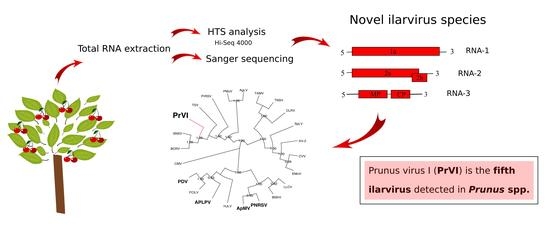
Identification and Sequence Analysis of a Novel Ilarvirus Infecting Sweet Cherry
Abstract
1. Introduction
2. Results
2.1. Analysis of the HTS Data
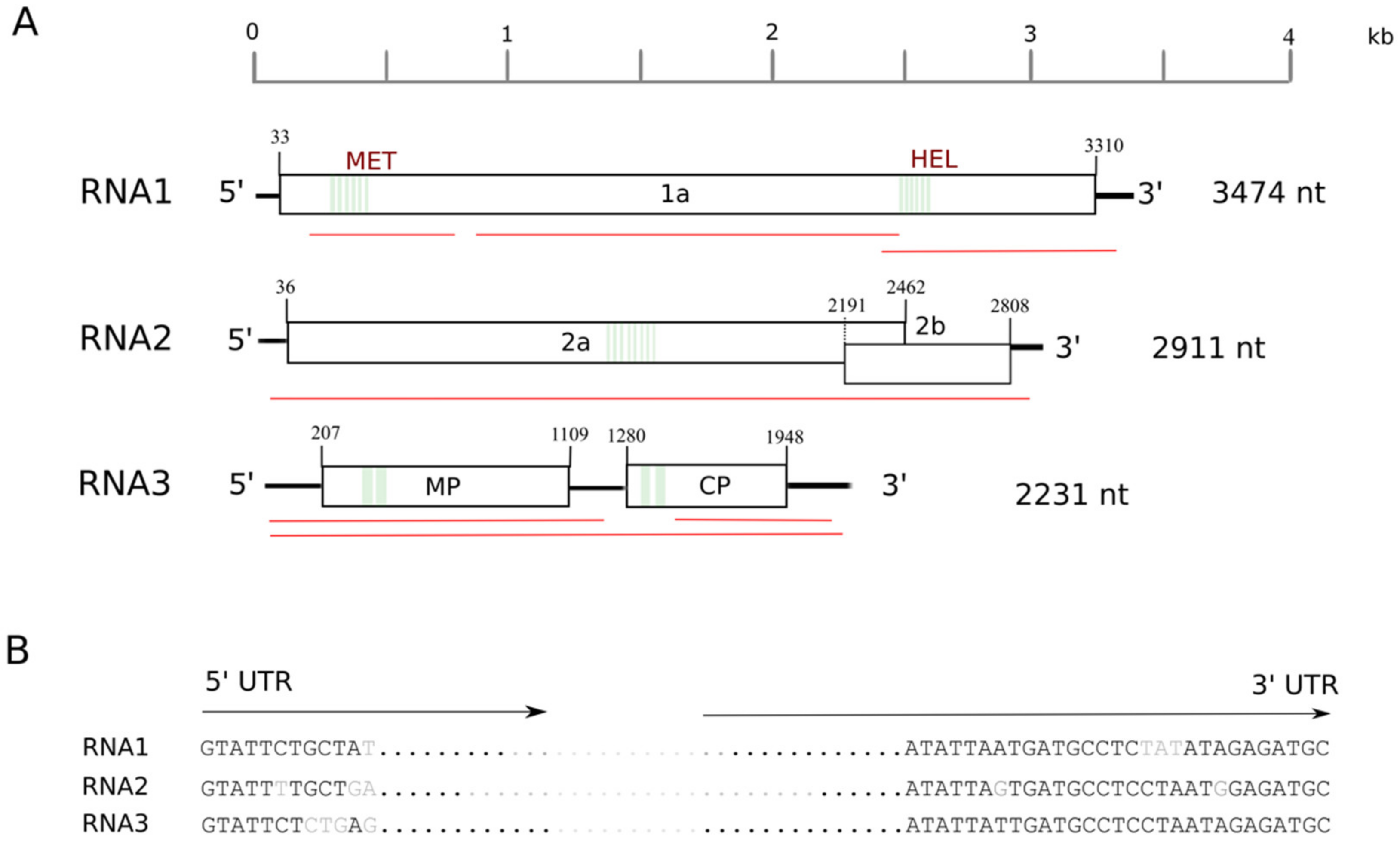
2.2. Genome Structure and Encoded Proteins of the New Ilarvirus
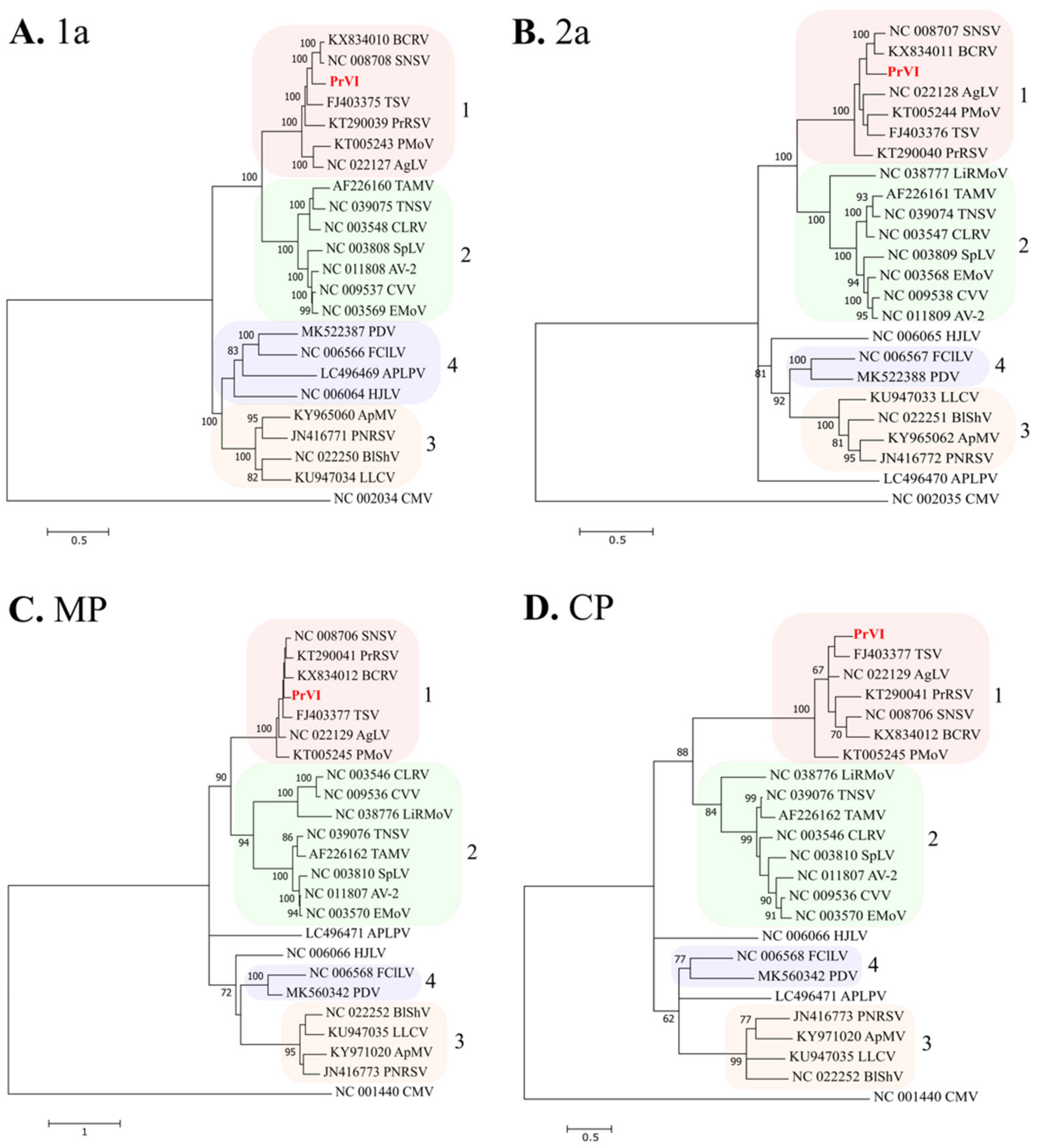
2.3. Nucleotide/Amino Acid Comparisons and Phylogenetic Analysis
2.4. Presence of PrVI in Prunus spp.
3. Discussion
4. Materials and Methods
4.1. High throughput Sequencing and Bioinformatics Analysis
4.2. Sanger Sequencing of the Novel Ilarvirus Genome
4.3. Sequence Analysis and Phylogenetic Reconstructions
4.4. Mini-Scale Survey for the Presence of PrVI in Greek Orchards
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Amari, K.; Sanchez-Pina, M.A.; Myrta, A.; Sanchez-Navarro, J.A. Ilarviruses of Prunus spp.: A continued concern for fruit trees. Phytopathology 2012, 102, 1108–1120. [Google Scholar] [CrossRef] [PubMed]
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Sanchez-Navvaro, J.A.; Scott, S.W. The Molecular Biology of Ilarviruses. Adv. Virus Res. 2013, 87, 139–181. [Google Scholar] [CrossRef] [PubMed]
- Shimura, H.; Masuta, C.; Yoshida, N.; Sueda, K.; Suzuki, M. The 2b protein of Asparagus virus 2 functions as an RNA silencing suppressor against systemic silencing to prove functional synteny with related cucumoviruses. Virology 2013, 442, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Katsiani, A.T.; Li, S.-F.; Zhou, J.; Demertzi, E.; Katis, N.I.; Maliogka, V.I. First report of cherry virus Turkey in sweet cherry in Greece. Plant Dis. 2020, 105, 235. [Google Scholar] [CrossRef]
- Koonin, E.V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 1992, 72, 2197–2206. [Google Scholar] [CrossRef] [PubMed]
- Bujarski, J.; Gallitelli, D.; García-Arenal, F.; Pallás, V.B.; Palukaitis, P.; Reddy, M.K.; Wang, A. ICTV Virus Taxonomy Profile: Bromoviridae. J. Gen. Virol. 2019, 100, 1206–1207. [Google Scholar] [CrossRef] [PubMed]
- Bol, J.F.; van Vloten-Doting, L.; Jaspars, E.M.J. A functional equivalence of top component a RNA and coat protein in the initiation of infection by alfalfa mosaic virus. Virology 1971, 46, 73–85. [Google Scholar] [CrossRef]
- Bol, J.F. Replication of alfamo- and ilarviruses: Role of the coat protein. Annu. Rev. Phytopathol. 2005, 43, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Jaspars, E.M.J. Genome activation in alfamo- and ilarviruses. Arch. Virol. 1999, 144, 843–863. [Google Scholar] [CrossRef] [PubMed]
- Ansel-McKinney, P.; Scott, S.W.; Swanson, M.; Ge, X.; Gehrke, L. A plant viral coat protein RNA binding consensus sequence contains a crucial arginine. EMBO J. 1996, 15, 5077–5084. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, F.; Vilar, M.; Perez-Paya, E.; Pallas, V. The coat protein of prunus necrotic ringspot virus specifically binds to and regulates the conformation of its genomic RNA. Virology 2003, 313, 213–223. [Google Scholar] [CrossRef]
- Fox, A. Reconsidering causal association in plant virology. Plant Pathol. 2020, 69, 956–961. [Google Scholar] [CrossRef]
- Masssart, S.; Candresse, T.; Gil, J.; Lacomme, C.; Predajna, L.; Ravnikar, M.; Reynard, J.S.; Rumbou, A.; Saldarelli, P.; Škorić, D.; et al. A framework for the evaluation of biosecurity, commercial, regulatory, and scientific impacts of plant viruses and viroids identified by NGS technologies. Front. Microbiol. 2017, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2013, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, D.L.; Church, D.M.; Federhen, S.; Lash, A.E.; Madden, T.L.; Pontius, J.U.; Schuler, G.D.; Schriml, L.M.; Sequeira, E.; Tatusova, T.A.; et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003, 31, 28–33. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Perrone, I.; Gribaudo, I. A Rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef]


| Prunus spp. | Collection Date | Geographic County | Total Number of Samples | Number of Samples Positive to PrVI | Accession Numbers |
|---|---|---|---|---|---|
| P. avium | 2009 | Imathia | 12 | 1 | MW591551 |
| 2014 | MW591550 | ||||
| Imathia | 10 | 2 | |||
| MW591552 | |||||
| 2019 | Imathia | 31 | 1 | MW591554 | |
| 2020 | Imathia | 15 | 0 | - 1 | |
| P. persica | 2013 | Arta | 12 | 0 | - |
| 2014 | Imathia | 11 | 0 | - | |
| 2019 | Imathia | 6 | 1 | MW591553 | |
| 2020 | Imathia | 29 | 0 | - | |
| P. domestica | 2020 | Imathia | 12 | 0 | - |
| SUM | 138 | 5 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orfanidou, C.G.; Xing, F.; Zhou, J.; Li, S.; Katis, N.I.; Maliogka, V.I. Identification and Sequence Analysis of a Novel Ilarvirus Infecting Sweet Cherry. Plants 2021, 10, 514. https://doi.org/10.3390/plants10030514
Orfanidou CG, Xing F, Zhou J, Li S, Katis NI, Maliogka VI. Identification and Sequence Analysis of a Novel Ilarvirus Infecting Sweet Cherry. Plants. 2021; 10(3):514. https://doi.org/10.3390/plants10030514
Chicago/Turabian StyleOrfanidou, Chrysoula G., Fei Xing, Jun Zhou, Shifang Li, Nikolaos I. Katis, and Varvara I. Maliogka. 2021. "Identification and Sequence Analysis of a Novel Ilarvirus Infecting Sweet Cherry" Plants 10, no. 3: 514. https://doi.org/10.3390/plants10030514
APA StyleOrfanidou, C. G., Xing, F., Zhou, J., Li, S., Katis, N. I., & Maliogka, V. I. (2021). Identification and Sequence Analysis of a Novel Ilarvirus Infecting Sweet Cherry. Plants, 10(3), 514. https://doi.org/10.3390/plants10030514