SNP-Based Analysis Reveals Authenticity and Genetic Similarity of Russian Indigenous V. vinifera Grape Cultivars
Abstract
:1. Introduction
2. Results and Discussion
2.1. Genetic Characterization of Russian Indigenous Cultivars
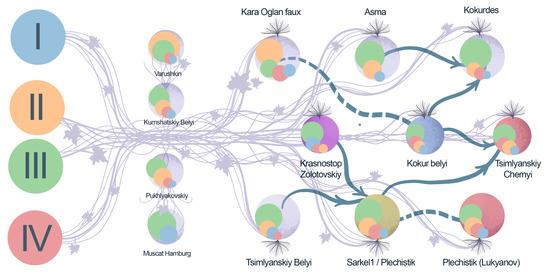
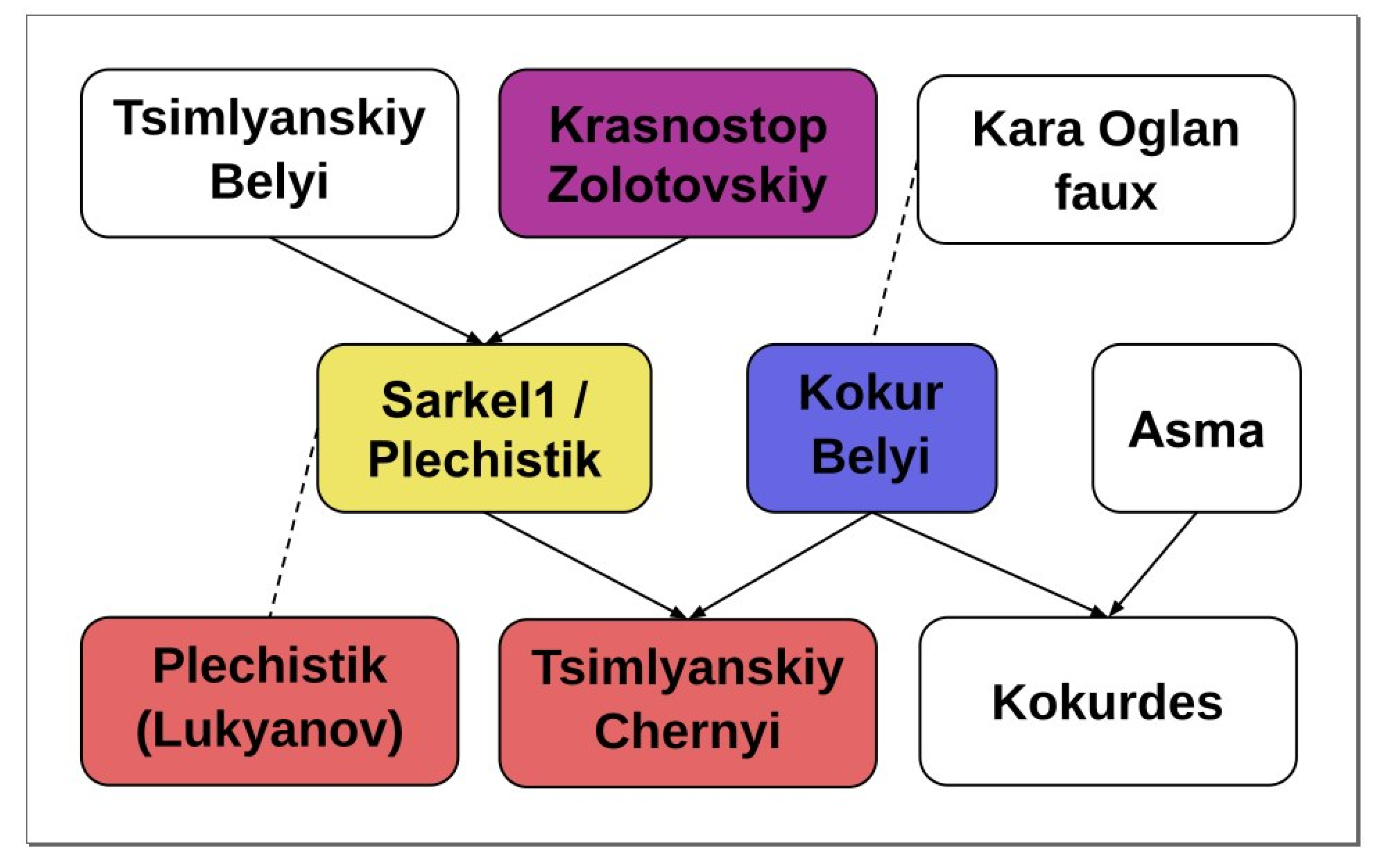
2.2. Parentage Analysis
2.3. Chlorotypes
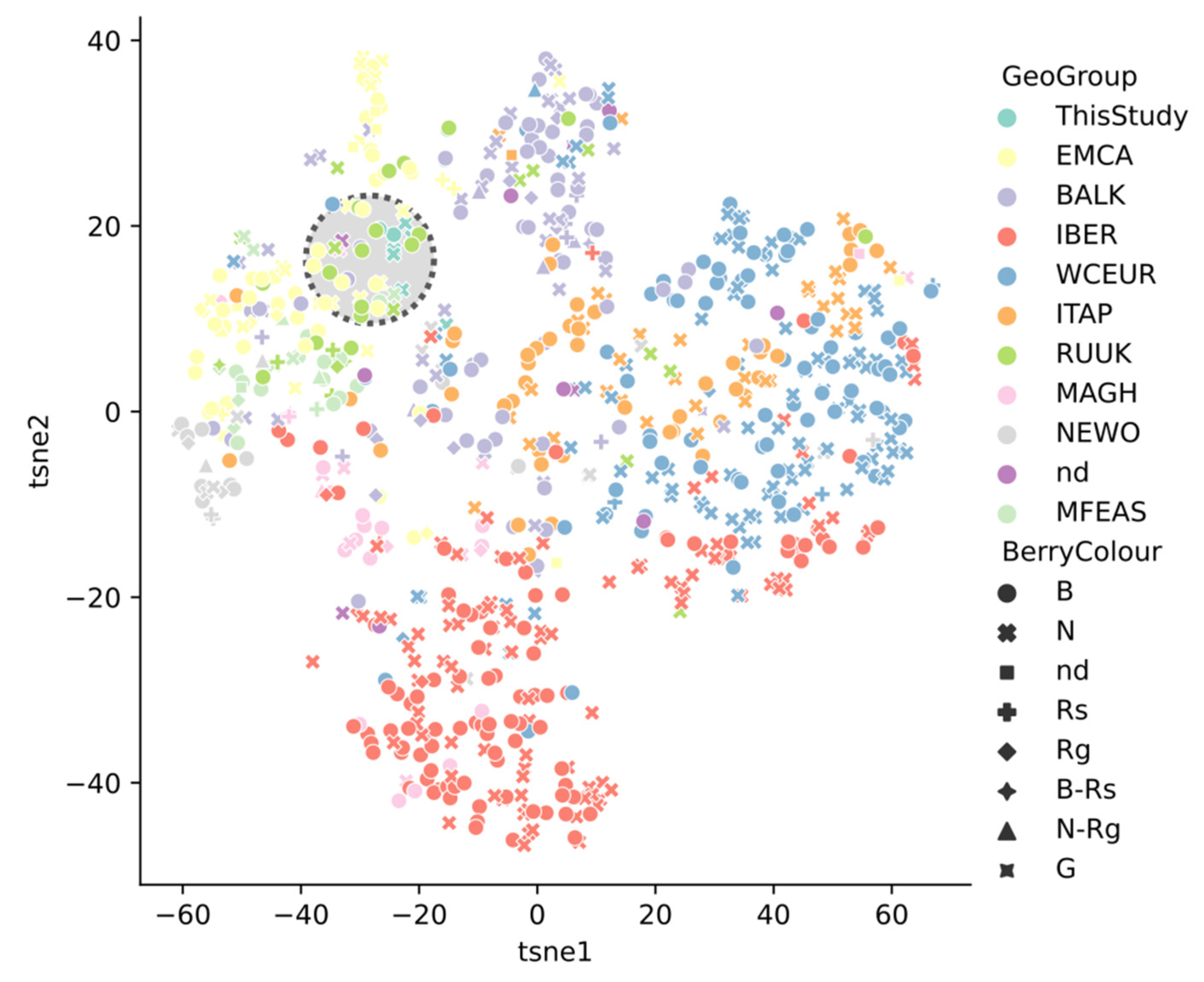
2.4. Grape Varieties Clustering
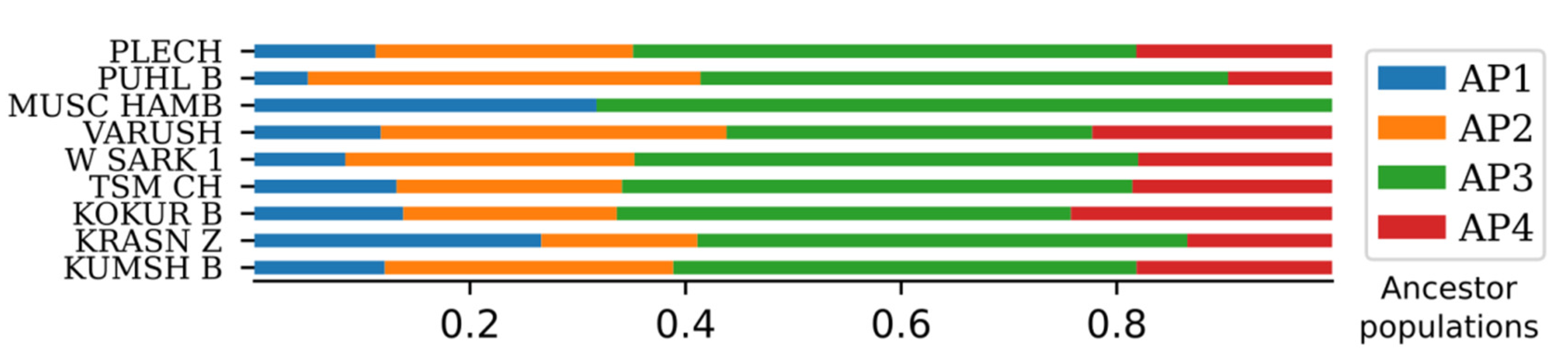
2.5. ADMIXTURE Analysis
3. Materials and Methods
3.1. Plants and Sampling
3.2. DNA Isolation
3.3. DNA Sequencing
3.4. NGS Data Processing and Genotyping
3.5. Population Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Cassagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The French–Italian Public Consortium for Grapevine Genome Characterization The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef]
- Zhou, Y.; Minio, A.; Massonnet, M.; Solares, E.; Lv, Y.; Beridze, T.; Cantu, D.; Gaut, B.S. The population genetics of structural variants in grapevine domestication. Nature 2019, 5, 965–979. [Google Scholar] [CrossRef]
- Bešlić, Z.T.; Korac, N.; Lorenzi, S.; Emanuelli, F.; Grando, M.S. Genetic characterization and relationships of traditional grape cultivars from Serbia. Vitis 2012, 51, 183–189. [Google Scholar]
- Žulj Mihaljević, M.; Maletić, E.; Preiner, D.; Zdunić, G.; Bubola, M.; Zyprian, E.; Pejić, I. Genetic Diversity, Population Structure, and Parentage Analysis of Croatian Grapevine Germplasm. Genes 2020, 11, 737. [Google Scholar] [CrossRef]
- Tomić, L.; Štajner, N.; Jovanović-Cvetković, T.; Cvetković, M.; Javornik, B. Identity and genetic relatedness of Bosnia and Herzegovina grapevine germplasm. Sci. Hortic. 2012, 143, 122–126. [Google Scholar] [CrossRef]
- Sargolzaei, M. Georgian Grapevine Cultivars: Ancient Biodiversity for Future Viticulture. Front. Plant Sci. 2021, 12, 630122. [Google Scholar] [CrossRef] [PubMed]
- This, P.; Boccacci, P.; Borrego, J.; Botta, R.; Costantini, L.; Crespan, M.; Dangl, G.S.; Eisenheld, C.; Ferreira-Monteiro, F. Development of a standard set of microsatellite reference alleles for identification of grape varieties. Theory Appl. Genet. 2004, 109, 1448–1458. [Google Scholar] [CrossRef]
- Laucou, V.; Launay, A.; Bacilieri, R.; Lacombe, T.; Adam-Blondon, A.-F.; Bérard, A.; Chauveau, A.; de Andrés, M.T.; Hausmann, L.; Ibáñez, J.; et al. Extended diversity analysis of cultivated grapevine Vitis vinifera with 10K genome-wide SNPs. PLoS ONE 2018, 13, e0192540. [Google Scholar] [CrossRef]
- Osorio, L. Russia’s Wine Revolution. Available online: https://www.wineintelligence.com/russias-wine-revolution (accessed on 5 October 2021).
- Bittner, S. Whites and Reds. A History of Wine in the Lands of Tsar and Commissar; Oxford University Press: New York, NY, USA, 2021; p. 3. [Google Scholar]
- Deloitte CIS Research Center Moscow. Consumer Activity Dynamics Amid Falling Real Household Incomes. Consumption in Russia. 2019. Available online: https://www2.deloitte.com/content/dam/Deloitte/ru/Documents/research-center/CBT-2019-EN.pdf (accessed on 5 October 2021).
- Volynkun, V.; Roshka, N.; Polulyakh, A. Viticulture and winemaking in Ukraine. In Caucasus and Northern Black Sea Region Ampelography; Julius Kühn Institut: Dresden, Germany, 2012. [Google Scholar]
- Troshin, L.P. Viticulture and winemaking in Russia. In Caucasus and Northern Black Sea Region Ampelography; Julius Kühn Institut: Dresden, Germany, 2012. [Google Scholar]
- Pallas, P.S. Bemerkungen auf Einer Reise in die Südlichen Statthalterschaften des Russischen Reichs in den Jahren 1793 und 1794; Martini: Leipzig, Saxony, 1803. [Google Scholar]
- Köppen, P.I. On Winemaking and Wine Trade in Russia; Typography, K. Kraya: St. Petersburg, Russia, 1832; pp. 118–147. (In Russian) [Google Scholar]
- Odart, A.-P. Ampélographie Universelle ou Traité des Cépages les plus Estimés dans tous les Vignobles de Quelque Renom; Chez Bixio, Editeur de la La Maison Rustique: Paris, France, 1845; pp. 250–256. [Google Scholar]
- Potebnia, A. Kokour Blanc. In Ampélographie; Masson et Cie: Paris, France, 1903; Volume 4, pp. 149–153. [Google Scholar]
- Ballas, M. Essay on Winemaking in Russia (The Caucasus and Crimea). Hist. Stat. Collect. Winemak. 1895–1903, 2, 137–138. [Google Scholar]
- Frolov-Bagreyev, A.M. (Ed.) Ampelography of the USSR; Pishchepromizdat: Moscow, USSR, 1946–1956. (In Russian) [Google Scholar]
- International Organization of Vine and Wine (OIV). Distribution of the World’s Grapevine Varieties. Focus OIV 2017. Available online: http://www.oiv.int/en/oiv-life/the-distribution-of-the-worlds-grapevinevarieties-new-oiv-study-available (accessed on 5 October 2021).
- Ministry of Agriculture of the Russian Federation. Register of grape plantations of the Russian Federation. Available online: http://opendata.mcx.ru/opendata/7708075454-vinogradniki (accessed on 5 October 2021). (In Russian).
- Ilnitskaya, E.T.; Tokmakov, S.V.; Suprun, E.T.; Ganich, V.A.; Naumova, L.G. Genetic similarity of the autochthonous grapevine varieties from don region revealed by SSR-analysis and main leaf ampelographic traits. Agric. Biol. (Sel’skhokhozyaistvennayaBiol.) 2016, 51, 60–67. [Google Scholar] [CrossRef]
- Popov, S. A Brief Outline of Viticulture and Winemaking in Don Host Oblast; Report Made at the Meeting of the Odessa Department of the Imperial Russian Horticultural Society; 22 January and 13 May 1888; (Original in Russian). [Google Scholar]
- Fedosov, D.; Guguchkina, T.; Antonenko, M.; Fedosova, A. Analysis of historical sources on the origins of Korur belyi cultivar. Sci. Work. Skfntssvv 2020, 29, 237–240. [Google Scholar] [CrossRef]
- Lacombe, T.; Boursiquot, J.-M.; Laucou, V.; Di Vecchi-Staraz, M.; Péros, J.-P.; This, P. Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.). Theory Appl. Genet. 2013, 126, 401–414. [Google Scholar] [CrossRef]
- Kara, Z.; Sabir, A.; Eker, Ö. Ancient Grape Vitis vinifera L. cv ‘Ekşi Kara in Anatolia. Selcuk. J. Agric. Food Sci. 2018, 32, 416–423. [Google Scholar] [CrossRef]
- Gorislavets, S.G.; Risovannaya, V.; Bacilieri, R.; Memetova, E.; Laucou, V. Genetic diversity of ancient grape cultivars of the Crimea region. Vitis 2015, 54, 37–41. [Google Scholar]
- Likhovskoy, V.V.; Zarmayev, A.A.; Polulyakh, A.A.; Volynkin, V.A.; Goroslavets, S.M.; Risovannaya, V.I.; Borisenko, M.N.; Sapsay, A.O. Ampelography of Crimean Indigenous and Local Grapes; Magarach Institute: Simferopol, Russia, 2018; p. 115. (In Russian) [Google Scholar]
- Arroyo‐García, R.; Ruiz‐Garcia, L.; Bolling, L.; Ocete, R.; López, M.A.; Arnold, C.; Ergul, A.; Söylemezo Lu, G.; Uzun, H.I.; Cabello, F.; et al. Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Mol. Ecol. 2006, 15, 3707–3714. [Google Scholar] [CrossRef] [Green Version]
- Nikoghosyan, M.; Schmidt, M.; Margaryan, K.; Loeffler-Wirth, H.; Arakelyan, A.; Binder, H. SOMmelier—Intuitive Visualization of the Topology of Grapevine Genome Landscapes Using Artificial Neural Networks. Genes 2020, 11, 817. [Google Scholar] [CrossRef]
- Imazio, S.; Maghradze, D.; De Lorenzis, G.; Bacilieri, R.; Laucou, V.; This, P.; Scienza, A.; Failla, O. From the Cradle of Grapevine Domestication: Molecular Overview and Description of Georgian Grapevine (Vitis Vinifera L.) Germplasm. Tree Genet. Genomes 2013, 9, 641–658. [Google Scholar] [CrossRef]
- Lo Picсolo, S.; Alfonzo, A.; Conigliaro, G.; Moschetti, G. A simple and rapid DNA extraction method from leaves of grapevine suitable for polymerase chain reaction analysis. Afr. J. Biotechnol. 2012, 11, 10305–10309. [Google Scholar] [CrossRef] [Green Version]
- BBMap Short Read Aligner, and Other Bioinformatic Tools. Available online: https://sourceforge.net/projects/bbmap (accessed on 5 October 2021).
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Melo, A.T.O.; Hale, I. ‘apparent’: A simple and flexible R package for accurate SNP-based parentage analysis in the absence of guiding information. BMC Bioinform. 2019, 20, 108. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Odart, A.-P. Ampélographie Universelle ou Traité des Cépages les plus Estimés dans tous les Vignobles de Quelque Renom; Libr. Dusacq: Paris, France, 1849; p. 492. [Google Scholar]
| ID, Cultivar Name | Read Pairs after Trimming, M | Bases after Trimming, B | Bases Mapped, B | Alignment Rate, % | Mean Coverage |
|---|---|---|---|---|---|
| KRASN_Z Krasnostop Zolotovskiy | 56.2 | 15.8 | 14.4 | 90.77 | 29.58 |
| KOKUR_B Kokur Belyi | 66.4 | 18.8 | 17.1 | 91.20 | 35.25 |
| TSM_CH Tsimlyanskiy Chernyi | 75.3 | 21.7 | 5.7 | 26.33 | 11.77 |
| W_SARK_1 Sarkel 1 (Wild grape) | 120.4 | 34.4 | 12.0 | 34.72 | 24.6 |
| VARYUSH Varyushkin | 51.9 | 14.6 | 13.1 | 89.97 | 26.97 |
| SIBIRK Sibirkovyi | 80.8 | 23.2 | 4.7 | 20.22 | 9.65 |
| KUMSH_B Kumshatskiy Belyi | 95.9 | 26.9 | 23.3 | 86.42 | 47.86 |
| PLECH Plechistik (Lukyanov) | 80.1 | 11.0 | 9.7 | 88.45 | 19.94 |
| PUHL_B Pukhlyakovskiy Belyi | 133,3 | 19.2 | 18.3 | 95.01 | 37.57 |
| MUSC_HMB Muscat Hamburg | 129.3 | 18.6 | 17.5 | 94.43 | 36.06 |
| IBS-Distance. | SNP Set Size | |
|---|---|---|
| 10 K | 527 | |
| median | 0.2572 | 0.3705 |
| mean | 0.2573 | 0.3687 |
| min | 0.1568 | 0.2372 |
| max | 0.3171 | 0.4573 |
| stdev | 0.0195 | 0.0272 |
| Offspring | Parent 1 | Parent 2 | Offspring-ExpProgeny Gower Distance, % | Mendellian Errors/Loci | Previously Reported |
|---|---|---|---|---|---|
| Muscat Hamburg | Schiava Grossa ~ Trollinger~Frankenthal * | Muscat d’Alexandrie | 0 | 0/133 | Lacombe et al., 2013 |
| Alphonse Lavallée | Muscat Hamburg | Gros Colman = Dodrelyabi | 0 | 0/112 | Lacombe et al., 2013 |
| Roumi Noir | Muscat Hamburg | Darkaia Noir = Coarna Neagra | 0.85 | 1/118 | Lacombe et al., 2013 |
| Misket Rusenski | Muscat Hamburg | Cardinal | 0 | 0/144 | Lacombe et al., 2013 |
| Italia | Muscat Hamburg | Bicane | 0 | 0/125 | Lacombe et al., 2013 |
| Kokurdes Belyi | Kokur Belyi | Asma | 0.77 | 1/130 | Lacombe et al., 2013 |
| Tsimlyanskiy Chernyi | Kokur Belyi | Sarkel-1/Wild grape ** | 3.17 | 4/126 | Lacombe et al., 2013 |
| Variety 1 | Variety 2 | Mendelian Errors/Loci | Previously Reported |
|---|---|---|---|
| Tsimlyanskiy Chernyi | Kumshatskiy Belyi | 5/155 | - |
| Sarkel 1/Wild Grape | Krasnostop Zolotovskiy | 0/141 | Lacombe et al., 2013 |
| Sarkel 1/Wild Grape | B00EQSE (Starinky) | 0/135 | Lacombe et al., 2013 |
| Sarkel 1/Wild Grape | B00EQSJ (Tzimliansky Belyi) | 1/137 | Lacombe et al., 2013 |
| Kokur Belyi | B00F6O0 (Kara oglan faux) | 3/153 | Lacombe et al., 2013 |
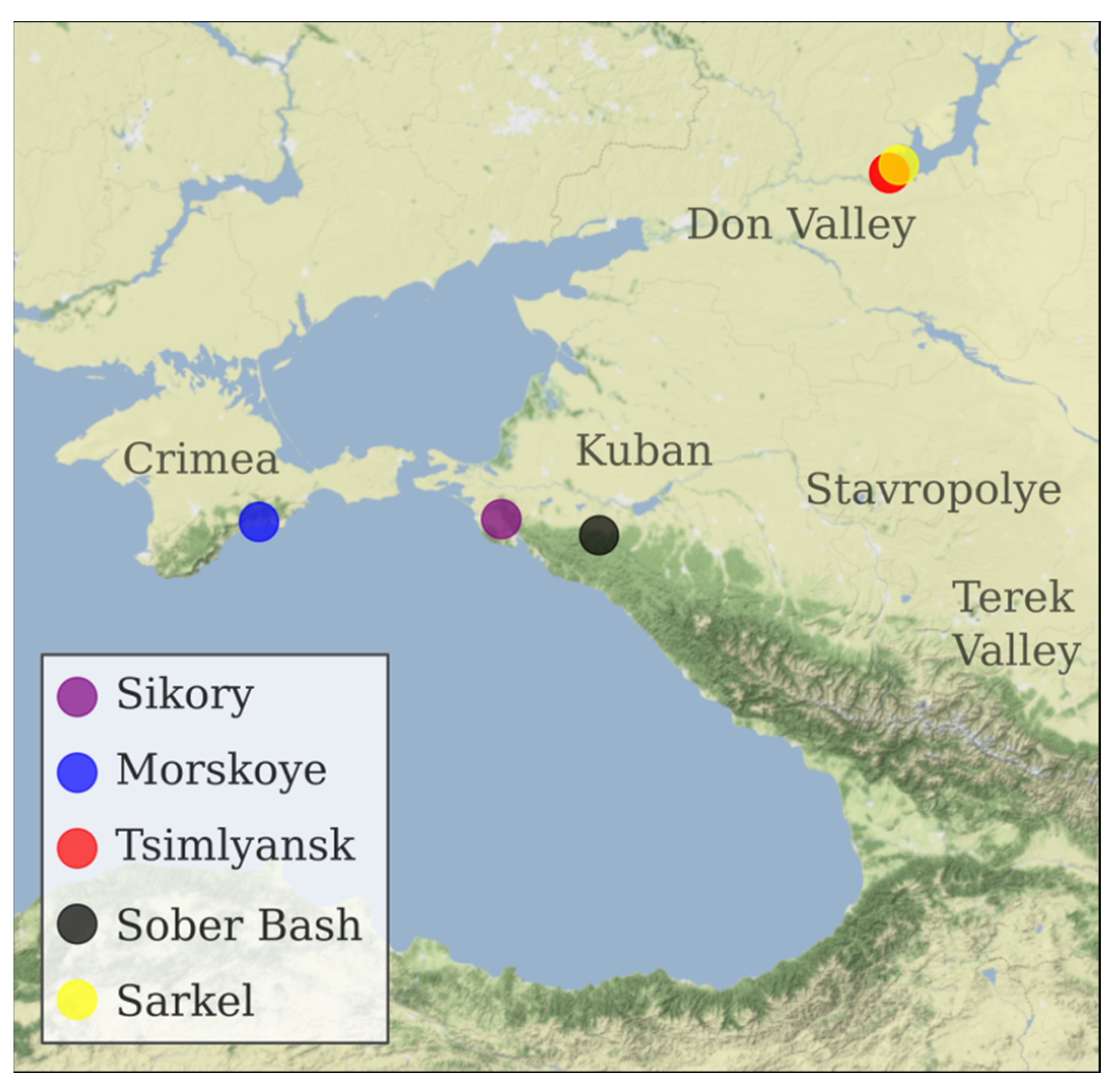
| Specimen ID | Variety | Berry Color | Origin |
|---|---|---|---|
| KRASN_Z | Krasnostop Zolotovskiy | Black | Sikory Estate, Novorossiysk, Krasnodar region (ZGU Kuban.Novorossiysk). Planted in 2014. |
| KOKUR_B | Kokur Belyi | White | Morskoye vineyard of Massandra winery, Sudak district, Republic of Crimea (ZGU Crimea). Planted in 1978. |
| TSM_CH | Tsimlyanskiy Chernyi | Black | Pavel Serikov vineyard, Tsimlyansk, Rostov region (ZGU Don Valley). Planted in 1983. |
| W_SARK_1 | Sarkel 1/Wild Grape | Black | Sarkel village, Tsimlyansk district, Rostov region. |
| VARYUSH | Varyushkin | Black | Sober Bash Vinery, Severskiy district, Krasnodar region (ZGU Kuban. Afips valley). Cuttings from A. Zarechenskiy nursery, Rostov region. Planted in 2013. |
| SIBIRK | Sibirkovyi | White | Pavel Serikov vineyard, Tsimlyansk, Rostov region (ZGU Don Valley). Planted in 1983. |
| KUMSH_B | Kumshatskiy Belyi | White | Pavel Serikov vineyard, Tsimlyansk, Rostov region (ZGU Don Valley). Planted in 1983. |
| PLECH | Plechistik | Black | Nikolay Lukyanov vineyard, Tsimlyansk, Rostov region (ZGU Don Valley). Planted in 1983. |
| PUHL_B | Pukhlyakovskiy Belyi | white | Nikolay Lukyanov vineyard, Tsimlyansk, Rostov region (ZGU Don Valley). Planted in 1983. |
| MUSC_HMB | Muscat Hamburg | black | Nikolay Lukyanov vineyard, Tsimlyansk, Rostov region (ZGU Don Valley). Planted in 1983. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedosov, D.Y.; Korzhenkov, A.A.; Petrova, K.O.; Sapsay, A.O.; Sharko, F.S.; Toshchakov, S.V.; Kolosova, A.A.; Bakhmutova, E.D.; Patrushev, M.V. SNP-Based Analysis Reveals Authenticity and Genetic Similarity of Russian Indigenous V. vinifera Grape Cultivars. Plants 2021, 10, 2696. https://doi.org/10.3390/plants10122696
Fedosov DY, Korzhenkov AA, Petrova KO, Sapsay AO, Sharko FS, Toshchakov SV, Kolosova AA, Bakhmutova ED, Patrushev MV. SNP-Based Analysis Reveals Authenticity and Genetic Similarity of Russian Indigenous V. vinifera Grape Cultivars. Plants. 2021; 10(12):2696. https://doi.org/10.3390/plants10122696
Chicago/Turabian StyleFedosov, Dmitriy Y., Aleksey A. Korzhenkov, Kristina O. Petrova, Alexey O. Sapsay, Fedor S. Sharko, Stepan V. Toshchakov, Adelina A. Kolosova, Elizaveta D. Bakhmutova, and Maxim V. Patrushev. 2021. "SNP-Based Analysis Reveals Authenticity and Genetic Similarity of Russian Indigenous V. vinifera Grape Cultivars" Plants 10, no. 12: 2696. https://doi.org/10.3390/plants10122696
APA StyleFedosov, D. Y., Korzhenkov, A. A., Petrova, K. O., Sapsay, A. O., Sharko, F. S., Toshchakov, S. V., Kolosova, A. A., Bakhmutova, E. D., & Patrushev, M. V. (2021). SNP-Based Analysis Reveals Authenticity and Genetic Similarity of Russian Indigenous V. vinifera Grape Cultivars. Plants, 10(12), 2696. https://doi.org/10.3390/plants10122696