Study of Inter- and Intra-varietal Genetic Variability in Grapevine Cultivars
Abstract
1. Introduction
2. Results and Discussion
2.1. Varietal Identification
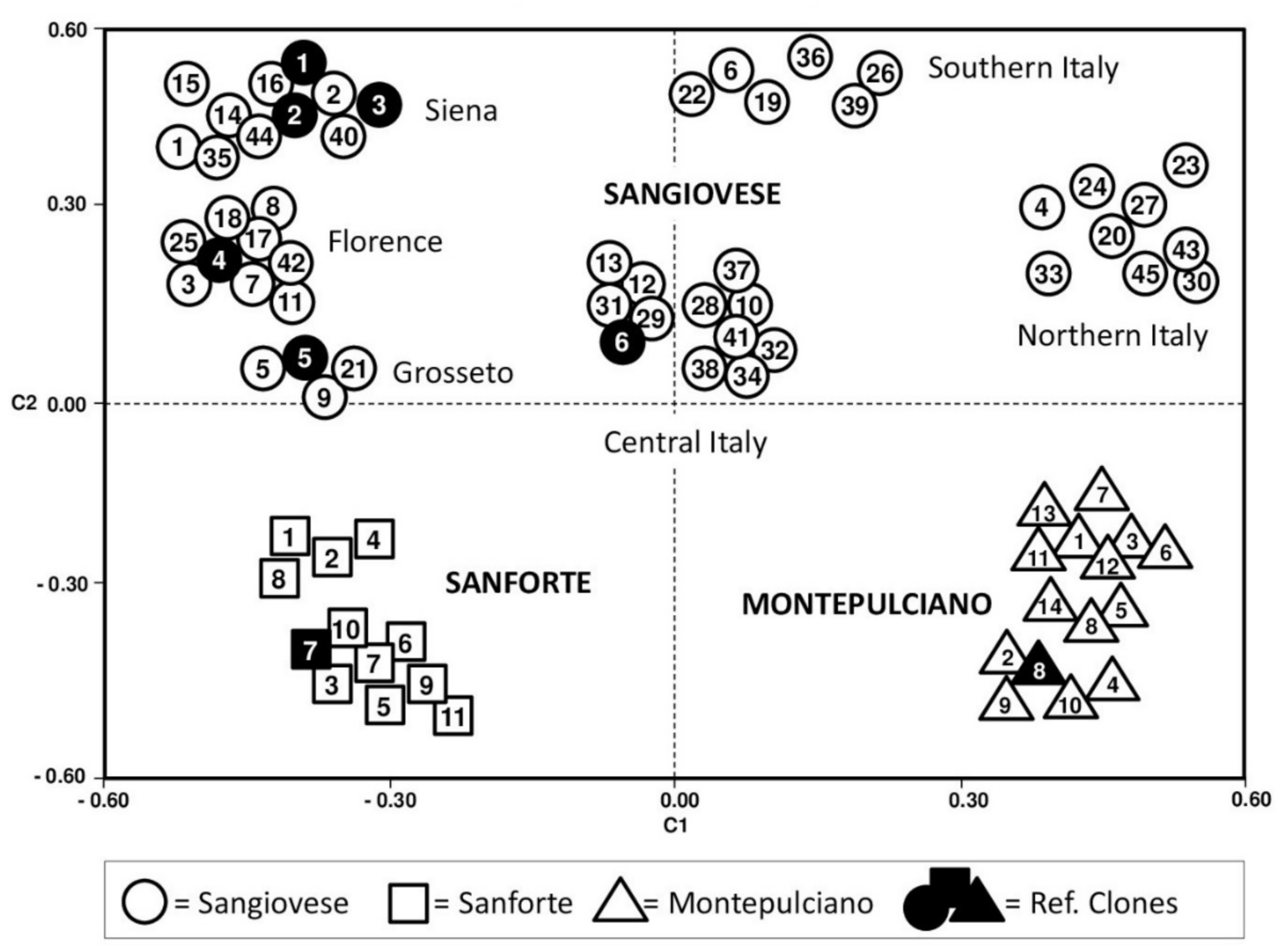
2.2. Intra-Varietal Analyses
2.3. Genetic Diversity
3. Materials and Methods
3.1. Plant Materials
3.2. DNA Extraction and Quantification
3.3. Varietal identification
3.4. Intra-Varietal Analyses
3.5. Genetic Similarity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jackson, R.S. Wine Science: Principles and Applications; Academic Press: Cambridge, MA, USA, 2008; ISBN 0080568742. [Google Scholar]
- Keller, M. The Science of Grapevines. Anatomy and Physiology; Academic Press: Burlington, MA, USA, 2010; ISBN 012374881X. [Google Scholar]
- Galet, P. Dictionnaire Encyclopédique des Cépages; Hachette: Paris, France, 2000; ISBN 2012363318. [Google Scholar]
- De Lorenzis, G.; Imazio, S.; Biagini, B.; Failla, O.; Scienza, A. Pedigree reconstruction of the Italian grapevine Aglianico (Vitis vinifera L.) from Campania. Mol. Biotechnol. 2013, 54, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Filippetti, I.; Intrieri, C.; Centinari, M.; Bucchetti, B.; Pastore, C. Molecular characterization of officially registered Sangiovese clones and of other Sangiovese-like biotypes in Tuscany, Corsica and Emilia-Romagna. Vitis—J. Grapevine Res. 2005, 44, 167–172. [Google Scholar]
- Pelsy, F. Molecular and cellular mechanisms of diversity within grapevine varieties. Heredity 2010, 104, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Vezzulli, S.; Leonardelli, L.; Malossini, U.; Stefanini, M.; Velasco, R.; Moser, C. Pinot blanc and Pinot gris arose as independent somatic mutations of Pinot noir. J. Exp. Bot. 2012, 63, 6359–6369. [Google Scholar] [CrossRef][Green Version]
- Strioto, D.K.; Pepineli, A.C.; Eisele, T.G.; Marinelli, G.C.; Mangolin, C.A.; Cantagali, L.B.; de Oliveira-Collet, S.A.; Maria de Fátima, P.S. Clonal propagation of cv. Italy grapes and the generation of genetic divergence among vineyards. Sci. Hortic. 2019, 244, 263–269. [Google Scholar] [CrossRef]
- Vondras, A.M.; Minio, A.; Blanco-Ulate, B.; Figueroa-Balderas, R.; Penn, M.A.; Zhou, Y.; Seymour, D.; Ye, Z.; Liang, D.; Espinoza, L.K.; et al. The genomic diversification of grapevine clones. BMC Genom. 2019, 20, 972. [Google Scholar] [CrossRef]
- Xie, H.; Konate, M.; Sai, N.; Tesfamicael, K.G.; Cavagnaro, T.; Gilliham, M.; Breen, J.; Metcalfe, A.; Stephen, J.R.; De Bei, R.; et al. Global DNA methylation patterns can play a role in defining terroir in grapevine (Vitis vinifera cv. Shiraz). Front. Plant Sci. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Varela, A.; Ibañez, V.N.; Alonso, R.; Zavallo, D.; Asurmendi, S.; Gomez Talquenca, S.; Marfil, C.F.; Berli, F.J. Vineyard environments influence Malbec grapevine phenotypic traits and DNA methylation patterns in a clone-dependent way. Plant Cell Rep. 2020, 40, 111–125. [Google Scholar] [CrossRef]
- OIV—International Organisation of Vine and Wine. Statistical Report on World Vitiviniculture. Available online: http://www.oiv.int/public/medias/5479/oiv-en-bilan-2017.pdf (accessed on 20 December 2021).
- OIV—International Organisation of Vine and Wine. Distribution of the World’s Grapevine Varieties; OIV: Paris, France, 2017; ISBN 9791091799898. [Google Scholar]
- Italian Catalogue of Grapevine Varieties—Catalogo Nazionale delle Varietà di Vite. Available online: http://catalogoviti.politicheagricole.it/catalogo.php (accessed on 20 December 2021).
- Sangiorgi, B.; Zinzani, G. Romagna sangiovese. Storia e Identità di un Famoso Vino e di un Antico Vitigno; Consorzio Vini di Romagna—Valfrido Editore: Faenza, Italy, 2017; ISBN 8894017877. [Google Scholar]
- Crespan, M.; Calò, A.; Giannetto, S.; Sparacio, A.; Storchi, P.; Costacurta, A. “Sangiovese” and “Garganega” are two key varieties of the Italian grapevine assortment evolution. Vitis—J. Grapevine Res. 2008, 47, 97–104. [Google Scholar] [CrossRef]
- Crespan, M.; Armanni, A.; Da Rold, G.; De Nardi, B.; Gardiman, M.; Migliaro, D. ‘Verdello’, ‘Verdicchio’ and ‘Verduschia’: An example of integrated multidisciplinary study to clarify grapevine cultivar identity. Adv. Hortic. Sci. 2013, 26, 92–99. [Google Scholar] [CrossRef]
- Mercenaro, L.; Usai, G.; Fadda, C.; Nieddu, G.; Caro, A. Cannonau Investigated by Fluorescence, Texture and Colorimetric Analysis. S. Afr. J. Enol. Vitic. 2016, 37, 67–78. [Google Scholar]
- Vignani, R.; Bowers, J.E.; Meredith, C.P. Microsatellite DNA polymorphism analysis of clones of Vitis vinifera “Sangiovese”. Sci. Hortic. 1996, 65, 163–169. [Google Scholar] [CrossRef]
- De Lorenzis, G.; Squadrito, M.; Rossoni, M.; Di Lorenzo, G.S.; Brancadoro, L.; Scienza, A. Study of intra-varietal diversity in biotypes of Aglianico and Muscat of Alexandria (Vitis vinifera L.) cultivars. Aust. J. Grape Wine Res. 2017, 23, 132–142. [Google Scholar] [CrossRef]
- Roach, M.J.; Johnson, D.L.; Bohlmann, J.; van Vuuren, H.J.J.; Jones, S.J.M.; Pretorius, I.S.; Schmidt, S.A.; Borneman, A.R. Population sequencing reveals clonal diversity and ancestral inbreeding in the grapevine cultivar Chardonnay. PLoS Genet. 2018, 14, e1007807. [Google Scholar] [CrossRef] [PubMed]
- Bergamini, C.; Caputo, A.R.; Gasparro, M.; Perniola, R.; Cardone, M.F.; Antonacci, D. Evidences for an alternative genealogy of “Sangiovese”. Mol. Biotechnol. 2013, 53, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Ramazzotti, S.; Filippetti, I.; Intrieri, C. Expression of genes associated with anthocyanin synthesis in red-purplish, pink, pinkish-green and green grape berries from mutated “Sangiovese” biotypes; A case study. Vitis—J. Grapevine Res. 2008, 47, 147–151. [Google Scholar]
- Di Rovasenda, G. Saggio di un’Ampelografia Universale; Tipografia Subalpina di Stefano Marino: Torino, Italy, 1877. [Google Scholar]
- Molon, G. Ampelografia; Ed. Hoepli: Milan, Italy, 1906; Volume II. [Google Scholar]
- Breviglieri, N.; Casini, E. Sangiovese. In I Principali Vitigni da Vino Coltivati in Italia; Ministero dell’Agricoltura e delle Foreste: Rome, Italy, 1965. [Google Scholar]
- Silvestroni, O.; Intrieri, C. Ampelometric assessment of clonal variability in the Sangiovese winegrape cultivar. In Proceedings of the Acta of the International Symposium on Clonal Selection; American Society for Enology and Viticulture, Portland, OR, USA, 20–21 June 1995; pp. 137–142. [Google Scholar]
- Calò, A.; Costacurta, A.; Egger, E.; Storchi, P.; Crespan, M.; Milani, N.; Sensi, E.; Carraro, R. Caratterizzazione molecolare, ampelografia e ampelometrica di 30 accessioni di Vitis vinifera L. riferibili al Sangiovese. In Proceedings of the Acta of International Symposium “Il Sangiovese”, Florence, Italy, 15–17 February 2000. [Google Scholar]
- Vignani, R.; Scali, M.; Masi, E.; Cresti, M. Genomic variability in Vitis vinifera L. “Sangiovese” assessed by microsatellite and non-radioactive AFLP test. Electron. J. Biotechnol. 2002, 5, 1–11. [Google Scholar] [CrossRef][Green Version]
- Falchini, D. Trattato di Agricoltura (Sec. XVIII); Republished by Edizioni all’Insegna del Giglio: Florence, Italy, 1994. [Google Scholar]
- Vitis International Variety Catalogue VIVC. Available online: https://www.vivc.de/ (accessed on 20 December 2021).
- Zombardo, A.; Storchi, P.; Valentini, P.; Ciofini, A.; Migliaro, D.; Crespan, M. Recovery, molecular characterization, and ampelographic assessment of marginal grapevine germplasm from southern umbria (Central Italy). Plants 2021, 10, 1539. [Google Scholar] [CrossRef]
- Di Vecchi Staraz, M.; Bandinelli, R.; Boselli, M.; This, P.; Boursiquot, J.M.; Laucou, V.; Lacombe, T.; Varès, D. Genetic structuring and parentage analysis for evolutionary studies in grapevine: Kin group and origin of the cultivar sangiovese revealed. J. Am. Soc. Hortic. Sci. 2007, 132, 514–524. [Google Scholar] [CrossRef]
- Odart, A.P. Ampélographie Universelle, ou Traité des Cépages les Plus Estimés dans tous les Vignobles de Quelque Renom; Dusacq Librairie Agricole de la Maison Rustique: Paris, France, 1845. [Google Scholar]
- Galet, P.; Morton, L.T.; Adams, L.D. A Practical Ampelography: Grapevine Identification; Cornell University Press: Ithaca, NY, USA, 1979; ISBN 9780801412400. [Google Scholar]
- Duchêne, E.; Legras, J.L.; Karst, F.; Merdinoglu, D.; Claudel, P.; Jaegli, N.; Pelsy, F. Variation of linalool and geraniol content within two pairs of aromatic and non-aromatic grapevine clones. Aust. J. Grape Wine Res. 2009, 15, 120–130. [Google Scholar] [CrossRef]
- Tedesco, G.; Scienca, A.; Villa, P.; Saino, N.; Magenes, S.; Ettori, C.; Gianazza, E. A chemotaxonomic investigation on Vitis vinifera 1. Within-cultivar population analysis. Vitis—J. Grapevine Res. 2015, 30, 71. [Google Scholar]
- Meneghetti, S.; Costacurta, A.; Morreale, G.; Calò, A. Study of intra-varietal genetic variability in grapevine cultivars by PCR-derived molecular markers and correlations with the geographic origins. Mol. Biotechnol. 2012, 50, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Sefc, K.M.; Lefort, F.; Grando, M.S.; Scott, K.D.; Steinkellner, H.; Thomas, M.R. Microsatellite Markers for Grapevine: A State of the Art. Mol. Biol. Biotechnol. Grapevine 2001, 433–463. [Google Scholar] [CrossRef]
- Crespo-Martínez, S.; Mayor, B.; Oneka, O.; Loidi, M.; Villa-Llop, A.; Marín, D.; Miranda, C.; Santesteban, L.G.; Urrestarazu, J. Recovery of ancient grapevine plant material in peri-urban areas. A case of success in Pamplona (Spain) leading to the recovery of cv. Berués. Sci. Hortic. 2022, 293, 110675. [Google Scholar] [CrossRef]
- This, P.; Jung, A.; Boccacci, P.; Borrego, J.; Botta, R.; Costantini, L.; Crespan, M.; Dangl, G.S.; Eisenheld, C.; Ferreira-Monteiro, F.; et al. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor. Appl. Genet. 2004, 109, 1448–1458. [Google Scholar] [CrossRef]
- Blaich, R.; Konradi, J.; Rühl, E.; Forneck, A. Assessing Genetic Variation among Pinot noir (Vitis vinifera L.) Clones with AFLP Markers. Am. J. Enol. Vitic. 2007, 58, 526–529. [Google Scholar]
- Böhm, A.; Zyprian, E. RAPD marker in grapevine (Vitis spp.) similar to plant retrotransposons. Plant Cell Rep. 1998, 17, 415–421. [Google Scholar] [CrossRef]
- Cipriani, G.; Marrazzo, M.T.; Di Gaspero, G.; Pfeiffer, A.; Morgante, M.; Testolin, R. A set of microsatellite markers with long core repeat optimized for grape (Vitis spp.) genotyping. BMC Plant Biol. 2008, 8, 127. [Google Scholar] [CrossRef]
- Cretazzo, E.; Meneghetti, S.; De Andrés, M.T.; Gaforio, L.; Frare, E.; Cifre, J. Clone differentiation and varietal identification by means of SSR, AFLP, SAMPL and M-AFLP in order to assess the clonal selection of grapevine: The case study of Manto Negro, Callet and Moll, autochthonous cultivars of Majorca. Ann. Appl. Biol. 2010, 157, 213–227. [Google Scholar] [CrossRef]
- D’Onofrio, C.; De Lorenzis, G.; Giordani, T.; Natali, L.; Scalabrelli, G.; Cavallini, A. Retrotransposon-based molecular markers in grapevine species and cultivars identification and phylogenetic analysis. Acta Hortic. 2009, 827, 45–52. [Google Scholar] [CrossRef]
- D’Onofrio, C.; Tumino, G.; Gardiman, M.; Crespan, M.; Bignami, C.; de Palma, L.; Barbagallo, M.G.; Muganu, M.; Morcia, C.; Novello, V.; et al. Parentage Atlas of Italian Grapevine Varieties as Inferred From SNP Genotyping. Front. Plant Sci. 2021, 11, 1–16. [Google Scholar] [CrossRef]
- Imazio, S.; Labra, M.; Grassi, F.; Winfield, M.; Bardini, M.; Scienza, A. Molecular tools for clone identification: The case of the grapevine cultivar “Traminer”. Plant Breed. 2002, 121, 531–535. [Google Scholar] [CrossRef]
- Meneghetti, S.; Costacurta, A.; Frare, E.; Da Rold, G.; Migliaro, D.; Morreale, G.; Crespan, M.; Sotés, V.; Calò, A. Clones identification and genetic characterization of garnacha grapevine by means of different PCR-derived marker systems. Mol. Biotechnol. 2011, 48, 244–254. [Google Scholar] [CrossRef]
- Owens, C.L. SNP detection and genotyping in Vitis. Acta Hortic. 2003, 603, 139–140. [Google Scholar] [CrossRef]
- Regner, F.; Wiedeck, E.; Stadlbauer, A. Differentiation and identification of White Riesling clones by genetic markers. Vitis 2000, 39, 103–107. [Google Scholar]
- Sensi, E.; Vignani, R.; Rohde, W.; Biricolti, S. Characterization of genetic biodiversity with Vitis vinifera L. Sangiovese and Colorino genotypes by AFLP and ISTR DNA marker technology. Vitis 1996, 35, 183–188. [Google Scholar]
- Istituto Nazionale di Statistica; ISTAT. Superfici vitate—Principali vitigni—Aggiornamento ISTAT 2010. Available online: http://www.inumeridelvino.it/tag/2010 (accessed on 20 December 2021).
- Meneghetti, S.; Calò, A.; Bavaresco, L. A strategy to investigate the intravarietal genetic variability in Vitis vinifera L. for clones and biotypes identification and to correlate molecular profiles with morphological traits or geographic origins. Mol. Biotechnol. 2012, 52, 68–81. [Google Scholar] [CrossRef]
- Crespan, M. The parentage of Muscat of Hamburg. Vitis 2003, 42, 193–197. [Google Scholar] [CrossRef]
- Bowers, J.E.; Dangl, G.S.; Meredith, C.P. Development and Characterization of Additional Microsatellite DNA Markers for Grape. Am. J. Enol. Vitic. 1999, 50, 243–246. [Google Scholar]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy. The Principles and Practice of Numerical Classification; WH Freeman and Company: San Francisco, CA, USA, 1973; ISBN 0-7167-0697-0. [Google Scholar]
- Castagna, A.; Csepregi, K.; Neugart, S.; Zipoli, G.; Večeřová, K.; Jakab, G.; Jug, T.; Llorens, L.; Martínez-Abaigar, J.; Martínez-Lüscher, J. Environmental plasticity of Pinot noir grapevine leaves: A trans-European study of morphological and biochemical changes along a 1500-km latitudinal climatic gradient. Plant. Cell Environ. 2017, 40, 2790–2805. [Google Scholar] [CrossRef]
- Duncan, E.J.; Gluckman, P.D.; Dearden, P.K. Epigenetics, plasticity, and evolution: How do we link epigenetic change to phenotype? J. Exp. Zool. Part B Mol. Dev. Evol. 2014, 322, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Lovisolo, C.; Perrone, I.; Carra, A.; Ferrandino, A.; Flexas, J.; Medrano, H.; Schubert, A. Drought-induced changes in development and function of grapevine (Vitis spp.) organs and in their hydraulic and non-hydraulic interactions at the whole-plant level: A physiological and molecular update. Funct. Plant Biol. 2010, 37, 98–116. [Google Scholar] [CrossRef]
- Dal Santo, S.; Tornielli, G.B.; Zenoni, S.; Fasoli, M.; Farina, L.; Anesi, A.; Guzzo, F.; Delledonne, M.; Pezzotti, M. The plasticity of the grapevine berry transcriptome. Genome Biol. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Venios, X.; Korkas, E.; Nisiotou, A.; Banilas, G. Grapevine responses to heat stress and global warming. Plants 2020, 9, 1754. [Google Scholar] [CrossRef]
- Haig, D. The (Dual) Origin of Epigenetics. Cold Spring Harb. Symp. Quant. Biol. 2004, LXIX, 67–70. [Google Scholar] [CrossRef]
- Fabres, P.J.; Collins, C.; Cavagnaro, T.R.; López, C.M.R. A concise review on multi-omics data integration for terroir analysis in Vitis vinifera. Front. Plant Sci. 2017, 8, 1–8. [Google Scholar] [CrossRef]
- Meneghetti, S.; Poljuha, D.; Frare, E.; Costacurta, A.; Morreale, G.; Bavaresco, L.; Calò, A. Inter- and intra-varietal genetic variability in Malvasia cultivars. Mol. Biotechnol. 2012, 50, 189–199. [Google Scholar] [CrossRef]
- Albertini, E.; Porceddu, A.; Marconi, G.; Barcaccia, G.; Pallottini, L.; Falcinelli, M. Microsatellite-AFLP for genetic mapping of complex polyploids. Genome 2003, 46, 824–832. [Google Scholar] [CrossRef][Green Version]



| Sample No. | Sample Name | Geographic Origin Region (Province) | No. | Sample Name | Geographic Origin Region (Province) |
|---|---|---|---|---|---|
| 1 | Sangiovese Chianti 1 | Tuscany (AR) | 40 | Sangiovese Emiliano | Emilia Romagna (RE) |
| 2 | Morellino di Toscana | Tuscany (FI) | 41 | Sangiovese Marchigiano | Marche (AP) |
| 3 | Morellino Precoce | Tuscany (FI) | 42 | Sanvicetro Marche | Marche (MC) |
| 4 | Sangiovese Brunello 1 | Tuscany (FI) | 43 | Sangiovese Perugino | Umbria (PG) |
| 5 | Sangiovese Chianti 2 | Tuscany (FI) | 44 | Brunello Abruzzese | Abruzzo (CH) |
| 6 | Sangiovese Chianti 3 | Tuscany (FI) | 45 | Sanvicetro Abruzzese | Abruzzo (CH) |
| 7 | Sangiovese Classico | Tuscany (FI) | 46 | Brunellino Rosso | Latium (FR) |
| 8 | Sangiovese Toscano 1 | Tuscany (FI) | 47 | Morellino Romano | Latium (FR) |
| 9 | Sanvicetro Fiorentino | Tuscany (FI) | 48 | Sanvicetro Laziale 1 | Latium (FR) |
| 10 | Sanvicetro Morellino | Tuscany (FI) | 49 | Brunello Montalcino 2 | Latium (RI) |
| 11 | Morellino di Scansano | Tuscany (GR) | 50 | Brunello di Frascati | Latium (RM) |
| 12 | Sangiovese Rosso | Tuscany (GR) | 51 | Sanvicetro Laziale 2 | Latium (VT) |
| 13 | Sangiovese Scansano 1 | Tuscany (GR) | 52 | Sangiovese Campano | Campania (BN) |
| 14 | Sangiovese Scansano 2 | Tuscany (GR) | 53 | Sangiovese Brunello 2 | Campania (SA) |
| 15 | Sangiovese Prugnolo | Tuscany (PT) | 54 | Morellino di Potenza | Basilicata (PZ) |
| 16 | Brunello di Montalcino | Tuscany (SI) | 55 | Sangiovese Calabro | Calabria (CS) |
| 17 | Brunello Montalcino 1 | Tuscany (SI) | 56 | Sangiovese Nostrano | Apulia (BA) |
| 18 | Montalcino Prugnolo | Tuscany (SI) | 57 | Sangiovese VCR 4 | Tuscany (FI) |
| 19 | Sangiovese Gaiole | Tuscany (SI) | 58 | Sangiovese VCR 108 | Tuscany (GR) |
| 20 | Sangiovese Grosso | Tuscany (SI) | 59 | Sangiovese BF 10 | Tuscany (SI) |
| 21 | Sangiovese Lungo | Tuscany (SI) | 60 | Sangiovese MI-BF 50 | Tuscany (SI) |
| 22 | Sangiovese Montepulciano | Tuscany (SI) | 61 | Sangiovese VCR 103 | Tuscany (SI) |
| 23 | Sangiovese Morellino | Tuscany (SI) | 62 | Sangiovese VCR 16 | Emilia Romagna (FC) |
| 24 | Sangiovese Senese | Tuscany (SI) | 63 | Sanvicetro F59 P6 C2 | Tuscany (FI) |
| 25 | Sangiovese Toscano 2 | Tuscany (SI) | 64 | Montepulciano di Toscana | Tuscany (AR) |
| 26 | Sanvicetro di Toscana | Tuscany (SI) | 65 | Montepulciano | Tuscany (FI) |
| 27 | Sanvicetro Toscano | Tuscany (SI) | 66 | Montepulciano Grosso | Tuscany (FI) |
| 28 | Sangiovese Rovigo | Veneto (RO) | 67 | Montepulciano Sangiovese | Tuscany (FI) |
| 29 | Sangiovese Susegana | Veneto (TV) | 68 | Montepulciano Toscano | Tuscany (SI) |
| 30 | Sangiovese Trevigiano | Veneto (TV) | 69 | Montepulciano Marche 1 | Marche (AP) |
| 31 | Morellino Berico | Veneto (VI) | 70 | Montepulciano Marche 2 | Marche (MC) |
| 32 | Sanvicetro Piccolo | Veneto (VI) | 71 | Montepulciano Abruzzese | Abruzzo (CH) |
| 33 | Morellino Gentile | Veneto (VR) | 72 | Montepulciano Classico 1 | Abruzzo (CH) |
| 34 | Sangiovese Trentino | Trentino Alto Adige (TN) | 73 | Montepulciano Rotondo | Abruzzo (CH) |
| 35 | Sangiovese Carsico | Friuli Venezia Giulia (TS) | 74 | Montepulciano Classico 2 | Abruzzo (PE) |
| 36 | Sangiovese Friulano | Friuli Venezia Giulia (UD) | 75 | Montepulciano Glabro | Abruzzo (PE) |
| 37 | Sanvicetro Emiliano | Emilia Romagna (FC) | 76 | Montepulciano Nobile | Abruzzo (TE) |
| 38 | Sangiovese Giove | Emilia Romagna (MO) | 77 | Montepulciano Teramano | Abruzzo (TE) |
| 39 | Sangiovese Montalcino | Emilia Romagna (PR) | 78 | Montepulciano Rauscedo 7 | Abruzzo (TE) |
| SSR locus | Sangiovese | Sanforte | Montepulciano | |||
|---|---|---|---|---|---|---|
| VVS2 | 133 | 133 | 133 | 151 | 133 | 145 |
| VVMD5 | 226 | 236 | 226 | 236 | 226 | 228 |
| VVMD7 | 239 | 263 | 239 | 249 | 249 | 249 |
| VVMD27 | 179 | 185 | 179 | 185 | 189 | 194 |
| VVMD28 | 237 | 247 | 239 | 239 | 237 | 247 |
| VrZAG62 | 193 | 195 | 195 | 201 | 189 | 199 |
| VrZAG79 | 242 | 258 | 250 | 258 | 250 | 250 |
| VMC6E1/ISV2 | 143 | 165 | 141 | 143 | 141 | 169 |
| VMC6F1/ISV3 | 139 | 139 | 131 | 139 | 139 | 145 |
| VMC6G1/ISV4 | 177 | 197 | 177 | 187 | 187 | 191 |
| VMCNG4b9 | 158 | 168 | 150 | 168 | 168 | 176 |
| Sample Name (Sample No.) | Genotype ID by SSR | Assigned Code (Origin) | Geographic Cluster |
|---|---|---|---|
| Sangiovese BF 10 (59) | Sangiovese | C-01 (SI) | Tuscany |
| Sangiovese MI-BF 50 (60) | Sangiovese | C-02 (SI) | Tuscany |
| Sangiovese VCR 103 (61) | Sangiovese | C-03 (SI) | Tuscany |
| Sangiovese VCR 4 (57) | Sangiovese | C-04 (FI) | Tuscany |
| Sangiovese VCR 108 (58) | Sangiovese | C-05 (GR) | Tuscany |
| Sangiovese Montepulciano (22) | Sangiovese | SG-01(SI) | Tuscany |
| Sangiovese Gaiole (19) | Sangiovese | SG-02 (SI) | Tuscany |
| Sangiovese Lungo (21) | Sangiovese | SG-03 (SI) | Tuscany |
| Sangiovese Scansano 1 (13) | Sangiovese | SG-05(GR) | Tuscany |
| Morellino di Toscana (2) | Sangiovese | SG-07 (FI) | Tuscany |
| Sanvicetro Morellino (10) | Sangiovese | SG-08 (FI) | Tuscany |
| Sangiovese Scansano 2 (14) | Sangiovese | SG-09 (GR) | Tuscany |
| Sangiovese Classico (7) | Sangiovese | SG-11 (FI) | Tuscany |
| Montalcino Prugnolo (18) | Sangiovese | SG-14 (SI) | Tuscany |
| Brunello di Montalcino (16) | Sangiovese | SG-15 (SI) | Tuscany |
| Sangiovese Toscano 2 (25) | Sangiovese | SG-16 (SI) | Tuscany |
| Sangiovese Chianti 2 (5) | Sangiovese | SG-17 (FI) | Tuscany |
| Sangiovese Chianti 3 (6) | Sangiovese | SG-18 (FI) | Tuscany |
| Morellino di Scansano (11) | Sangiovese | SG-21 (GR) | Tuscany |
| Morellino Precoce (3) | Sangiovese | SG-25 (FI) | Tuscany |
| Sangiovese Chianti 1 (1) | Sangiovese | SG-29 (AR) | Tuscany |
| Sanvicetro Toscano (27) | Sangiovese | SG-35 (SI) | Tuscany |
| Sangiovese Prugnolo (15) | Sangiovese | SG-39 (PT) | Tuscany |
| Brunello Montalcino 1 (17) | Sangiovese | SG-40 (SI) | Tuscany |
| Sangiovese Brunello 1 (4) | Sangiovese | SG-42 (FI) | Tuscany |
| Sangiovese Grosso (20) | Sangiovese | SG-44 (SI) | Tuscany |
| Sangiovese Carsico (35) | Sangiovese | SG-04 (TS) | Northern Italy |
| Sangiovese Rovigo (28) | Sangiovese | SG-20 (RO) | Northern Italy |
| Morellino Gentile (33) | Sangiovese | SG-23 (VR) | Northern Italy |
| Sangiovese Friulano (36) | Sangiovese | SG-24 (UD) | Northern Italy |
| Sangiovese Trevigiano (30) | Sangiovese | SG-27 (TV) | Northern Italy |
| Sangiovese Susegana (29) | Sangiovese | SG-30 (TV) | Northern Italy |
| Sangiovese Trentino (34) | Sangiovese | SG-33 (TN) | Northern Italy |
| Sanvicetro Piccolo (32) | Sangiovese | SG-43 (VI) | Northern Italy |
| Morellino Berico (31) | Sangiovese | SG-45 (VI) | Northern Italy |
| Sangiovese VCR 16 (62) | Sangiovese | C-06 (FC) | Central Italy |
| Sangiovese Emiliano (40) | Sangiovese | SG-12 (RE) | Central Italy |
| Sangiovese Montalcino (39) | Sangiovese | SG-13 (PR) | Central Italy |
| Brunellino Rosso (46) | Sangiovese | SG-28 (FR) | Central Italy |
| Sanvicetro Laziale 2 (51) | Sangiovese | SG-32 (VT) | Central Italy |
| Brunello Montalcino 2 (49) | Sangiovese | SG-34 (RI) | Central Italy |
| Brunello di Frascati (50) | Sangiovese | SG-37 (RM) | Central Italy |
| Morellino Romano (47) | Sangiovese | SG-38 (FR) | Central Italy |
| Brunello Abruzzese (44) | Sangiovese | SG-10 (CH) | Central Italy |
| Sangiovese Marchigiano (41) | Sangiovese | SG-31 (AP) | Central Italy |
| Sangiovese Perugino (43) | Sangiovese | SG-41 (PG) | Central Italy |
| Sangiovese Brunello 2 (53) | Sangiovese | SG-06 (SA) | Southern Italy |
| Morellino di Potenza (54) | Sangiovese | SG-19 (PZ) | Southern Italy |
| Sangiovese Calabro (55) | Sangiovese | SG-22 (CS) | Southern Italy |
| Sangiovese Campano (52) | Sangiovese | SG-26 (BN) | Southern Italy |
| Sangiovese Nostrano (56) | Sangiovese | SG-36 (BA) | Southern Italy |
| Sanvicetro F59 P6 C2 (63) | Sanforte | C-07 (FI) | Tuscany |
| Sanvicetro di Toscana (26) | Sanforte | SF-01 (SI) | Tuscany |
| Sangiovese Toscano 1 (8) | Sanforte | SF-02 (FI) | Tuscany |
| Sanvicetro Fiorentino (9) | Sanforte | SF-03 (FI) | Tuscany |
| Sangiovese Rosso (12) | Sanforte | SF-04 (GR) | Tuscany |
| Sangiovese Morellino (23) | Sanforte | SF-08 (SI) | Tuscany |
| Sangiovese Senese (24) | Sanforte | SF-10 (SI) | Tuscany |
| Sanvicetro Abruzzese (45) | Sanforte | SF-06 (CH) | Central Italy |
| Sanvicetro Marche (42) | Sanforte | SF-05 (MC) | Central Italy |
| Sangiovese Giove (38) | Sanforte | SF-07 (MO) | Central Italy |
| Sanvicetro Emiliano (37) | Sanforte | SF-09 (FC) | Central Italy |
| Sanvicetro Laziale 1 (48) | Sanforte | SF-11 (FR) | Central Italy |
| Montepulciano Rauscedo 7 (78) | Montepulciano | C-08 (TE) | Abruzzo |
| Montepulciano Glabro (75) | Montepulciano | M-02 (PE) | Abruzzo |
| Montepulciano Classico 1 (72) | Montepulciano | M-04 (CH) | Abruzzo |
| Montepulciano Abruzzese (71) | Montepulciano | M-05 (CH) | Abruzzo |
| Montepulciano Classico 2 (74) | Montepulciano | M-08 (PE) | Abruzzo |
| Montepulciano Teramano (77) | Montepulciano | M-09 (TE) | Abruzzo |
| Montepulciano Nobile (76) | Montepulciano | M-10 (TE) | Abruzzo |
| Montepulciano Rotondo (73) | Montepulciano | M-14 (CH) | Abruzzo |
| Montepulciano Marche 1 (69) | Montepulciano | M-07 (AP) | Marche |
| Montepulciano Marche 2 (70) | Montepulciano | M-12 (MC) | Marche |
| Montepulciano Grosso (66) | Montepulciano | M-01 (FI) | Tuscany |
| Montepulciano di Toscana (64) | Montepulciano | M-03 (AR) | Tuscany |
| Montepulciano Toscano (68) | Montepulciano | M-06 (SI) | Tuscany |
| Montepulciano (65) | Montepulciano | M-11 (FI) | Tuscany |
| Montepulciano Sangiovese (67) | Montepulciano | M-13 (FI) | Tuscany |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zombardo, A.; Meneghetti, S.; Morreale, G.; Calò, A.; Costacurta, A.; Storchi, P. Study of Inter- and Intra-varietal Genetic Variability in Grapevine Cultivars. Plants 2022, 11, 397. https://doi.org/10.3390/plants11030397
Zombardo A, Meneghetti S, Morreale G, Calò A, Costacurta A, Storchi P. Study of Inter- and Intra-varietal Genetic Variability in Grapevine Cultivars. Plants. 2022; 11(3):397. https://doi.org/10.3390/plants11030397
Chicago/Turabian StyleZombardo, Alessandra, Stefano Meneghetti, Giacomo Morreale, Antonio Calò, Angelo Costacurta, and Paolo Storchi. 2022. "Study of Inter- and Intra-varietal Genetic Variability in Grapevine Cultivars" Plants 11, no. 3: 397. https://doi.org/10.3390/plants11030397
APA StyleZombardo, A., Meneghetti, S., Morreale, G., Calò, A., Costacurta, A., & Storchi, P. (2022). Study of Inter- and Intra-varietal Genetic Variability in Grapevine Cultivars. Plants, 11(3), 397. https://doi.org/10.3390/plants11030397







