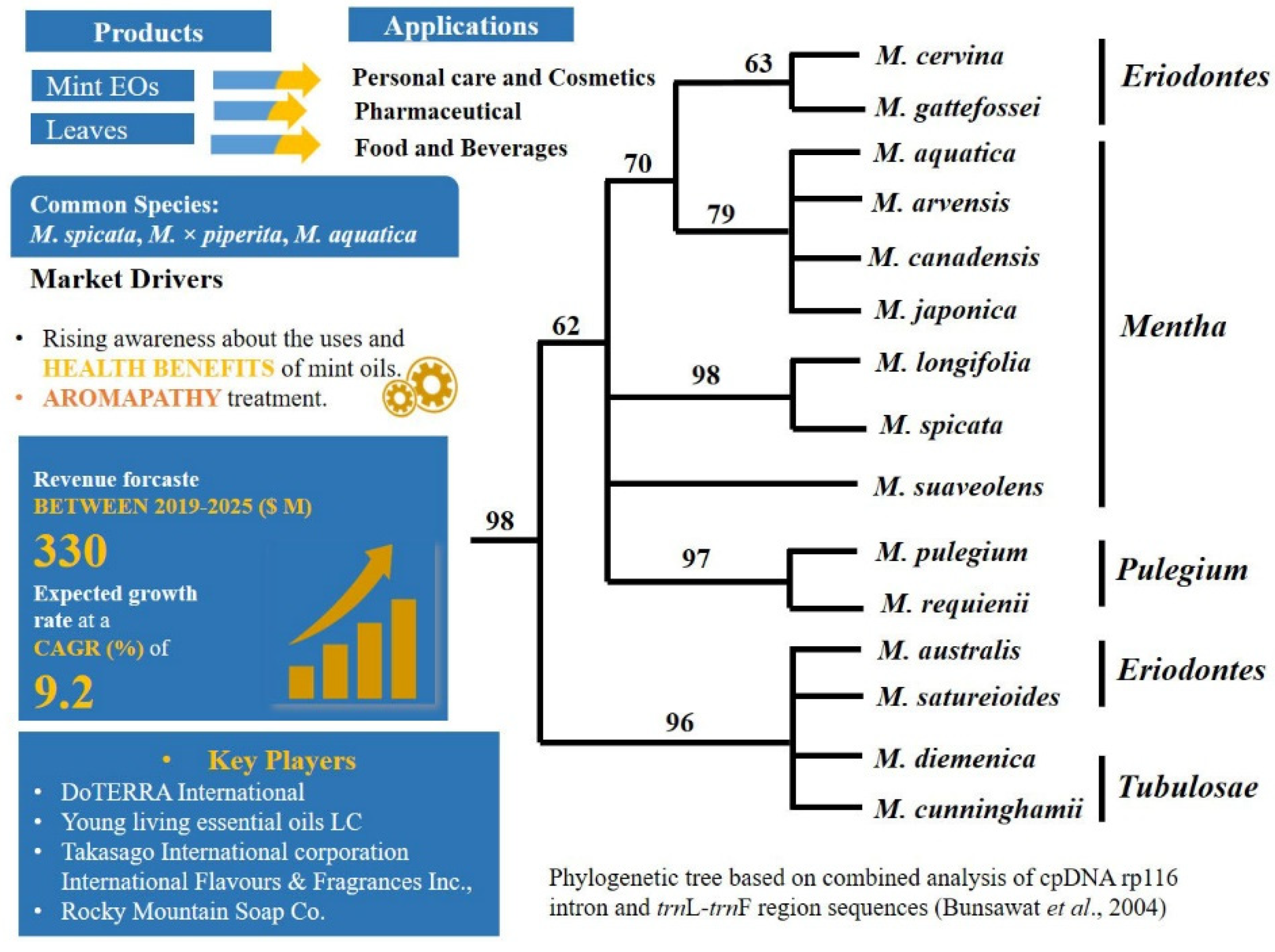
- (i)
- Hybridization or cryptic taxa. Hybridization and polyploidy have indeed most likely played important roles during speciation in mints, which forms one reason the number of taxonomically valid species is a subject of controversy [153,154].The complex genomic networks of taxa with porous genomes, cause phenotypic mosaics that behave dynamically [155]. Indeed, plasticity is highly known in Mentha [156,157], which confounds morphological identification. Complex morphological, chemical, and molecular diversity in mints have already been described in many studies, e.g., [126,131,158,159,160,161,162,163,164,165,166]. Despite the enormous amount of data gathered, however, there is still need of taxonomic revisions within the genus. In the recent revised phylogenetic analysis [131] the origin of M. spicata as hybrid was not supported and hidden cryptic taxa were detected in the genus.
- (ii)
- Selection of chemical markers. Carvone, a characteristic compound produced by M. spicata is also produced by different species from different plant families [144]. Chemical markers such as carvone in spearmint, and menthofuran and menthol in peppermint are used in practice for authentication of oils regardless of their sources [49].
2.2. Two-Tier Trade Variety Selection for the Fragrance and Horticulture Industries: Lavandula L.—Ocimeae: Nepetioideae
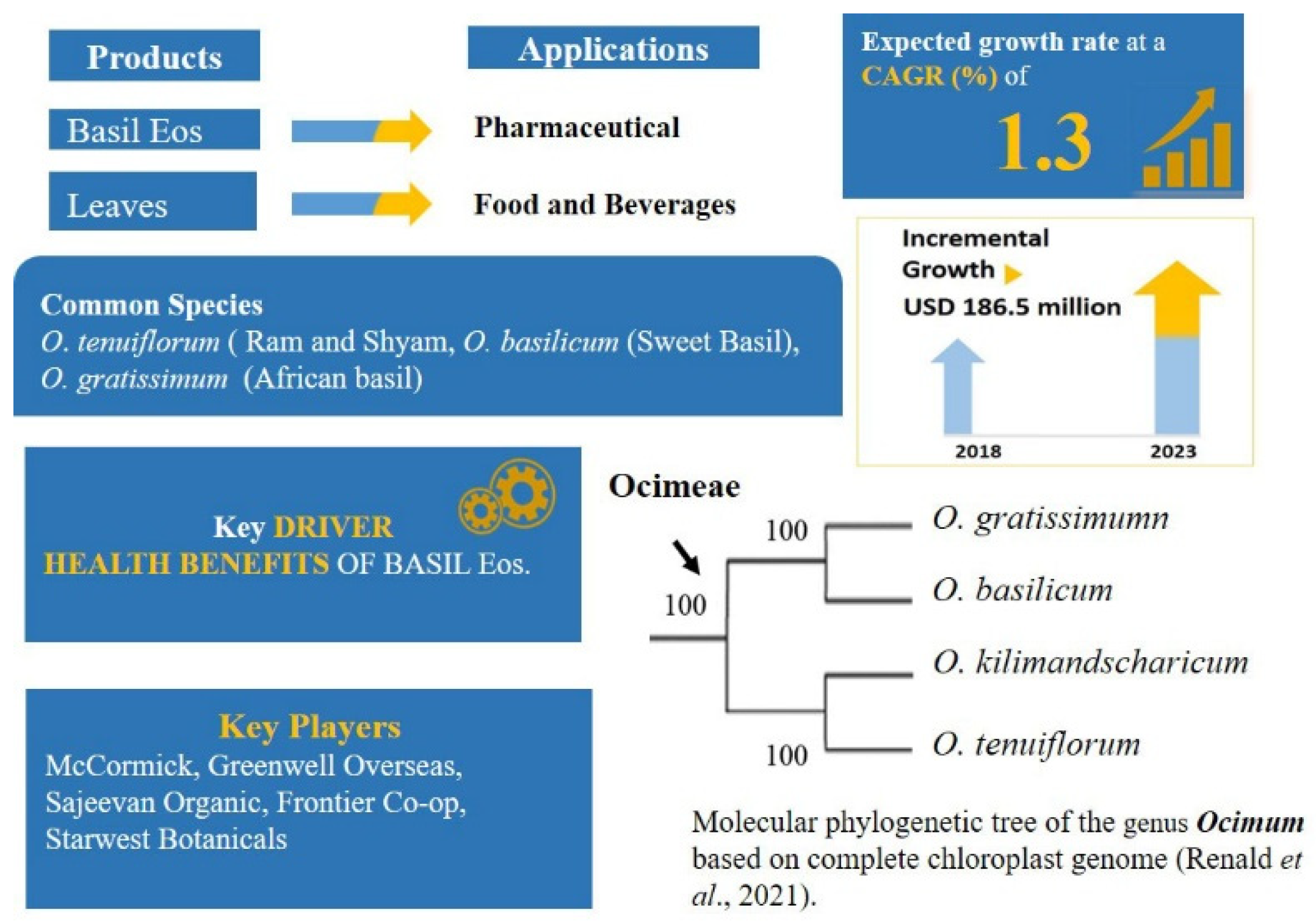
2.3. The Diaspora of People and Plants: Ocimum L.—Ocimeae: Nepetioideae
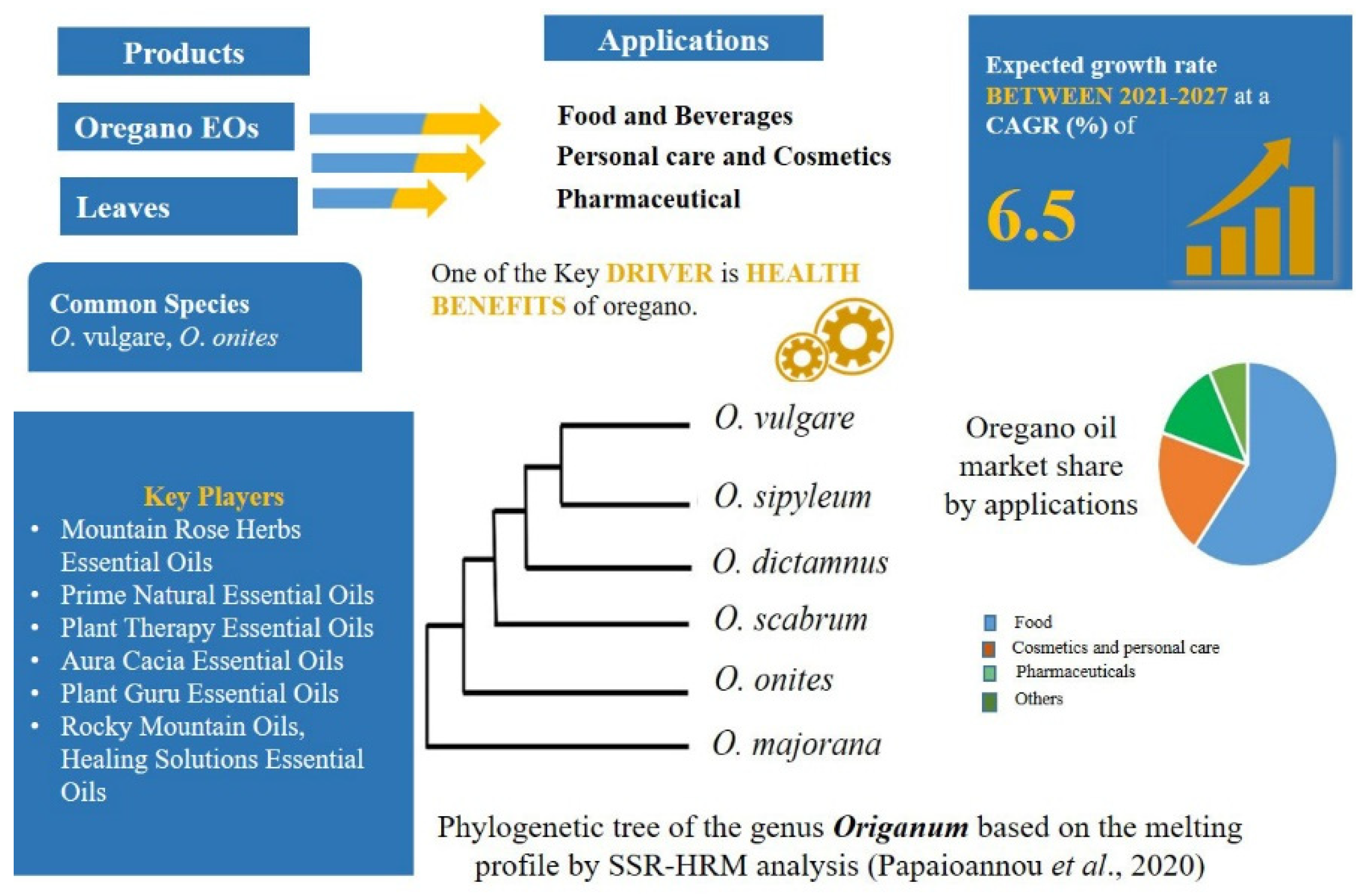
2.4. Demands of High-Quality Herbal Products in the Food Market: Origanum L.—Mentheae: Nepetioideae
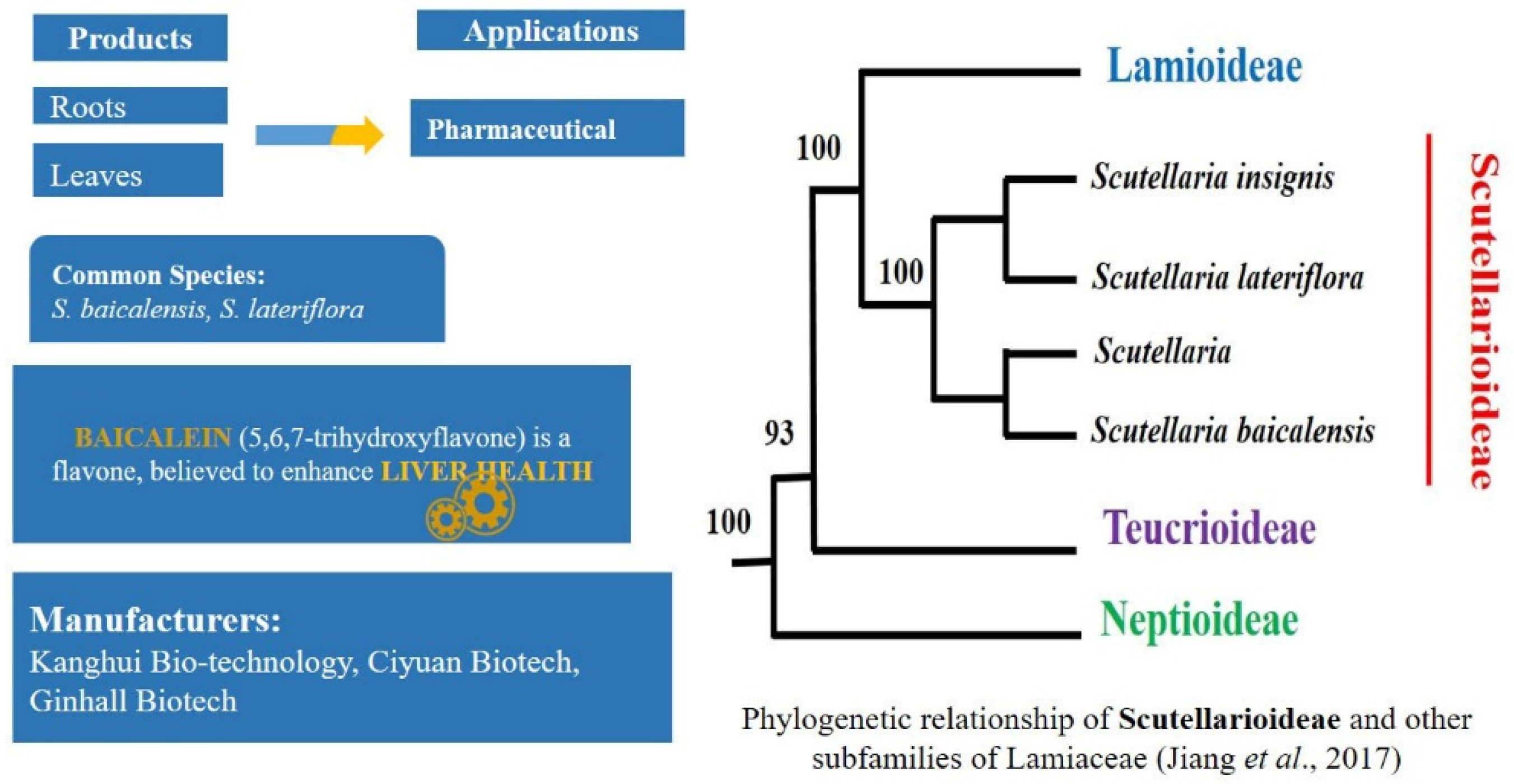
2.5. Rising Demand of Natural Products in Pharma Market: Scutellaria L.—Scutellarioideae
2.6. High Utilisation of Functional or Superfood Food and Complex Taxonomy: Salvia L.—Mentheae: Nepetioideae
3. Evolving DNA Barcoding Technologies
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaufman, J.; Kalaitzandonakes, N. The economic potential of plant-made pharmaceuticals in the manufacture of biologic pharmaceuticals. J. Commer. Biotechnol. 2011, 17, 173–182. [Google Scholar] [CrossRef]
- Srivastava, P.K.; Pandey, A.K. Natural products and derivatives: Biological and pharmacological activities. Biochem. Cell. Arch. 2015, 15, 1–38. [Google Scholar]
- Leja, K.B.; Czaczyk, K. The industrial potential of herbs and spices—A mini review. Acta Sci. Pol. Technol. Aliment. 2016, 15, 353–365. [Google Scholar] [CrossRef] [Green Version]
- Allkin, B. Useful Plants—Medicines: At Least 28,187 Plant Species Are Currently Recorded as Being of Medicinal Use. In State of the World’s Plants 2017; Willis, K.J., Ed.; Royal Botanic Gardens Kew: London, UK, 2017. [Google Scholar]
- González-Minero, F.J.; Bravo-Díaz, L. The use of plants in skin-care products, cosmetics and fragrances: Past and present. Cosmetics 2018, 5, 50. [Google Scholar] [CrossRef] [Green Version]
- Marranzano, M.; Rosa, L.R.; Malaguarnera, M.; Palmeri, R.; Tessitori, M.; Barbera, A.C. Polyphenols: Plant Sources and Food Industry Applications. Curr. Pharm. Des. 2018, 24, 4125–4130. [Google Scholar] [CrossRef] [PubMed]
- Anand, U.; Jacobo-Herrera, N.; Altemimi, A.; Lakhssassi, N. A comprehensive review on medicinal plants as antimicrobial therapeutics: Potential avenues of biocompatible drug discovery. Metabolites 2019, 9, 258. [Google Scholar] [CrossRef] [Green Version]
- Faccio, G. Plant complexity and cosmetic innovation. iScience 2020, 23, 101358. [Google Scholar] [CrossRef]
- Salmerón-Manzano, E.; Garrido-Cardenas, J.A.; Manzano-Agugliaro, F. Worldwide research trends on medicinal plants. Int. J. Environ. Res. Public Health 2020, 17, 3376. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M.; Snader, K.M. Natural products as sources of new drugs over the period 1981–2002. J. Nat. Prod. 2003, 66, 1022–1037. [Google Scholar] [CrossRef]
- Li-Weber, M. New therapeutic aspects of flavones: The anticancer properties of Scutellaria and its main active constituents Wogonin, Baicalein and Baicalin. Cancer Treat. Rev. 2009, 35, 57–68. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 2013, 1830, 3670–3695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, H.; Ma, Q.; Ye, L.; Piao, G. The traditional medicine and modern medicine from natural products. Molecules 2016, 21, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowan, M.M. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 1999, 12, 564–582. [Google Scholar] [CrossRef] [Green Version]
- Jassim, S.A.; Naji, M.A. Novel antiviral agents: A medicinal plant perspective. J. Appl. Microbiol. 2003, 95, 412–427. [Google Scholar] [CrossRef] [Green Version]
- Ganjhu, R.K.; Mudgal, P.P.; Maity, H.; Dowarha, D.; Devadiga, S.; Nag, S.; Arunkumar, G. Herbal plants and plant preparations as remedial approach for viral diseases. Virusdisease 2015, 26, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Hussain, H.; Green, I.R.; Ali, I.; Khan, I.A.; Ali, Z.; Al-Sadi, A.M.; Ahmed, I. Ursolic acid derivatives for pharmaceutical use: A patent review (2012–2016). Expert Opin. Ther. Pat. 2017, 27, 1061–1072. [Google Scholar] [CrossRef]
- Parham, S.; Kharazi, A.Z.; Bakhsheshi-Rad, H.R.; Nur, H.; Ismail, A.F.; Sharif, S.; Rama Krishna, S.; Berto, F. Antioxidant, antimicrobial and antiviral properties of herbal materials. Antioxidants 2020, 9, 1309. [Google Scholar] [CrossRef]
- Brochot, A.; Guilbot, A.; Haddioui, L.; Roques, C. Antibacterial, antifungal, and antiviral effects of three essential oil blends. Microbiologyopen 2017, 6, e00459. [Google Scholar] [CrossRef]
- Astani, A.; Reichling, J.; Schnitzler, P. Screening for antiviral activities of isolated compounds from essential oils. Evid. Based Complement. Altern. Med. 2011, 2011, 253643. [Google Scholar] [CrossRef] [Green Version]
- Silva, J.K.R.; Figueiredo, P.L.B.; Byler, K.G.; Setzer, W.N. Essential oils as antiviral agents. Potential of essential oils to treat SARS-CoV-2 infection: An in-silico investigation. Int. J. Mol. Sci. 2020, 21, 3426. [Google Scholar] [CrossRef]
- Petrovska, B.B. Historical review of medicinal plants’ usage. Pharmacogn. Rev. 2012, 6, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, B.; Marasini, B.P.; Rayamajhee, B.; Bhattarai, B.R.; Lamichhane, G.; Khadayat, K.; Adhikari, A.; Khanal, S. Potential roles of medicinal plants for the treatment of viral diseases focusing on COVID-19: A review. Phytother. Res. 2020, 1, 15. [Google Scholar] [CrossRef]
- Benarba, B.; Pandiella, A. Medicinal plants as sources of active molecules against COVID-19. Front. Pharmacol. 2020, 11, 1189. [Google Scholar] [CrossRef] [PubMed]
- Joshi, J.A.; Puthiyedath, R. Outcomes of Ayurvedic care in a COVID-19 patient with hypoxia—A Case Report. J. Ayurveda Integr. Med. 2020, 13, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Dong, L.; Yang, L.; Chen, D.; Peng, C. Potential drugs for the treatment of the novel coronavirus pneumonia (COVID-19) in China. Virus Res. 2020, 286, 198057. [Google Scholar] [CrossRef] [PubMed]
- Silveira, D.; Prieto-Garcia, J.M.; Boylan, F.; Estrada, O.; Fonseca-Bazzo, Y.M.; Jamal, C.M.; Magalhães, P.O.; Pereira, E.O.; Tomczyk, M.; Heinrich, M. COVID-19: Is there evidence for the use of herbal medicines as adjuvant symptomatic therapy? Front. Pharmacol. 2020, 11, 581840. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Chaurasia, J.P.; Khan, R.; Dhand, C.; Verma, S. Role of medicinal plants of traditional use in recuperating devastating COVID-19 situation. Med. Aromat. Plants 2020, 9, 359. [Google Scholar]
- Wen, W.; Su, W.; Tang, H.; Le, W.; Zhang, X.; Zheng, Y.; Liu, X.; Xie, L.; Li, J.; Ye, J.; et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, B.; Lv, J.T.; Sa, R.N.; Zhang, X.M.; Lin, Z.J. The clinical benefits of Chinese patent medicines against COVID-19 based on current evidence. Pharmacol. Res. 2020, 157, 104882. [Google Scholar] [CrossRef] [PubMed]
- WHO Traditional Medicine Strategy: 2014–2023. Available online: https://www.who.int/publications/i/item/9789241506096 (accessed on 7 October 2021).
- WHO Global Report on Traditional and Complementary Medic. Available online: https://apps.who.int/iris/rest/bitstreams/1217520/retrieve (accessed on 15 March 2021).
- Market Research Future Report. Available online: https://www.globenewswire.com/news-release/2019/04/03/1796359/0/en/Herbal-Medicine-Market-Value-to-Surpass-USD-129-Billion-Revenue-Mark-by-2023-at-5-88-CAGR-Predicts-Market-Research-Future.html (accessed on 21 August 2021).
- Directive 2004/24/Ecof The European Parliament and of the Council of 31 March 2004. Official Journal of the European Union. 136/85. Available online: https://eur-lex.europa.eu/legal-content/en/ALL/?uri=CELEX%3A32004L0024 (accessed on 13 November 2021).
- Advice on Regulating Herbal Medicines and Practitioners. Available online: https://www.gov.uk/government/publications/advice-on-regulating-herbal-medicines-and-practitioners (accessed on 27 March 2021).
- Aliki, X.; Ganopoulos, I.; Kalivas, A.; Osathanunkul, M.; Chatzopoulou, P.; Athanasios, T.; Madesis, P. Multiplex HRM analysis as a tool for rapid molecular authentication of nine herbal teas. Food Control 2015, 60, 10. [Google Scholar]
- Gottardi, D.; Bukvicki, D.; Prasad, S.; Tyagi, A.K. Beneficial effects of spices in food preservation and safety. Front. Microbiol. 2016, 7, 1394. [Google Scholar] [CrossRef] [Green Version]
- Bondi, M.; Lauková, A.; de Niederhausern, S.; Messi, P.; Papadopoulou, C. Natural preservatives to improve food quality and safety. J. Food Qual. 2017, 2017, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Srirama, R.; Kumar, J.U.S.; Seethapathy, G.S.; Newmaster, S.G.; Ragupathy, S.; Ganeshaiah, K.N.; Shaanker, R.U.; Ravikanth, G. Species adulteration in the herbal trade: Causes, consequences and mitigation. Drug Saf. 2017, 40, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, B.; Vaidya, A.D.B.; Chorghade, M. Ayurveda and natural products drug discovery. Curr. Sci. 2004, 86, 789–799. [Google Scholar]
- Yadav, M.; Chatterji, S.; Gupta, S.K.; Watal, G. Preliminary phytochemical screening of six medicinal plants used in traditional medicine. Int. J. Pharm. Pharm. Sci. 2020, 6, 539–542. [Google Scholar]
- Ghosh, D. Quality issues of herbal medicines: Internal and external factors. Int. J. Complement. Altern. Med. 2018, 11, 67–69. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Li, C.G.; Chang, D.; Bensoussan, A. Current Status and Major Challenges to the Safety and Efficacy Presented by Chinese Herbal Medicine. Medicines 2019, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Upton, R.; David, B.; Gafner, S.; Glasl, S. Botanical ingredient identification and quality assessment: Strengths and limitations of analytical techniques. Phytochem. Rev. 2020, 19, 1157–1177. [Google Scholar] [CrossRef] [Green Version]
- Balekundri, A.; Mannur, V. Quality control of the traditional herbs and herbal products: A review. Future J. Pharm. Sci. 2020, 6, 67. [Google Scholar] [CrossRef]
- Parveen, I.; Gafner, S.; Techen, N.; Murch, S.J.; Khan, I.A. DNA barcoding for the identification of botanicals in herbal medicine and dietary supplements: Strengths and limitations. Planta Med. 2016, 82, 1225–1235. [Google Scholar] [CrossRef] [Green Version]
- Raclariu, A.C.; Heinrich, M.; Ichim, M.C.; de Boer, H. Benefits and limitations of DNA barcoding and metabarcoding in herbal product authentication. Phytochem.Anal. 2018, 29, 123–128. [Google Scholar] [CrossRef]
- de Boer, H.J.; Ichim, M.C.; Newmaster, S.G. DNA barcoding and pharmacovigilance of herbal medicines. Drug Saf. 2015, 38, 611–620. [Google Scholar] [CrossRef]
- Adulteration of Essential Oils and Detection Techniques. Available online: https://www.linkedin.com/pulse/adulteration-essential-oils-detection-techniques-dr-sudhi-mestri (accessed on 10 May 2021).
- Gao, J.; Yin, W.; Corcoran, O. From Scutellariabarbata to BZL101 in cancer patients: Phytochemistry, Pharmacology, and Clinical Evidence. Nat. Prod. Commun. 2019, 14, 1–12. [Google Scholar]
- Schlick-Steiner, B.C.; Steiner, F.M.; Seifert, B.; Stauffer, C.; Christian, E.; Crozier, R.H. Integrative taxonomy: A multisource approach to exploring biodiversity. Annu. Rev. Entomol. 2010, 55, 421–438. [Google Scholar] [CrossRef]
- Pawar, R.S.; Handy, S.M.; Cheng, R.; Shyong, N.; Grundel, E. Assessment of the authenticity of herbal dietary supplements: Comparison of chemical and DNA barcoding methods. Planta Med. 2017, 83. [Google Scholar] [CrossRef] [Green Version]
- Kazi, T.; Hussain, N.; Bremner, P.; Slater, A.; Howard, C. The application of a DNAbased identification technique to over-the-counter herbal medicines. Fitoterapia 2013, 87, 27–30. [Google Scholar] [CrossRef]
- Mishra, P.; Kumar, A.; Nagireddy, A.; Mani, D.N.; Shukla, A.K.; Tiwari, R.; Sundaresan, V. DNA barcoding: An efficient tool to overcome authentication challenges in the herbal market. PlantBiotechnol. J. 2016, 14, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.M.; Brodnicke, O.B.; Evankow, A.M.; Ferreira, A.O.; Fontes, J.T.; Hansen, A.K.; Jensen, M.R.; Kalaycı, T.E.; Leeper, A.; Patil, S.K.; et al. The future of DNA barcoding: Reflections from early career researchers. Diversity 2021, 13, 313. [Google Scholar] [CrossRef]
- Newmaster, S.G.; Grguric, M.; Shanmughanandhan, D.; Ramalingam, S.; Ragupathy, S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013, 11, 222. [Google Scholar] [CrossRef] [Green Version]
- Ichim, M.C. The DNA-based authentication of commercial herbal products reveals their globally widespread adulteration. Front. Pharmacol. 2019, 10, 1227. [Google Scholar] [CrossRef] [PubMed]
- Sgamma, T.; Lockie-Williams, C.; Kreuzer, M.; Williams, S.; Scheyhing, U.; Koch, E.; Slater, A.; Howard, C. DNA barcoding for industrial quality assurance. Planta Med. 2017, 83, 1117–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nehal, N.; Choudhary, B.; Nagpure, A.; Gupta, R.K. DNA barcoding: A modern age tool for detection of adulteration in food. Crit. Rev. Biotechnol. 2021, 41, 767–791. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollingsworth, P.M. Refining the DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2011, 108, 9451–9452. [Google Scholar] [CrossRef] [Green Version]
- Kress, W.J.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef] [Green Version]
- Chase, M.W.; Salamin, N.; Wilkinson, M.; Dunwell, J.M.; Kesanakurthi, R.P.; Haider, N.; Savolainen, V. Land plants and DNA barcodes: Short-term and long-term goals. Philos. Trans. 2005, 360, 1889–1895. [Google Scholar] [CrossRef] [Green Version]
- Kress, W.J.; Erickson, D.L. DNA barcodes: Genes, genomics, and bioinformatics. Proc. Natl. Acad. Sci. USA 2007, 105, 2761–2762. [Google Scholar] [CrossRef] [Green Version]
- Lahaye, R.; Savolainen, V.; Duthoit, S.; Maurin, O.; van der Bank, M. A test of psbK-psbI and atpF-atpH as potential plant DNA barcodes using the flora of the Kruger National Park (South Africa) as a model system. Nat. Preced. 2008, 1, 1–21. [Google Scholar] [CrossRef]
- Newmaster, S.G.; Fazekas, A.J.; Steeves, R.A.D.; Janovec, J. Testing candidate plant barcode regions in the Myristicaceae. Mol. Ecol.Resour. 2008, 8, 480–490. [Google Scholar] [CrossRef]
- CBOL Plant Working Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Flower, A.; Ritchie, A.; Liuc, J.; Molassiotis, A.; Yue, H.; Lewithe, G. Oral Chinese herbal medicine (CHM) as an adjuvant treatment during chemotherapy for non-small cell lung cancer: A systematic review. Lung Cancer 2010, 68, 137–145. [Google Scholar] [CrossRef]
- China Plant BOL Group; Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [PubMed] [Green Version]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA Barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Henry, R.J.; Rossetto, M.; Wang, Y.; Chen, S. Plant DNA barcoding: From gene to genome. Biol. Rev. 2015, 90, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.R.; Zhang, Y.H.; Nakamura, K.; Guan, B.C.; Qiu, Y.X. Developing DNA barcodes for species identification in Podophylloideae (Berberidaceae). J. Syst. Evol. 2014, 52, 487–499. [Google Scholar] [CrossRef]
- Buddhachat, K.; Osathanunkul, M.; Madesis, P.; Chomdej, S.; Ongchai, S. Authenticity analyses of Phyllanthus amarus using barcoding coupled with HRM analysis to control its quality for medicinal plant product. Gene 2015, 573, 84–90. [Google Scholar] [CrossRef]
- Feng, S.; Jiang, M.; Shi, Y.; Jiao, K.; Shen, C.; Lu, J.; Ying, Q. Application of the ribosomal DNA ITS2 region of Physalis (Solanaceae): DNA barcoding and phylogenetic study. Front. Plant Sci. 2016, 7, 1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umdale, S.D.; Kshirsagar, P.R.; Lekhak, M.M.; Gaikwaid, N.B. Molecular authentication of the traditional medicinal plant “Lakshman Booti” (Smithiaconferta Sm.) and its adulterants through DNA barcoding. Pharmacogn. Mag. 2017, 13, S224–S229. [Google Scholar]
- Yu, J.; Wu, X.; Liu, C.; Newmaster, S.; Ragupathy, S.; Kress, W.J. Progress in the use of DNA barcodes in the identification and classification of medicinal plants. Ecotoxicol. Environ. Saf. 2021, 208, 111691. [Google Scholar] [CrossRef]
- Vivas, C.V.; Moraes, R.C.S.; Alves-Araújo, A.; Alves, M.; Mariano-Neto, E.; Berg, C.V.; Gaiotto, F.A. DNA barcoding in Atlantic Forest plants: What is the best marker for Sapotaceae species identification? Genet. Mol. Biol. 2014, 37, 662–670. [Google Scholar] [CrossRef]
- Bolson, M.; de Camargo Smidt, E.; Brotto, M.L.; Silva-Pereira, V. ITS and trnH-psbA as efficient DNA Barcodes to identify threatened commercial woody angiosperms from southern Brazilian Atlantic rainforests. PLoS ONE 2015, 10, e0143049. [Google Scholar]
- Duan, H.; Wang, W.; Zeng, Y.; Guo, M.; Zhou, Y. The screening and identification of DNA barcode sequences for Rehmannia. Sci. Rep. 2019, 9, 17295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.J.; Wang, X.; Wang, J.; Su, N.; Zhang, L.; Ma, Y.; Chang, Z.; Zhao, L.; Potter, D. Efficient identification of Pulsatilla (Ranunculaceae) using DNA barcodes and micro-morphological characters. Front. Plant Sci. 2019, 10, 1196. [Google Scholar] [CrossRef] [PubMed]
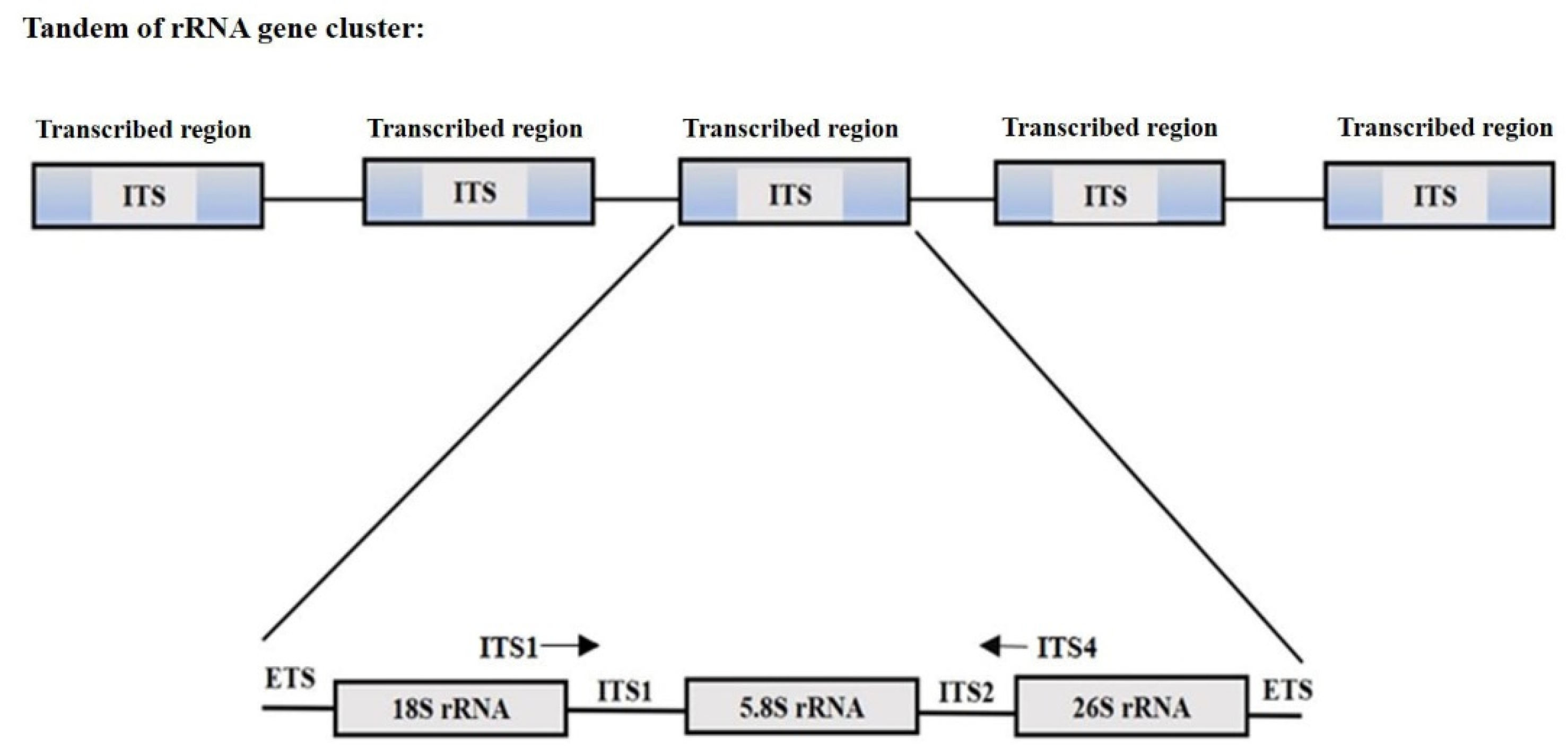
- Potapova, T.A.; Gerton, J.L. Ribosomal DNA and the nucleolus in the context of genome organization. Chromosome Res. 2019, 27, 109–127. [Google Scholar] [CrossRef] [PubMed]
- Banchi, E.; Ametrano, C.G.; Greco, S.; Stanković, D.; Muggia, L.; Pallavicini, A. PLANiTS: A curated sequence reference dataset for plant ITS DNA metabarcoding. Database 2020, 155, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Besse, P.; Silva, D.D.; Michel Grisoni, M. Plant DNA barcoding principles and limits: A case study in the genus Vanilla. Methods Mol. Biol. 2020, 2222, 131–148. [Google Scholar]
- Masiero, E.; Banik, D.; Abson, J.; Greene, P.; Slater, A.; Sgamma, T. Molecular verification of the UK national collection of cultivated Liriope and Ophiopogon plants. Plants 2020, 9, 558. [Google Scholar] [CrossRef]
- Jobes, D.V.; Thien, L.B. A conserved motif in the 5.8S ribosomal RNA (rRNA) gene is a useful diagnostic marker for plant internal transcribed spacer (ITS) sequences. PlantMol. Biol. Rep. 1997, 15, 326–334. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990. [Google Scholar]
- Manter, D.K.; Vivanco, J.M. Use of the ITS primers, ITSF and ITS4, to characterize fungal abundance and diversity in mixed-template samples by qPCR and length heterogeneity analysis. J. Microbiol. Methods 2007, 7, 7–14. [Google Scholar] [CrossRef]
- Cheng, T.; Xu, C.; Lei, L.; Li, C.; Zhang, Y.; Zhou, S. Barcoding the kingdom Plantae: New PCR primers for ITS regions of plants with improved universality and specificity. Mol. Ecol. Resour. 2016, 6, 38–49. [Google Scholar] [CrossRef]
- Badotti, F.; de Oliveira, F.S.; Garcia, C.F.; Vaz, A.B.M.; Fonseca, P.L.C.; Nahum, L.A.; Oliveira, G.; Góes-Neto, A. Effectiveness of ITS and sub-regions as DNA barcode markers for the identification of Basidiomycota (Fungi). BMC Microbiol. 2017, 7, 42. [Google Scholar] [CrossRef] [Green Version]
- Zheng, R.; Wang, W.; Tan, J.; Xu, H.; Zhan, R.; Chen, W. An investigation of fungal contamination on the surface of medicinal herbs in China. Chin.Med. 2017, 12, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Soltis, P.S.; Daniel, L.; Nickrent, D.L.; Leigh, A.; Johnson, L.A.; Hahn, W.J.; Hoot, S.B.; Sweere, J.A.; Kuzoff, R.K.; Kron, K.A.; et al. Angiosperm phylogeny inferred from 8S ribosomal DNA sequences. Ann. Mo. Bot. Gard. 1997, 84, 1–49. [Google Scholar] [CrossRef]
- Kim, W.J.; Moon, B.C.; Yang, S.; Han, K.S.; Choi, G.; Lee, A.Y. Rapid authentication of the herbal medicine plant species Aralia continentalis Kitag. and Angelica biserrata C.Q. Yuan and R.H. Shan using ITS2 sequences and multiplex-SCAR markers. Molecules 2016, 21, 270. [Google Scholar] [CrossRef] [Green Version]
- Veldman, S.; Ju, Y.; Otieno, J.N.; Abihudi, S.; Posthouwer, C.; Gravendeel, B.; van Andel, T.R.; de Boer, H.J. DNA barcoding augments conventional methods for identification of medicinal plant species traded at Tanzanian markets. J. Ethnopharmacol. 2020, 250, 112495. [Google Scholar] [CrossRef]
- Rai, P.D.; Bellampalli, R.; Dobriyal, R.M.; Agarwal, A.; Satyamoorthy, K.; Anantha Narayana, D.A. DNA barcoding of authentic and substitute samples of herb of the family Asparagaceae and Asclepiadaceae based on the ITS2 region. J. Ayurveda Integr. Med. 2012, 3, 36–40. [Google Scholar]
- Zhao, S.; Chen, X.; Song, J.; Pang, X.; Chen, S. Internal transcribed spacer 2 barcode: A good tool for identifying Acanthopanacis cortex. Front. Plant Sci. 2015, 6, 840. [Google Scholar] [CrossRef] [Green Version]
- Awad, M.; Fahmy, R.M.; Mosa, K.A.; Helmy, M.; El-Feky, F.A. Identification of effective DNA barcodes for Triticum plants through chloroplast genomewide analysis. Comput. Biol. Chem. 2017, 71, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Fadzil, N.F.; Wagiran, A.; Salleh, F.M.; Abdullah, S.; Izham, N.H.M. Authenticity testing and detection of Eurycoma longifolia in commercial herbal products using bar-high resolution melting analysis. Genes 2018, 9, 408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, C.D.; Carr, T.G.; Harris, S.A.; Hughes, C.E. Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Mol. Phylogenet. Evol. 2003, 29, 435–455. [Google Scholar] [CrossRef] [PubMed]
- Howard, C.; Lockie-Williams, C.; Slater, A. Applied barcoding: The practicalities of DNA testing for herbals. Plants 2020, 9, 1150. [Google Scholar] [CrossRef]
- Harley, R.M.; Atkins, S.; Budantsev, A.L.; Cantino, P.D.; Conn, B.J.; Grayer, R.; Harley, M.M.; De Kok, R.; Krestovskaja, T.; Morales, R.; et al. Labiatae. In The Families and Genera of Vascular Plants; Kubitzki, K., Kadereit, J.W., Eds.; Springer: Berlin, Germany, 2004; Volume 7, pp. 167–275. [Google Scholar]
- Carović-Stanko, K.; Petek, M.; Grdiša, M.; Pintar, J.; Bedeković, D.; Ćustić, M.H.; Satovic, Z. Medicinal plants of the family Lamiaceae as functional foods—A review. Czech J. Food Sci. 2016, 34, 377–390. [Google Scholar] [CrossRef] [Green Version]
- Mamadalieva, N.Z.; Akramov, D.K.; Ovidi, E.; Tiezzi, A.; Nahar, L.; Azimova, S.S.; Sarker, S.D. Aromatic medicinal plants of the Lamiaceae family from Uzbekistan: Ethnopharmacology, essential oils composition, and biological activities. Medicines 2017, 4, 8. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, B.M. Basil oil—Perfume. Flavor 1992, 17, 47–50. [Google Scholar]
- Ascensão, L.; Marques, N.; Pais, M.S. Glandular trichomes on vegetative and reproductive organs of Leonotisleonurus (Lamiaceae). Ann. Bot. 1995, 75, 619–626. [Google Scholar] [CrossRef]
- Giuliani, C.; Maleci-Bini, L. Insight into the structure and chemistry of glandular trichomes of Labiatae, with emphasis on subfamily Lamioideae. Plant Syst. Evol. 2008, 276, 199–208. [Google Scholar] [CrossRef]
- Puškárová, A.; Bučková, M.; Kraková, L.; Pangallo, D.; Kozics, K. The antibacterial and antifungal activity of six essential oils and their cyto/genotoxicity to human HEL 12469 cells. Sci. Rep. 2017, 7, 8211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozin, B.; Mimica-Dukic, N.; Simin, N.; Anackov, G. Characterization of the volatile composition of essential oils of some Lamiaceae spices and the antimicrobial and antioxidant activities of the entire oils. J. Agric. Food Chem. 2006, 54, 1822–1828. [Google Scholar] [CrossRef] [PubMed]
- Birkett, M.A.; Hassanali, A.; Hoglund, S.; Pettersson, J.; Pickett, J.A. Repellent activity of catmint, Nepeta cataria, and iridoid nepetalactone isomers against Afro-tropical mosquitoes, ixodid ticks and red poultry mites. Phytochemistry 2011, 72, 109–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chadwick, M.; Trewin, H.; Gawthrop, F.; Wagstaff, C. Sesquiterpenoids lactones: Benefits to plants and people. Int. J. Mol. Sci. 2013, 14, 12780–12805. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.P.; Li, B.; Olmstead, R.G.; Cantino, P.D.; Liu, E.D.; Xiang, C.L. Phylogenetic placement of the enigmatic genus Holocheila (Lamiaceae) inferred from plastid DNA sequences. Taxon 2014, 63, 355–366. [Google Scholar] [CrossRef]
- Aburjai, T.; Natsheh, F.M. Plants used in cosmetics. Phytother. Res. 2003, 17, 987–1000. [Google Scholar] [CrossRef]
- Grassmann, J.; Elstner, E.F. Essential oils. Properties and uses. In Encyclopedia of Food Science and Nutrition, 2nd ed.; Caballero, B., Trugo, L., Finglas, P., Eds.; Elsevier: New York, NY, USA, 2003; pp. 2177–2184. [Google Scholar]
- Burt, S. Essential oils: Their antibacterial properties and potential applications in foods—A review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef] [PubMed]
- Edris, A.E. Pharmaceutical and therapeutic potentials of essential oils and their individual volatile constituents: A review. Phytother. Res. 2007, 21, 308–323. [Google Scholar] [CrossRef]
- Ozkan, M. Glandular and eglandular hairs of Salvia recognita Fisch. & Mey. (Lamiaceae) in Turkey. Bangladesh J. Bot. 2008, 37, 93–95. [Google Scholar]
- Rattray, R.D.; Van Wyk, B.E. The Botanical, Chemical and Ethnobotanical Diversity of Southern African Lamiaceae. Molecules 2021, 26, 3712. [Google Scholar] [CrossRef] [PubMed]
- Jurges, G.; Sahi, V.; Rodriguez, D.R.; Reich, E.; Bhamra, S.; Howard, C.; Slater, A.; Nick, P. Product authenticity versus globalisation—The Tulsi case. PLoS ONE 2018, 13, e0207763. [Google Scholar] [CrossRef]
- Leyva-López, N.; Gutiérrez-Grijalva, E.P.; Vazquez-Olivo, G.; Heredia, J.B. Essential Oils of Oregano: Biological Activity beyond Their Antimicrobial Properties. Molecules 2017, 22, 989. [Google Scholar] [CrossRef] [Green Version]
- Foster, S. Adulteration of Skullcap with American Germander; American Botanical Council: Austin, TX, USA, 2016. [Google Scholar]
- World Checklist of Lamiaceae (Mentha). Available online: http://wcsp.science.kew.org/namedetail.do?name_id=124319 (accessed on 5 May 2020).
- Savithri, B.; Priti, M.; Sushil, K.; Anil, K. Mentha species: In vitro regeneration and genetic Transformation. Mol. Biol. Today 2002, 3, 11–23. [Google Scholar]
- Pandey, A.K.; Rai, M.K.; Acharya, D. Chemical composition and antimycotic activity of the essential oils of corn mint (Mentha arvensis) and lemon grass (Cymbopogon flexuosus) against human pathogenic fungi. Pharm. Biol. 2003, 41, 421–425. [Google Scholar] [CrossRef]
- Gulluce, M.; Sahin, F.; Sokmen, M. Antimicrobial and antioxidant properties of the essential oils and methanol extract from Mentha longifolia L. ssp. longifolia. Food Chem. 2007, 103, 1449–1456. [Google Scholar] [CrossRef]
- Nickavar, B.; Alinaghi, A.; Kamalinejad, M. Evaluation of the Antioxidant Properties of Five Mentha Species. Iran. J. Pharm. Sci. 2008, 7, 203–209. [Google Scholar]
- Morton, J.K. The chromosome numbers of the British Menthae. Warsonia 1956, 3, 244–252. [Google Scholar]
- Harley, R.M.; Brighton, C.A. Chromosome numbers in the genus Mentha, L. Bot. J. Linn. Soc. 1977, 74, 71–96. [Google Scholar] [CrossRef]
- Gobert, V.; Moja, S.; Colson, M.; Taberlet, P. Hybridization in the section Mentha (Lamiaceae) inferred from AFLP markers. Am. J. Bot. 2002, 89, 2017–2023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tucker, A.; Chambers, H.L. Mentha canadensis L. (Lamiaceae): A relict amphidiploid from the lower tertiary. Taxon 2002, 51, 703. [Google Scholar] [CrossRef]
- Testing Hypotheses of Hybridization in Mentha Spicata and M. Canadensis Using Molecular Data. Available online: https://digitalcommons.murraystate.edu/postersatthecapitol/2005/WKU/5/ (accessed on 2 November 2021).
- Mint Essential Oil Market Size Worth $330.02 Million by 2025. CAGR: 9.2%: Grand View Research, Inc. Available online: https://www.prnewswire.com/news-releases/mint-essential-oil-market-size-worth-330-02-million-by-2025--cagr-9-2-grand-view-research-inc-300971922.html (accessed on 26 September 2021).
- Heylen, O.C.G.; Debortoli, N.; Marescaux, J.; Olofsson, J.K. A Revised Phylogeny of the Mentha spicata Clade Reveals Cryptic Species. Plants 2021, 10, 819. [Google Scholar] [CrossRef]
- Snoussi, M.; Noumi, E.; Trabelsi, N.; Flamini, G.; Papetti, A.; De Feo, V. Menthaspicata Essential Oil: Chemical Composition, Antioxidant and Antibacterial Activities against Planktonic and Biofilm Cultures of Vibrio spp. Strains. Molecules 2015, 20, 14402–14424. [Google Scholar] [CrossRef] [PubMed]
- Wilson, L. Spices and Flavoring Crops: Leaf and Floral Structures. In Encyclopedia of Food and Health, 1st ed.; Caballero, B., Fingal, P., Toldra, F., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 84–92. [Google Scholar]
- Khanuja, S.P.S.; Shasany, A.K.; Srivastava, A.; Kumar, S. Assessment of genetic relationships in Mentha species. Euphytica 2000, 111, 121–125. [Google Scholar] [CrossRef]
- Shaikh, S.; Yaacob, H.B.; Rahim, Z.H. A Prospective role in treatment of major illnesses and potential benefits as a safe insecticide and natural food preservative of mint (Mentha spp.): A Review. Asian J. Biomed. Pharm. Sci. 2014, 4, 1–12. [Google Scholar] [CrossRef]
- Kumar, K.V.; Patra, D. Alteration in yield and chemical composition of essential oil of Mentha piperita L. plant: Effect of fly ash amendments and organic wastes. Ecol. Eng. 2012, 47, 237–241. [Google Scholar] [CrossRef]
- Prakash, O.; Chandra, M.; Pant, A.K.; Rawat, D.S. Mint (Mentha spicata L.) Oils. In Essential Oils in Food Preservation, Flavor and Safety; Preedy, V.R., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 561–572. [Google Scholar]
- Salehi, B.; Stojanović-Radić, Z.; Matejić, J.; Sharopov, F.; Antolak, H.; Kręgiel, D.; Sen, S.; Sharifi-Rad, M.; Acharya, K.; Sharifi-Rad, R.; et al. Plants of Genus Mentha: From Farm to Food Factory. Plants 2018, 4, 70. [Google Scholar] [CrossRef] [Green Version]
- Pandey, J.; Verma, R.; Singh, S. Suitability of aromatic plants for phytoremediation of heavy metal contaminated areas: A review. Int. J. Phytoremediat. 2019, 21, 1–14. [Google Scholar] [CrossRef]
- Schmidt, E.; Bail, S.; Buchbauer, G.; Stoilova, I.; Atanasova, T.; Stoyanova, A.; Krastanov, A.; Jirovetz, L. Chemical composition, olfactory evaluation and antioxidant effects of essential oil from Mentha × piperita. Nat. Prod. Commun. 2009, 4, 1107–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afridi, M.S.; Ali, J.; Abbas, S.; Rehman, S.U.; Khan, F.A.; Khan, M.A.; Shahid, M. Essential oil composition of Mentha piperita L. And its antimicrobial effects against common human pathogenic bacterial and fungal strains. Pharmacologyonline 2016, 3, 90–97. [Google Scholar]
- Bhattacharya, S. Cultivation of Essential Oils. In Essential Oils in Food Preservation, Flavor and Safety; Preedy, V.R., Ed.; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Dionísio, A.; Molina, G.; Carvalho, D.; Santos, R.; Bicas, J.; Pastore, G. Natural flavourings from biotechnology for foods and beverages. In Natural Food Additives, Ingredients and Flavourings; Baines, D., Seal, R., Eds.; Woodhead: Sawston, UK, 2012. [Google Scholar]
- Morcia, C.; Tumino, G.; Ghizzoni, R.; Terzi, V. Carvone (Mentha spicata L.) oils. In Essential Oils in Food Preservation, Flavor and Safety; Preedy, V.R., Ed.; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Kokkini, S.; Karousou, R.; Hanlidou, E. Herbs of the Labiatae. In Encyclopedia of Food Sciences and Nutrition; Caballero, B., Ed.; Academic Press: Oxford, UK, 2003; pp. 3082–3090. [Google Scholar]
- Taylan, O.; Cebi, N.; Sagdic, O. Rapid screening of Mentha spicata essential oil and l-menthol in Mentha piperita essential oil by ATR-FTIR spectroscopy coupled with multivariate analyses. Foods 2021, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Mattia, F.D.; Bruni, I.; Galimberti, A.; Cattaneo, F.; Casiraghi, M.; Labra, M. A comparative study of different DNA barcoding markers for the identification of some members of Lamiaceae. Food Res. Int. 2011, 44, 693–702. [Google Scholar] [CrossRef]
- Attiya, J.; Bin, G.; Bilal, H.A.; Zabta, K.S.; Tariq, M. Phylogenetics of selected Mentha species on the basis of rps8, rps11 and rps14 chloroplast genes. J. Med. PlantRes. 2012, 6, 30–36. [Google Scholar]
- Thakur, V.V.; Tiwari, S.; Tripathi, N.; Tiwari, G.; Sapre, S. DNA barcoding and phylogenetic analyses of Mentha species using rbcL sequences. Ann. Phytomed. 2016, 5, 59–62. [Google Scholar]
- Ahmed, S.M. Molecular identification of Lavendula dentata L.; Mentha longifolia (L.) Huds. and Mentha × piperita L. by DNA barcodes. Bangladesh J. Plant Taxon 2018, 25, 149–157. [Google Scholar] [CrossRef]
- Wang, K.; Li, L.; Hua, Y.; Zhao, M.; Li, S.; Sun, H.; Lv, Y.; Wang, Y. The complete chloroplast genome of Mentha spicata, an endangered species native to South Europe. Mitochondrial DNA B 2017, 2, 907–909. [Google Scholar] [CrossRef] [Green Version]
- Vining, K.J.; Johnson, S.R.; Ahkami, A.; Lange, I.; Parrish, A.N.; Trapp, S.C.; Croteau, R.B.; Straub, S.C.K.; Pandelova, I.; Lange, B.M. Draft genome sequence of Mentha longifolia and development of resources for mint cultivar improvement. Mol. Plant 2017, 10, 323–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saric-Kundalic, B.; Fialova, S.; Dobes, C.; Ölzant, S.; Tekelova, A.D.; Grancal, D.; Reznicek, G.; Saukel, J. Multivariate numerical taxonomy of Mentha species, hybrids, varieties and cultivars. Sci. Pharm. 2009, 77, 851–876. [Google Scholar] [CrossRef]
- Mogosan, C.; Vostinaru, O.; Oprean, R.; Heghes, C.; Filip, L.; Balica, G.; Moldovan, R.I. A comparative analysis of the chemical composition, anti-inflammatory, and antinociceptive effects of the essential oils from three species of mentha cultivated in Romania. Molecules 2017, 22, 263. [Google Scholar] [CrossRef] [PubMed]
- Harder, L.D.; Strelin, M.M.; Clocher, I.C.; Kulbaba, M.W.; Aizen, M.A. The dynamic mosaic phenotypes of flowering plants. New Phytol. 2019, 224, 1021–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, V.R.; Lal, R.K. Genotype × environment interaction, genetic variability and inheritance pattern in breeding lines including varieties/cultivars of menthol mint (Mentha arvensis L.). J. Med. Aromat. Plant Sci. 2020, 42, 145–156. [Google Scholar]
- Mishra, A.; Jain, P.; Lal, R.K.; Dhawan, S.S. Trichomes and Yield Traits in Mentha arvensis: Genotype Performance and Stability Evaluation. J. Herbs SpicesMed. Plants 2018, 24, 1–14. [Google Scholar] [CrossRef]
- Bunsawatt, J.; Elliott, N.E.; Hertweck, K.L.; Sproles, E.; Alice, L.A. Phylogenetics of Mentha (Lamiaceae): Evidence from Chloroplast DNA Sequences. Syst. Bot. 2004, 29, 959–964. [Google Scholar] [CrossRef]
- Shasany, A.K.; Shukla, A.K.; Soni, G.; Khanuja, S.P.S. Suman. AFLP analysis for genetic relationships among Mentha species. PlantGenet. Resour. Newsl. 2005, 144, 14–19. [Google Scholar]
- Celenk, S.; Dirmenci, T.; Malyer, H.; Bicakci, A. A palynological study of the genus Nepeta L. (Lamiaceae). Plant Syst. Evol. 2008, 276, 105–123. [Google Scholar] [CrossRef]
- Chengyuan, L.; Wei-Lin, L.; Yong, Z.; Bo Heng, X.; Xiao-Yue, W. Study on Morphological Diversity of Mentha HaplocalyxBriq. Med. Plant 2011, 2, 1–3. [Google Scholar]
- Hanafy, D.M.; Prenzler, P.D.; Hill, R.A.; Burrows, G.E. Leaf micromorphology of 19 Mentha Taxa. Aust. J. Bot. 2019, 67, 463. [Google Scholar] [CrossRef]
- Vining, K.J.; Pandelova, I.; Hummer, K.E.; Bassil, N.V.; Contreras, R.; Neill, K.; Chen, H.; Parrish, A.N.; Lange, B.M. Genetic diversity survey of Mentha aquatica L. and Mentha suaveolens Ehrh., mint crop ancestors. Genet. Resour. CropEvol. 2019, 66, 825–845. [Google Scholar] [CrossRef]
- Bokić, B.S.; Rat, M.M.; Kladar, N.V.; Anačkov, G.T.; Božin, B.N. Chemical diversity of volatile compounds of mints from southern part of Pannonian Plain and Balkan Peninsula—New Data. Chem.Biodivers. 2020, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mamadalieva, N.Z.; Hidayat, H.; Jianbo, X. Recent advances in genus Mentha: Phytochemistry, antimicrobial effects, and food applications. Food Front. 2020, 1, 435–458. [Google Scholar] [CrossRef]
- Moshrefi-Araghi, A.; Nemati, H.; Azizi, M.; Moshtaghi, N.; Shoor, M. Study of genetic diversity of some genotypes of Iranian wild mint (Menthalongifolia L.) using ISSRmarker and its correlation with dry yield and essential oil content. Agric. Biotechnol. J. 2020, 12, 117–140. [Google Scholar]
- Stoeckle, M.Y.; Hebert, P.D.N. Bar Code of Life: DNA tags help classify animals. Sci.Am. 2008, 299, 82–88. [Google Scholar]
- Flavor and Fragrances Market Worldwide—Statistics & Facts. Available online: https://www.statista.com/topics/6300/flavor-and-fragrances-market-worldwide (accessed on 21 November 2021).
- United Kingdom Exports of Essential Oils, Perfumes, Cosmetics, Toiletries. Available online: https://tradingeconomics.com/united-kingdom/exports/essential-oils-perfumes-cosmetics-toileteries (accessed on 15 July 2021).
- Satyal, P.; Setzer, W.N. Adulteration Analysis in Essential Oils. In Essential Oil Research; Malik, S., Ed.; Springer: Cham, Switzerland, 2019. [Google Scholar]
- Ng, T.B.; Fang, E.F.; Bekhit, A.E.-D.A.; Wong, J.H. Methods for the Characterization, Authentication, and Adulteration of Essential Oils. In Essential Oils in Food Preservation, Flavor and Safety; Preedy, V.R., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 11–17. [Google Scholar]
- Sarkic, A.; Stappen, I. Essential oils and their single compounds in cosmetics—A critical review. Cosmetics 2018, 5, 11. [Google Scholar] [CrossRef] [Green Version]
- Dubnicka, M.; Cromwell, B.; Levine, M. Investigation of the adulteration of essential oils by GC-MS. Curr. Anal. Chem. 2020, 16, 965–969. [Google Scholar] [CrossRef]
- Paton, A.J.; Springate, D.; Suddee, S.; Otieno, D.; Grayer, R.J.; Harley, M.M.; Willis, F.; Simmonds, M.S.J.; Powell, M.P.; Savolainen, V. Phylogeny and evolution of basils and allies (Ocimeae, Labiatae) based on three plastid DNA regions. Mol. Phylogenet. Evol. 2004, 31, 277–299. [Google Scholar] [CrossRef]
- Alan, P. The Genus Lavandula. Botanical Magazine Monograph; Kew Bulletin: Heidelberg, Germany, 2005; p. 160. [Google Scholar]
- World Checklist of Lamiaceae (Lavender). Available online: http://wcsp.science.kew.org/namedetail.do?name_id=108966 (accessed on 19 July 2020).
- Giray, F.H. An analysis of world lavender oil markets and lessons for Turkey. J. Essent. Oil Bear. Plants 2018, 21, 1612–1623. [Google Scholar] [CrossRef]
- Benabdelkader, T.; Guitton, Y.; Pasquier, B.; Magnard, J.L.; Jullien, F.; Kameli, A.; Legendre, L. Functional characterization of terpene synthases and chemotypic variation in three lavender species of section Stoechas. Physiol. Plant 2015, 153, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Moja, S.; Guitton, Y.; Nicolè, F.; Legendre, L.; Pasquier, B.; Upson, T.; Jullien, F. Genome size and plastid trnK-matK markers give new insights into the evolutionary history of the genus Lavandula, L. Plant Biosyst. 2016, 150, 1216–1224. [Google Scholar] [CrossRef]
- Lavender Oil Market Size 2021 by Top Countries Data, Industry Analysis by Regions, Revenue, Share, Development, Tendencies and Forecast to 2026. Available online: https://www.yournewsnet.com/story/44196805/lavender-oil-market-size-2021-by-top-countries-data-industry-analysis-by-regions-revenue-share-development-tendencies-and-forecast-to-2026 (accessed on 5 October 2021).
- Wells, R.; Truong, F.; Adal, A.M.; Sarker, L.; Mahmoud, S. Lavandula Essential Oils: A Current Review of Applications in Medicinal, Food, and Cosmetic Industries of Lavender. Nat. Prod. Commun. 2018, 13, 1403–1417. [Google Scholar] [CrossRef] [Green Version]
- Gören, A.C.; Topçu, G.; Bilsel, G.; Bilsel, M.; Aydoğmuş, Z.; Pezzuto, J.M. The chemical constituents and biological activity of essential oil of Lavandula stoechas ssp. stoechas. Z. Naturforsch. C 2002, 57, 797–800. [Google Scholar] [CrossRef]
- Hajhashemi, V.; Ghannadi, A.; Sharif, B. Anti-inflammatory and analgesic properties of the leaf extracts and essential oil of Lavandula angustifolia Mill. J. Ethnopharmacol. 2003, 89, 67–71. [Google Scholar] [CrossRef]
- Koulivand, P.H.; Ghadiri, M.K.; Gorji, A. Lavender and the Nervous System. Evid. Based Complement. Altern. Med. 2013, 2013, 681304. [Google Scholar] [CrossRef] [Green Version]
- Pokajewicz, K.; Białoń, M.; Svydenko, L.; Fedin, R.; Hudz, N. Chemical composition of the essential oil of the new cultivars of Lavandula angustifolia Mill. Bred in Ukraine. Molecules 2021, 26, 5681. [Google Scholar] [CrossRef]
- Cavanagh, H.C.; Jenny, W. Biological activities of lavender essential oil. Phytother. Res. 2002, 16, 301–308. [Google Scholar] [CrossRef]
- Woronuk, G.; Demissie, Z.; Rheault, M.; Mahmoud, S. Biosynthesis and therapeutic properties of Lavandula essential oil constituents. Planta Med. 2011, 77, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Bejar, E. Adulteration of English Lavender (Lavandula angustifolia) Essential Oil; Botanical Adulterants Prevention Bulletin: Austin, TX, USA, 2020. [Google Scholar]
- Satyal, P.; Sorensen, A.C. Authentication of lavender essential oil: Commercial essential oil samples and validity of standard specifications. Int. J. Prof. Hol. Aromather. 2016, 5, 17–22. [Google Scholar]
- Hind, K.R.; Adal, A.M.; Upson, T.M.; Mahmoud, S.S. An assessment of plant DNA barcodes for the identification of cultivated Lavandula (Lamiaceae) taxa. Biocatal. Agric. Biotechnol. 2018, 16, 459–466. [Google Scholar] [CrossRef]
- Theodoridis, S.; Stefanaki, A.; Tezcan, M.; Aki, C.; Kokkini, S.; Vlachonasios, K.E. DNA barcoding in native plants of the Labiatae (Lamiaceae) family from Chios Island (Greece) and the adjacent Çeşme-Karaburun Peninsula (Turkey). Mol. Ecol. Resour. 2012, 12, 620–633. [Google Scholar] [CrossRef] [PubMed]
- Soares, S.; Grazina, L.; Costa, J.; Amaral, J.S.; Oliveira, M.B.P.P.; Mafra, I. Botanical authentication of lavender (Lavandula spp.) honey by a novel DNA-barcoding approach coupled to high resolution melting analysis. Food Control 2018, 86, 367–373. [Google Scholar] [CrossRef] [Green Version]
- Pieroni, A.; Vandebroek, I. Travelling Cultures and Plants the Ethnobiology and Ethnopharmacy of Human Migrations; Berghahn Books Ltd.: New York, NY, USA, 2009. [Google Scholar]
- Sahoo, Y.; Pattnaik, S.K.; Chand, P.K. In vitro clonal propagation of an aromatic medicinal herb Ocimumbasilicum L. (sweet basil) by axillary shoot proliferation. In Vitro Cell. Dev. Biol.-Plant 1997, 33, 293–296. [Google Scholar] [CrossRef]
- Rastogi, S.; Meena, S.; Bhattacharya, A.; Ghosh, S.; Shukla, R.K.; Sangwan, N.S.; Lal, R.K.; Gupta, M.M.; Lavania, U.C.; Gupta, V.; et al. De novo sequencing and comparative analysis of holy and sweet basil transcriptomes. BMC Genom. 2014, 15, 588. [Google Scholar] [CrossRef] [Green Version]
- World Checklist of Lamiaceae (Ocimum). Available online: http://wcsp.science.kew.org/namedetail.do?name_id=136790 (accessed on 27 August 2020).
- Sudha, G.C.; Seeni, S. In vitro propagation and field establishment of Adhatodabeddomei C. B. Clarke, a rare medicinal plant. Plant Cell Rep. 1994, 13, 203–207. [Google Scholar]
- Pandey, A.K.; Singh, P.; Tripathi, N.N. Chemistry and bioactivities of essential oils of some Ocimum species: An overview. Asian Pac. J. Trop. Biomed. 2014, 4, 682–694. [Google Scholar] [CrossRef] [Green Version]
- Global Basil Essential Oil Market 2019–2023. Increasing Demand for Organic and Cold-Pressed Basil Essential Oil to Boost Growth | Technavio. Available online: https://www.businesswire.com/news/home/20190411005319/en/Global-Basil-Essential-Oil-Market-2019-2023-Increasing-Demand-for-Organic-and-Cold-Pressed-Basil-Essential-Oil-to-Boost-Growth-Technavio (accessed on 12 October 2021).
- Mali, P. Comparison of Methyl Eugenol Levels and Eugenol O-Methyltransferase Gene Structure in Different Ocimum Plant Species. Ph.D. Thesis, De Montfort University, Leicester, UK, 2018. [Google Scholar]
- Uma, D.P. Radioprotective, anticarcinogenic and antioxidant properties of the Indian holy basil, Ocimum sanctum (Tulasi). Indian J. Exp. Biol. 2001, 39, 185–190. [Google Scholar]
- Gupta, S.K.; Prakash, J.; Srivastava, S. Validation of claim of Tulsi, Ocimum sanctum Linn as a medicinal plant. Indian J. Exp. Biol. 2002, 40, 765–773. [Google Scholar]
- Rastogi, S.; Kalra, A.; Gupta, V.; Khan, F.; Lal, R.K.; Tripathi, A.K.; Parameswaran, S.; Gopalakrishnan, C.; Ramaswamy, G.; Shasany, A.K. Unravelling the genome of Holy basil: An “incomparable” “elixir of life” of traditional Indian medicine. BMC Genom. 2015, 16, 413. [Google Scholar] [CrossRef] [Green Version]
- Tangpao, T.; Chung, H.H.; Sommano, S.R. Aromatic Profiles of Essential Oils from Five Commonly Used Thai Basils. Foods 2018, 7, 175. [Google Scholar] [CrossRef] [Green Version]
- Mondal, S.; Mirdha, B.R.; Mahapatra, S.C. The science behind sacredness of Tulsi (Ocimum sanctum Linn.). Indian J. Physiol. Pharmacol. 2009, 53, 291–302. [Google Scholar] [PubMed]
- Rahman, S.; Islam, R.; Kamruzzaman, M.; Alam, M.; Jamal, M.A. Ocimum sanctum L.: A Review of Phytochemical and Pharmacological Profile. Am. J. Drug Discov. Dev. 2011, 154, 455–460. [Google Scholar]
- Silva, M.G.D.; Matos, F.J.D.; Lopes, P.R.O.; Silva, F.O.; Holanda, M.T. Composition of essential oils from three Ocimum species obtained by steam and microwave distillation and supercritical CO2 extraction. Arkivoc 2004, 2004, 66–71. [Google Scholar] [CrossRef] [Green Version]
- Zheljazkov, V.D.; Cantrell, C.L.; Evans, W.B.; Ebelhar, M.W.; Coker, C. Yield and composition of Ocimumbasilicum L. and Ocimum sanctum L. grown at four Locations. HortScience 2008, 43, 737–741. [Google Scholar] [CrossRef] [Green Version]
- Joshi, R.K. Chemical Composition, In Vitro Antimicrobial and Antioxidant Activities of the Essential Oils of Ocimumgratissimum, O. Sanctum and Their Major Constituents. Indian J. Pharm. Sci. 2013, 75, 457–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- British Pharmacopoeia Commission. Deoxyribonucleic Acid (DNA) Based Identification Techniques for Herbal Drugs; British Pharmacopoeia Appendix XI V; British Pharmacopoeia Commisssion: London, UK, 2017. [Google Scholar]
- Bhamra, S.; Heinrich, M.; Howard, C.; Johnson, M.; Slater, A. DNA authentication of tulsi (Ocimumtenuiflorum) using the nuclear ribosomal internal transcribed spacer (ITS) and the chloroplast intergenic spacer trnH-psbA. Planta Med. 2015, 81, 20. [Google Scholar] [CrossRef]
- British Pharmacopoeia Commission. DNA Barcoding as a Tool for Botanical Identification of Herbal Drugs; British Pharmacopoeia Supplementary Chapter SC VII D; British Pharmacopoeia Commission: London, UK, 2018. [Google Scholar]
- Ríos-Rodríguez, D.; Sahi, V.P.; Nick, P. Authentication of holy basil using markers relating to a toxicology-relevant compound. Eur. Food Res. Technol. 2021, 247, 2485–2497. [Google Scholar] [CrossRef]
- Khalid, K.; Hendawy, S.F.; El-Gezawy, E. Ocimumbasilicum L. production under organic farming. Res. J. Agric. Biol. Sci. 2006, 2, 25–32. [Google Scholar]
- Christina, V.; Arunachalam, A. Nucleotide based validation of Ocimum species by evaluating three candidate barcodes of the chloroplast region. MolEcolResour 2003, 14, 60–68. [Google Scholar]
- Fügel, R.; Carle, R.; Schieber, A. Quality and authenticity control of fruit purées, fruit preparations and jams—A review. Trends Food Sci. Technol. 2005, 16, 433–441. [Google Scholar] [CrossRef]
- Kurz, C.; Leitenberger, M.; Carle, R.; Schieber, A. Evaluation of fruit authenticity and determination of the fruit content of fruit products using FTNIR spectroscopy of cell wall components. Food Chem. 2010, 119, 806–812. [Google Scholar] [CrossRef]
- Klaudija, C.S.; Zlatko, L.; Olivera, P.; Strikić, F.; Kolak, I.; Milos, M.; Satovic, Z. Molecular and chemical characterization of the most widespread Ocimum species. Plant Syst. Evol. 2012, 249, 253–262. [Google Scholar]
- Mafra, I.; Ferreira, I.; Oliveira, M. Food authentication by PCR-based methods. Eur. Food Res. Technol. 2008, 227, 649–665. [Google Scholar] [CrossRef]
- Nieto, G. Biological Activities of three essential oils of the Lamiaceae family. Medicines 2017, 4, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Global Oregano Oil Market Outlook. Available online: https://www.expertmarketresearch.com/reports/oregano-oil-market (accessed on 18 July 2021).
- Skoula, M.; Harborne, J.B. Taxonomy and Chemistry. In Oregano: The Genera Origanum and Lippia; Kintzios, S.E., Ed.; Taylor and Francis CRC Press: Boca Raton, FL, USA, 2002; pp. 67–108. [Google Scholar]
- World Checklist of Lamiaceae (Origanum). Available online: http://wcsp.science.kew.org/namedetail.do?name_id=143731 (accessed on 11 August 2020).
- Stefanaki, A.; Andel, T. Mediterranean aromatic herbs and their culinary use. In Aromatic Herbs in Foods Bioactive Compounds, Processing, and Application; Galanakis, C., Ed.; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Tucker, A.O.; DeBaggio, T. The Big Book of Herbs: A Comprehensive Illustrated Reference to Herbs of Flavor and Fragrance; Interweave Press: Loveland, CO, USA, 2000. [Google Scholar]
- Vannozzi, A.; Lucchin, M.; Barcaccia, G. cpDNA Barcoding by Combined End-Point and Real-Time PCR Analyses to Identify and Quantify the Main Contaminants of Oregano (Origanum vulgare L.) in Commercial Batches. Diversity 2018, 10, 98. [Google Scholar] [CrossRef] [Green Version]
- Singletary, K. Oregano: Overview of the literature on health benefits. Nutr. Today 2010, 45, 129–138. [Google Scholar] [CrossRef] [Green Version]
- Toncer, O.; Karaman, S.; Diraz, E. An annual variation in essential oil composition of Origanum syriacum from Southeast Anatolia of Turkey. J. Med. Plants Res. 2010, 4, 1059–1064. [Google Scholar]
- Lòpez, C.; Martin-Sánchez, A.; Fuentes-Zaragoza, E.; Viuda-Martos, M.; Fernández-López, J.; Sendra, E.; Sayas, E.; Pérez-Álvarez, J. Role of oregano (Origanum vulgare) essential oil as a surface fungus inhibitor on fermented sausages: Evaluation of its effect on microbial and physicochemical characteristics. J. Food Prot. 2012, 75, 104–111. [Google Scholar]
- Alwafa, R.A.; Mudalal, S.; Mauriello, G. Origanum syriacum L. (Za’atar), from Raw to Go: A Review. Plants 2021, 10, 1001. [Google Scholar] [CrossRef] [PubMed]
- Fleishe, A.; Sneer, N. Oregano spices and Origanum chemotypes. J. Sci. Food Agric. 1982, 33, 441–446. [Google Scholar] [CrossRef]
- Hazzit, M.; Baaliouamer, A.; Faleiro, M.L.; Miguel, M.C. Composition of the Essential Oils of Thymus and Origanum Species from Algeria and Their Antioxidant and Antimicrobial Activities. J. Agric. Food Chem. 2006, 54, 6314–6321. [Google Scholar] [CrossRef]
- Kordali, S.; Cakir, A.; Ozer, H.; Çakmakçı, R.; Kesdek, M.; Mete, E. Antifungal, phytotoxic and insecticidal properties of essential oil isolated from Turkish Origanumacutidens and its three components, carvacrol, thymol and p-cymene. Bioresour. Technol. 2008, 99, 8788–8795. [Google Scholar] [CrossRef] [PubMed]
- Figueredo, G.; Chalchat, J.C.; Pasquier, B. Studies of Mediterranean oregano populations IX: Chemical composition of essential oils of seven species of oregano of various origins. J. Essent. Oil Res. 2006, 18, 411–415. [Google Scholar] [CrossRef]
- Napoli, E.; Giovino, A.; Carrubba, A.; How Yuen Siong, V.; Rinoldo, C.; Nina, O.; Ruberto, G. Variations of Essential Oil Constituents in Oregano (Origanum vulgare subsp. viridulum (= O. heracleoticum) over Cultivation Cycles. Plants 2020, 9, 1174. [Google Scholar] [CrossRef]
- Marieschi, M.; Torelli, A.; Poli, F.; Sacchetti, G.; Bruni, R. RAPD-based method for the quality control of Mediterranean oregano and its contribution to pharmacognostic techniques. J. Agric. Food Chem. 2009, 57, 1835–1840. [Google Scholar] [CrossRef] [PubMed]
- Marieschi, M.; Torelli, A.; Bianchi, A.; Bruni, R. Detecting Saturejamontana L. and Origanummajorana L. by means of SCAR-PCR in commercial samples of Mediterranean oregano. Food Control 2011, 22, 542–548. [Google Scholar] [CrossRef]
- Black, C.; Haughey, S.A.; Chevallier, O.P.; Galvin-King, P.; Elliott, C.T. A comprehensive strategy to detect the fraudulent adulteration of herbs: The oregano approach. Food Chem. 2016, 210, 551–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bejar, E. Adulteration of Oregano Herb, and Essential Oil of Oregano; Botanical Adulterants Prevention Bulletin: Austin, TX, USA, 2019. [Google Scholar]
- Węglarz, Z.; Kosakowska, O.; Przybył, J.L.; Pióro-Jabrucka, E.; Bączek, K. The Quality of Greek Oregano (O. vulgare L. Subsp. hirtum (Link) Ietswaart) and Common Oregano (O. vulgare L. Subsp. vulgare) Cultivated in the Temperate Climate of Central Europe. Foods 2020, 9, 1671. [Google Scholar] [CrossRef]
- Ekor, M. The growing use of herbal medicines: Issues relating to adverse reactions and challenges in monitoring safety. Front. Pharmacol. 2014, 4, 177. [Google Scholar] [CrossRef] [Green Version]
- Papaioannou, C.; Zeliou, K.; Trigas, P.; Papasotiropoulos, V. High Resolution Melting (HRM) Genotyping in the Genus Origanum: Molecular Identification and Discrimination for Authentication Purposes. Biochem. Genet. 2020, 58, 725–737. [Google Scholar] [CrossRef]
- Yaprak, A.; Mehmet, C. A new natural hybrid of Scutellaria (Lamiaceae) from Turkey. Phytotaxa 2011, 29, 51–55. [Google Scholar]
- Zhao, T.; Tang, H.; Xie, L.; Zheng, Y.; Ma, Z.; Sun, Q. Scutellariabaicalensis Georgi (Lamiaceae): A review of its traditional uses, botany, phytochemistry, pharmacology and toxicology. J. Pharm. Pharmacol. 2019, 71, 1353–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brock, C.; Whitehouse, J.; Tewfik, I.; Towell, T. American Skullcap (Scutellarialateriflora): A randomised, double-blind placebo-controlled crossover study of its effects on mood in healthy volunteers. Phytother. Res. 2014, 28, 692–698. [Google Scholar] [CrossRef]
- Dong, Q.; Chu, F.; Wu, C.; Huo, Q.; Gan, H.; Li, X.; Liu, H. Scutellariabaicalensis Georgi extract protects against alcohol induced acute liver injury in mice and affects the mechanism of ER stress. Mol. Med. Rep. 2016, 13, 3052–3062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Li, P.; Liu, S.; Liu, Q.; Li, Y.; Sun, Y.; He, C.; Xiao, P. Traditional uses, ten-years research progress on phytochemistry and pharmacology, and clinical studies of the genus Scutellaria. J. Ethnopharmacol. 2021, 265, 113198. [Google Scholar] [CrossRef]
- Cheng, L.; Han, M.; Yang, L.; Li, Y.; Sun, Z.; Zhang, T. Changes in the physiological characteristics and baicalin biosynthesis metabolism of Scutellariabaicalensis Georgi under drought stress. Ind. Crops Prod. 2018, 122, 473–482. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, D.; Wang, N.; Li, S.; Tanb, H.-Y.; Feng, Y. Polyphenols of Chinese skullcap roots: From chemical profiles to anticancer effects. R. Soc. Chem. 2019, 9, 25518–25532. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Xiao, H.; Chen, B.; Bian, Z.; Kwan, H.Y. The advantages of using Scutellariabaicalensis and its flavonoids for the management of non-viral hepatocellular carcinoma. J. Funct. Foods 2020, 78, 104389. [Google Scholar] [CrossRef]
- Shang, X.; He, X.; He, X.; Li, M.; Zhang, R.; Fan, P.; Zhang, Q.; Jia, Z. The genus Scutellaria an ethnopharmacological and phytochemical review. J. Ethnopharmacol. 2010, 128, 279–313. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.N.; Downey, F.; Ng, C.K.Y. Comparative analysis of bioactive phytochemicals from Scutellariabaicalensis, Scutellarialateriflora, Scutellariaracemosa, Scutellaria tomentosa and Scutellariawrightii by LC-DAD-MS. Metabolomics 2017, 7, 446–453. [Google Scholar] [CrossRef]
- Mamadalieva, N.Z.; Herrmann, F.; El-Readi, M.Z.; Tahrani, A.; Hamoud, R.; Egamberdieva, D.R.; Azimova, S.S.; Wink, M. Flavonoids in Scutellariaimmaculata and S. ramosissima(Lamiaceae) and their biological activity. J. Pharm. Pharmacol. 2011, 63, 1346–1357. [Google Scholar] [CrossRef]
- He, L.; Sun, F.; Wang, Y.; Zhu, J.; Fang, J.; Zhang, S.; Yu, Q.; Gong, Q.; Ren, B.; Xiang, X.; et al. HMGB1 exacerbates bronchiolitis obliterans syndrome via RAGE/NF-κB/HPSE signaling to enhance latent TGF-β release from ECM. Am. J. Transl. Res. 2016, 8, 1971–1984. [Google Scholar] [PubMed]
- Zhao, Q.; Chen, X.Y.; Martin, C. Scutellariabaicalensis, the golden herb from the garden of Chinese medicinal plants. Sci. Bull. 2016, 61, 1391–1398. [Google Scholar] [CrossRef] [Green Version]
- Georgieva, Y.; Katsarova, M.; Gercheva, K.; Bozov, P.; Dimitrova, S. HPLC analysis of flavonoids from Scutellariaaltissima. Bulg. Chem. Commun. 2019, 51, 119–123. [Google Scholar]
- Makino, T.; Hishida, A.; Goda, Y.; Mizukami, H. Comparison of the major flavonoid content of S. baicalensis, S. lateriflora, and their commercial products. J. Nat. Med. 2008, 62, 294–299. [Google Scholar] [CrossRef]
- Sun, J.; Chen, P. A flow-injection mass spectrometry fingerprinting method for authentication and quality assessment of Scutellarialateriflora-based dietary supplements. Anal. Bioanal. Chem. 2011, 401, 1577–1584. [Google Scholar] [CrossRef]
- Sandasi, M.; Vermaak, I.; Chen, W.; Viljoen, A.M. Skullcap and Germander: Preventing Potential Toxicity through the Application of Hyperspectral Imaging and Multivariate Image Analysis as a Novel Quality Control Method. Planta Med. 2014, 80, 1329–1339. [Google Scholar] [CrossRef] [Green Version]
- Gafner, S. Skullcap Adulteration Laboratory Guidance Document; American Botanical Council: Austin, TX, USA, 2015. [Google Scholar]
- Zhao, F.; Chen, Y.P.; Salmaki, Y.; Drew, B.T.; Wilson, T.C.; Scheen, A.C.; Celep, F.; Bräuchler, C.; Bendiksby, M.; Wang, Q.; et al. An updated tribal classification of Lamiaceae based on plastomephylogenomics. BMC Biol. 2021, 19, 2. [Google Scholar] [CrossRef]
- Jiang, D.; Zhao, Z.; Zhang, T.; Zhong, W.; Liu, C.; Yuan, Q.; Huang, L. Chloroplast Genome Sequence of Scutellariabaicalensis Provides Insight into Intraspecific and Interspecific Chloroplast Genome Diversity in Scutellaria. Genes 2017, 8, 227. [Google Scholar] [CrossRef]
- Gafner, S.; Bergeron, C.; Batcha, L.L.; Reich, J.; Arnason, J.T.; Burdette, J.E.; Pezzuto, J.M.; Angerhofer, C.K. Inhibition of [3H]- LSD binding to 5-HT7 receptors by flavonoids from Scutellarialateriflora. J. Nat. Prod. 2003, 66, 535–537. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, P.R.; Slavoff, S.A.; Grundel, E.; White, K.D.; Mazzola, E.; Koblenz, D.; Rader, J.I. Isolation and characterisation of selected germander diterpenoids from authenticated Teucrium chamaedrys and T. canadense by HPLC, HPLC-MS and NMR. Phytochem. Anal. 2006, 17, 243–250. [Google Scholar] [CrossRef]
- Lin, L.-Z.; Harnly, J.M.; Upton, R. Comparison of the phenolic component profiles of skullcap (Scutellarialateriflora) and germander (Teucrium canadense and T. chamaedrys), a potentially hepatotoxic adulterant. Phytochem. Anal. 2009, 20, 298–306. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, X.; Su, W.; Zhang, G.; Zhou, R. DNA barcodes for discriminating the medicinal plant Scutellariabaicalensis (Lamiaceae) and its adulterants. Biol. Pharm. Bull. 2011, 34, 1198–1203. [Google Scholar] [CrossRef] [Green Version]
- Walker, J.B.; Sytsma, K.J.; Treutlein, J.; Wink, M. Salvia (Lamiaceae) is not monophyletic: Implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. Am. J. Bot. 2004, 91, 1115–1125. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.K.; Wang, Q.; Huang, Y.B. Species diversity and distribution of Salvia (Lamiaceae). Biodivers. Sci. 2015, 23, 3–10. [Google Scholar]
- Drew, B.; González-Gallegos, J.G.; Xiang, C.L.; Kriebel, R.; Drummond, C.P.; Walked, J.B.; Sytsma, K.J. Salvia united: The greatest good for the greatest number. Taxon 2017, 66, 133–145. [Google Scholar] [CrossRef] [Green Version]
- Will, M.; Classen-Bockhoff, R. Time to split Salvia s.l. (Lamiaceae)—New insights from old world Salvia phylogeny. Mol. Phylogenet. Evol. 2017, 109, 33–58. [Google Scholar] [CrossRef]
- Hu, G.X.; Takano, A.; Drew, B.T.; Liu, E.D.; Soltis, D.E.; Soltis, P.S.; Peng, H.; Xiang, C.L. Phylogeny and staminal evolution of Salvia (Lamiaceae, Nepetoideae) in East Asia. Ann. Bot. 2018, 122, 649–668. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, H.X.; Wang, L.; Wang, T.; Yang, R.W.; Wang, X.L.; Zhou, Y.H.; Ding, C.B.; Zhang, L. Potential use of DNA barcoding for the identification of Salvia based on cpDNA and nrDNA sequences. Gene 2013, 528, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Joshi, A.; Kothari, S.L.; Kachhwaha, S.; Purohit, S. Medicinal, nutritional and industrial applications of Salvia species: A revisit. Int. J. Pharm. Sci. Rev. Res. 2017, 43, 27–37. [Google Scholar]
- Gao, C.; Wu, C.; Zhang, Q.; Zhao, X.; Wu, M.; Chen, R.; Zhao, Y.; Li, Z. Characterization of chloroplast genomes from two Salvia medicinal plants and gene transfer among their mitochondrial and chloroplast genomes. Front. Genet. 2020, 11, 574962. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.Q. Salviamiltiorrhiza:Chemicalandpharmacologicalreviewofamedicinalplant. J. Med. Plants Res. 2010, 425, 2813–2820. [Google Scholar]
- Ullah, R.; Nadeem, M.; Khalique, A.; Imran, M.; Mehmood, S.; Javid, A.; Hussain, J. NutritionalandtherapeuticperspectivesofChia (Salviahispanica L.): A review. J. Food Sci. Technol. 2016, 53, 1750–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, N.M.; Yeap, S.K.; Ho, W.Y.; Beh, B.K.; Tan, S.W.; Tan, S.G. Thepromisingfutureofchia, Salviahispanica L. J. Biomed. Biotechnol. 2012, 2012, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Hrnčič, M.K.; Ivanovski, M.; Cör, D.; Knez, Z. ChiaSeeds (Salviahispanica L.): An Overview-Phytochemical Profile, IsolationMethods, and Application. Molecules 2019, 25, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felemban, L.F.; Attar, A.M.A.; Zeid, I.M.A. MedicinalandNutraceuticalBenefitsofChiaSeed(Salviahispanica). J. Pharm. Res. Int. 2021, 32, 15–26. [Google Scholar] [CrossRef]
- Reisfield, A.S. Thebotany of Salviadivinorum (Labiateae). Sida Contrib. Bot. 1993, 15, 349–366. [Google Scholar]
- Clebsch, B. The New Book of Salvias: Sages for Every Garden; Timber Press: Portland, ME, USA, 2008. [Google Scholar]
- Claßen-Bockhoff, R.; Wester, P.; Tweraser, E. Thestaminallevermechanismin Salvia L.(Lamiaceae)—A review. Plant Biol. 2008, 8, 33–41. [Google Scholar]
- Bentham, G. LabiatarumGeneraetSpecies; Ridgeway: London, UK, 1836. [Google Scholar]
- Briquet, J. Salvia. In Die NaturlichenPflanzenfamiliennebst IhrerGattungenund WichtigerenArten; Engler, A., Prantl, K., Eds.; WilhelmEngelmann: Leipzig, Germany, 1897; pp. 183–287. [Google Scholar]
- Epling, C. The Californiasalvias. A review of Salvia, section Audibertia. Ann. Mo. Bot. Gard. 1938, 25, 95–188. [Google Scholar] [CrossRef]
- Epling, C. A RevisionofSalvia, SubgenusCalosphace; VerlagdesRepertoriums: Berlin, Germany, 1939. [Google Scholar]
- Pobedimova, E.G. Salvia. In Floraofthe U.S.S.R; Shishkin, B.K., Ed.; Izdatel’stvoAkademiiNauk: Moscow, Russia, 1954; pp. 154–255. [Google Scholar]
- Wu, C.Y. Salvia. In FloraReipublicaePopularisSinicae; Wu, C.Y., Ed.; SciencePress: Beijing, China, 1977; pp. 70–196. [Google Scholar]
- Murata, G.; Yamazaki, T. Salvia. In FloraofJapan; Iwatsuki, K., Yamazaki, T., Boufford, D., Ohba, H., Eds.; Kodansha: Tokyo, Japan, 1993; pp. 302–307. [Google Scholar]
- Kriebel, R.; Drew, B.; Drummond, C.; González-Gallegos, J.; Celep, F.; Malik, M.; Rose, J.; Xiang, C.; Hu, G.; Walker, J.; et al. Tracking temporal shifts in area, biomes, and pollinators in the radiation of Salvia (sages) across continents: Leveraging anchored hybrid enrichment and targeted sequence data. Am. J. Bot. 2019, 106, 573–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Ma, P.F.; Li, H.T.; Hu, G.X.; Li, D.Z. Comparative plastomic analysis and insights into the phylogeny of Salvia (Lamiaceae). Plant Divers. 2020, 43, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Galvin-King, P.; Haughey, S.A.; Montgomery, H.; Elliott, C.T. The rapid detection of sage adulteration using Fourier Transform Infra-Red (FTIR) Spectroscopy and chemometrics. J. AOAC Int. 2019, 102, 354–362. [Google Scholar] [CrossRef]
- Rodríguez, S.D.; Gagneten, M.; Farroni, A.E.; Percibaldi, N.M.; Buera, M.P. FT-IR and untargeted chemometric analysis for adulterant detection in chia and sesame oils. Food Control 2019, 105, 78–85. [Google Scholar] [CrossRef]
- Tulukcu, E.; Cebi, N.; Sagdic, O. Chemical fingerprinting of seeds of some Salvia species in Turkey by using GC-MS and FTIR. Foods 2019, 8, 118. [Google Scholar] [CrossRef] [Green Version]
- Bielecka, M.; Pencakowski, B.; Stafiniak, M.; Jakubowski, K.; Rahimmalek, M.; Gharibi, S.; Matkowski, A.; Slusarczyk, S. Metabolomics and DNA-Based Authentication of Two Traditional Asian Medicinal and Aromatic Species of Salvia subg. Perovskia. Cells 2021, 10, 112. [Google Scholar] [CrossRef]
- Dormontt, E.E.; van Dijk, K.; Bell, K.L.; Biffin, E.; Breed, M.F.; Byrne, M.; Caddy-Retalic, S.; Encinas-Viso, F.; Nevill, P.G.; Shapcott, A.; et al. Advancing DNA Barcoding and Metabarcoding Applications for Plants Requires Systematic Analysis of Herbarium Collections—An Australian Perspective. Front. Ecol. Evol. 2018, 6, 134. [Google Scholar] [CrossRef] [Green Version]
- Kress, W.J. Plant DNA barcodes: Applications today and in the future. J. Syst. Evol. 2017, 55, 291–307. [Google Scholar] [CrossRef] [Green Version]
- Conserving Genetic Diversity of the Endangered Texas Endemic, Big Red Sage, Salvia pentstemonoides. Available online: https://www.publicgardens.org/events/conserving-genetic-diversity-endangered-texas-endemic-big-red-sage-salvia-pentstemonoides (accessed on 11 December 2021).
- Zhou, X.; Zhang, Z.C.; Huang, Y.B.; Xiao, H.W.; Wu, J.J.; Qi, Z.C.; Wei, Y.K. Conservation Genomics of Wild Red Sage (Salvia miltiorrhiza) and Its Endangered Relatives in China: Population Structure and Interspecific Relationships Revealed from 2b-RAD Data. Front. Genet. 2021, 12, 688323. [Google Scholar] [CrossRef]
- IUCN Red List of Threatened Species. Available online: https://www.iucnredlist.org/species/139603526/139603531 (accessed on 10 December 2021).
- Batovska, J.; Cogan, N.O.; Lynch, S.E.; Blacket, M.J. Using Next-Generation Sequencing for DNA Barcoding: Capturing Allelic Variation in ITS2. G3 Genes Genom. Genet. 2017, 7, 19–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, M.; Szabo, C.; Ford, C.; Yarom, Y.; Croxford, A.E.; Camp, A.; Gooding, P. Replacing Sanger with Next Generation Sequencing to improve coverage and quality of reference DNA barcodes for plants. Sci. Rep. 2017, 7, 46040. [Google Scholar] [CrossRef] [Green Version]
- Hebert, P.D.N.; Braukmann, T.W.A.; Prosser, S.W.J.; Ratnasingham, S.; deWaard, J.R.; Ivanova, N.V.; Janzen, D.H.; Hallwachs, W.; Naik, S.; Sones, J.E.; et al. A Sequel to Sanger: Amplicon sequencing that scales. BMC Genom. 2018, 19, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fopa, F.B.; Brunel, D.; Bérard, A.; Rivoal, J.B.; Gallois, P.; Le-Paslier, M.C.; Bouverat-Bernier, J.P. Quick and efficient approach to develop genomic resources in orphan species: Application in Lavandula angustifolia. PLoS ONE 2020, 15, e0243853. [Google Scholar]
- Lukas, B.; Novak, J. Thecompletechloroplastgenomeof Origanumvulgare L.(Lamiaceae). Gene 2013, 528, 163–169. [Google Scholar] [CrossRef] [PubMed]
- He, S.L.; Yang, Y.; Tian, Y. Characteristic and phylogenetic analyses of chloroplast genome for Mentha haplocalyx (Lamiaceae). Mitochondrial DNA B 2020, 5, 2099–2100. [Google Scholar] [CrossRef] [PubMed]
- Huaizhu, L.; Bai, L.; Bai, J.; Wang, P.; Zhou, C.; Lingling, D.; Jiang, J.; Liu, J.; Wang, Q. The complete chloroplast genome sequence of Thymus mongolicus (Labiatae), a special spice plant. Mitochondrial DNA B 2020, 5, 2597–2598. [Google Scholar] [CrossRef]
- Balaji, R.; Ravichandiran, K.; Tanuja, P.M. The complete chloroplast genome of Ocimumgratissimum from India—A medicinal plant in the Lamiaceae. Mitochondrial DNA B 2021, 6, 948–950. [Google Scholar] [CrossRef]
- Yesuthason, R.S.; Balaji, R.; Tanuja, P.M. The complete chloroplast genome and phylogenetic analysis of Ocimumkilimandscharicum Gurke (Camphor Basil) from India. Mitochondrial DNA B 2021, 6, 2164–2165. [Google Scholar]
- Ma, L. The complete chloroplast genome sequence of the fragrant plant Lavandula angustifolia (Lamiaceae). Mitochondrial DNA B 2018, 3, 135–136. [Google Scholar] [CrossRef] [Green Version]
- Yen, L.T.; Park, J. The complete chloroplast genome sequence of Origanum majorana L. Mitochondrial DNA B 2021, 6, 1224–1225. [Google Scholar] [CrossRef]
- Zhang, C.; Xia, P.; Wu, R.; Mans, D. The complete chloroplast genome of Scutellariameehanioides (Lamiaceae) from Shaanxi Province, China. Mitochondrial DNA B 2021, 6, 1685–1686. [Google Scholar] [CrossRef]
- Little, D.P. A DNA mini-barcode for land plants. Mol. Ecol. Resour. 2014, 14, 437–446. [Google Scholar] [CrossRef]
- Meusnier, I.; Singer, G.A.; Landry, J.F.; Donal A Hickey, D.A.; Hebert, P.D.N.; Hajibabaei, M. A universal DNA mini-barcode for biodiversity analysis. BMC Genom. 2008, 9, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, X.; Su, X.; Chen, X.; Zhao, H.; Bo, C.; Xu, J.; Bai, H.; Ning, K. Biological ingredient analysis of traditional Chinese medicine preparation based on high-throughput sequencing: The story for LiuweiDihuang Wan. Sci. Rep. 2014, 4, 5147. [Google Scholar] [CrossRef]
- Bruno, A.; Sandionigi, A.; Agostinetto, G.; Bernabovi, L.; Frigerio, J.; Casiraghi, M.; Labra, M. Food Tracking Perspective: DNA Metabarcoding to Identify Plant Composition in Complex and Processed Food Products. Genes 2019, 10, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.; Liu, Y.; Wang, X.; Wei, X.; Han, J. DNA Mini-Barcoding: A derived barcoding method for herbal molecular identification. Front. Plant Sci. 2019, 10, 987. [Google Scholar] [CrossRef] [PubMed]
- Raime, K.; Krjutškov, K.; Remm, M. Method for the identification of plant DNA in food using alignment-free analysis of sequencing reads: A case study on Lupin. Front. Plant Sci. 2020, 11, 646. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nazar, N.; Howard, C.; Slater, A.; Sgamma, T. Challenges in Medicinal and Aromatic Plants DNA Barcoding—Lessons from the Lamiaceae. Plants 2022, 11, 137. https://doi.org/10.3390/plants11010137
Nazar N, Howard C, Slater A, Sgamma T. Challenges in Medicinal and Aromatic Plants DNA Barcoding—Lessons from the Lamiaceae. Plants. 2022; 11(1):137. https://doi.org/10.3390/plants11010137
Chicago/Turabian StyleNazar, Nazia, Caroline Howard, Adrian Slater, and Tiziana Sgamma. 2022. "Challenges in Medicinal and Aromatic Plants DNA Barcoding—Lessons from the Lamiaceae" Plants 11, no. 1: 137. https://doi.org/10.3390/plants11010137
APA StyleNazar, N., Howard, C., Slater, A., & Sgamma, T. (2022). Challenges in Medicinal and Aromatic Plants DNA Barcoding—Lessons from the Lamiaceae. Plants, 11(1), 137. https://doi.org/10.3390/plants11010137





