Abstract
In this review, we explore how ecological concepts may help assist with applying microbial biocontrol agents to oomycete pathogens. Oomycetes cause a variety of agricultural diseases, including potato late blight, apple replant diseases, and downy mildew of grapevine, which also can lead to significant economic damage in their respective crops. The use of microbial biocontrol agents is increasingly gaining interest due to pressure from governments and society to reduce chemical plant protection products. The success of a biocontrol agent is dependent on many ecological processes, including the establishment on the host, persistence in the environment, and expression of traits that may be dependent on the microbiome. This review examines recent literature and trends in research that incorporate ecological aspects, especially microbiome, host, and environmental interactions, into biological control development and applications. We explore ecological factors that may influence microbial biocontrol agents’ efficacy and discuss key research avenues forward.
1. Introduction
Host-associated microbiota are integral parts of organismal biology and have a strong impact on host physiology [1,2]. The microbiome, which includes the microbiota, their collective genetic material, as well as their collective functions and properties [3,4], represents a novel resource for disease prevention and treatment in plant and animal health [5,6,7]. It is increasingly examined in a context of ecological and evolutionary concepts [8,9]. The inclusion of a comprehensive microbiome perspective is revolutionizing a number of fields, including medicine, agriculture, forestry, and evolutionary biology [10,11,12,13,14]. Within human medicine, recent advancements in microbiome research are changing the understanding of certain diseases and methods to treat them [15,16]. In order to establish and maintain a healthy microbiome, human disease treatments now consider ecological principles regarding microbial colonization and community establishment to treat diseases without antibiotics [17,18,19]. New efforts are driving the use of naturally occurring microbiota to manipulate the microbiome for the maintenance of weight, mental health, and reduction of certain diseases [20,21,22,23]. Similar to the development of perspectives within human health fields, microbiota-based treatments are increasingly considered as viable sources for disease control in plants and may help to decrease synthetic inputs [6,11,24]. This review explores challenges associated with oomycete diseases in plants, their relationship to the microbiome, and possible avenues of using the microbiome to help suppress diseases.
The oomycetes are a mostly pathogenic class, of which 60% of known species are plant pathogens [25], but members also parasitize fish, crustaceans, insects, humans, and other vertebrates [26,27,28]. The lineage has independently evolved pathogenicity in plants and other organisms several times, and it includes members with diverse lifestyles ranging from asymptomatic endophytes to obligate biotrophic pathogens [25,29]. Members of the oomycetes cause some of the most problematic plant diseases, including Phytophthora infestans (potato late blight), Plasmopara viticola (grapevine downy mildew), and Pythium spp. (cause of damping off and root rot), and can severely reduce agricultural output [30,31]. Due to global climate change, oomycetes may become more problematic as their geographic distribution shifts polewards [32].
Oomycetes are more closely related to brown algae and diatoms than true fungi, and they differ physiologically from fungi [26,33]. Even though they have superficial similarities, the structural differences between fungal and oomycete pathogens mean that many fungicides do not work against them [33]. Cellulose and -glucans primarily compose the cell walls of oomycetes, distinguishing them from the chitin-rich cell walls of fungi [34]. Oomycete lineages, however, differ from each other in the compositions of carbohydrates, including N-acetylglucosamine (the monomer of chitin) within their cell wall [35]. For example, N-acetylglucosamine is absent in the cell wall of some oomycete lineages such as Phytophthora, but it composes nearly 10% in the cell wall of other oomycetes, such as those within the Aphanomyces genus [35]. Oomycetes are also heterogeneous in their ability to biosynthesize sterols [36,37]. A number of oomycetes, such as P. infestans, are sterol auxotrophs, making fungicides targeting sterol production ineffective against them [36,38].
Due to the unique challenges related to the physiology of oomycetes, it is desirable to obtain an arsenal of control strategies, including microbial biocontrol agents (MBCAs), which can provide alternatives to chemical-based plant protection products. MBCAs can work against oomycete pathogens through indirect mechanisms, such as plant priming and induced resistance, or direct mechanisms through chemical defenses, such as enzymatic, volatile organic compounds, antibiotic production, competition, and hyperparasitism, as well as combinations of the above modes [39,40,41,42,43]. Microbiome manipulations (see Box 1) through the addition of synthetic communities may offer treatment options for oomycete pathogens, potentially reducing a reliance on chemical fungicides. However, a number of barriers exist to the adoption of MBCAs for widespread commercial use, and there has been limited success of MBCAs in the field compared to experimental setups in laboratory environments [44,45]. While theory gives some information regarding circumstances under which biocontrol agents might work [46], their applicability in the field depends on our understanding of ecological dynamics in natural populations. The evaluation of MBCAs’ ecological characteristics, including their distribution, culturability, phenotypic plasticity, effectiveness, and persistence in natural conditions, can help dictate when and where they are appropriate to use [47,48,49,50]. This review aims to identify and discuss relevant ecological dynamics that may influence the effectiveness of microbiome-based treatments against pathogenic oomycetes. Additionally, we identify studies that consider the microbiome in the context of the biocontrol of oomycete plant pathogens and for which ecological processes are relevant.
2. Ecological Processes and Their Effects on the Introduction and Success of Microbial Biocontrol Agents
2.1. Community Assembly Processes and Context
Community assembly processes, which are series of events that affect species’ identity and abundance within a community, may influence the establishment and expression of MBCAs on plants. The habitats for microorganisms on the above-ground part of the plant, the phyllosphere, or below-ground part of the plant, the rhizosphere, have different characteristics, ecological constraints, and abiotic and biotic factors, such as UV, wind, rain, insect vectors, and human activities, that can affect how microbial communities establish [51,52]. Plant phyllosphere and rhizosphere community structure, including diversity, richness, and the presence of particular species, has a role in determining the community invasibility or the outcome of the introduction of additional species [53,54,55,56]. Recent work focused on utilizing the plant microbiome to combat plant diseases has largely consisted of engineering particular microbiomes through inorganic or organic amendments or direct applications of microorganisms. Soil amendments consisting of inorganic material (i.e., lime and vermiculite) or organic material (i.e., biochar, organic waste, and manure) are added to encourage the establishment of a favorable microbiome through the construction of a suitable microhabitat [57,58,59]. Such organic amendments that help recruit and/or retain disease suppressive or growth promoting microorganisms can be considered prebiotics [58,59,60]. The direct applications of derived or engineered communities to shift the microbiome can be considered more like a probiotic treatment [60,61,62,63]. The success of pre- and probiotics are driven by the ecological dynamics of the system. The contribution of both deterministic (niche driven) and stochastic (neutral) processes to community assembly [64] may help explain the success of biocontrol organisms from various sources in different situations. In this section, the following ecological concepts and their potential effect on the establishment and success of MBCAs are explored: priority effects, genotype- and environment-dependent expression, species interactions, competition, and niche use.
Box 1. Glossary of terms used to describe ecological processes or concepts that may aid in the understanding of or affect biocontrol microorganism establishment.
| Colonization/Establishment: The ability of an organism to start a new population in a novel or uncolonized habitat. |
| Community invasibility: The ability for a species or population to establish itself and grow in a habitat occupied by a community of different organisms. |
| Dispersal: The movement of organisms among and within habitats and habitat patches. |
| Ecological drift: The change of species abundances within a community over time due to stochastic processes. |
| Environmental filters: The selection of a subset of species that can withstand the abiotic conditions of an environment and determines a community. |
| Metacommunity: An interconnected community of multiple species that is spread across different habitat patches. |
| Metapopulation: An interconnected population of one species that is spread across different habitat patches. |
| Neutral theory in ecology: Simplest model possible for biogeographical patterns of diversity in which all species are first assumed to be equally capable of competing. Neutral drift and random dispersal are main factors that shape community assembly. |
| Niche theory: The distribution of species due to their n-dimensional hypervolume or the space corresponding to species’ requirements (habitat, environmental conditions, food, etc.). This framework is contrary to the neutral theory and suggests certain species are better suited for particular environments and community assembly processes are deterministic. |
| Patch dynamics: The interconnectedness of populations and communities across mosaic landscapes composed of heterogeneous patches or habitats. The distribution, size, and interconnectedness of patches have an effect of biodiversity maintenance. |
| Phenotypic plasticity: The variation of phenotypic traits observed due to differences in environmental conditions. Variation in phenotypic plasticity gives rise to genotype by environment (G×E) interactions. |
| Priority effects: The occurrence of earlier arrivals to a habitat having an advantage for establishment compared to later arrivals during community assembly. |
| Reaction norm: Pattern of a genotype’s trait expression across different environments. The slope of the reaction norm gives information regarding how responsive a trait is to environmental variation. |
| Source-sink dynamics: An aspect of patch dynamics where some high quality patches represent sources of species or populations while other poor-quality patches represent sinks. |
2.1.1. Priority Effects
Priority effects appear to influence the plant microbiome [65,66,67], so that early arrivals are better able to colonize and establish themselves on plants. Priority effects are borne out in the results of soil swap experiments, which show that the soil influences the bacteria within the phyllosphere [68]. The importance of priority effects and the early establishment of the microbiome indicate that seed or soil treatments may be a useful tool for microbiome manipulations [69,70]. The plant microbiome has been described as having a source to sink gradient from the ground up [71], meaning that soil serves as a reservoir for microbial colonization of above-ground plant parts. These source–sink dynamics of plant microbiota may also correspond to the role that soil can play in disease suppression or induction [72,73,74,75]. Priority effects can influence the composition of the community by resulting in alternative stable states or different final community outcomes, ultimately affecting function or epidemiological outcome [67,76]. By capitalizing on the influence of priority effects, microbial inputs at an early plant stage may have large effects in community assembly and on disease protection [69]. Community assembly can also be governed by multiple processes, including host genotype [77,78] and its interaction with priority effects [66]. Such assembly dynamics can, in turn, influence the community invasibility and dictate the establishment and success of the biocontrol agent [79].
2.1.2. Community Dynamics, Resource Competition, and Niche Space
Once a community is established, some species may act as keystone or hub species and have an outsized effect on the microbiome. The oomycete pathogen Albugo laibachii has such a disproportionate influence on community assembly in infected plants because it suppresses other microbes on the plants [80]. In addition to changing the diversity of the microbial community, an infection can also alter the composition of the microbial community. An oomycete infection of Phytophthora parasitica in tomatoes increased the relative abundance of members of the bacterial phylum Bacteroidetes [81], and their growth was thought to be promoted due to increased pectin degradation caused by the infection. Aside from an infection shifting community dynamics, a loss of community members can also affect disease suppression. The loss of relatively rare bacterial species has been shown to reduce antifungal volatile organic compounds (VOCs)—secondary bacterial metabolites that can move through air or water and can inhibit the growth of pathogenic organisms, suggesting that microbial community interactions may drive volatile production and affect the biocontrol activity of the community [82]. Such interactions among community members and pathogens that modulate gene expression have implications for biocontrol success. For example, the biological control agent Pseudomonas fluorescens In5 increases expression of secondary metabolites in the presence of the plant fungal pathogen Rhizoctonia solani, but not the oomycete pathogen Pythium aphanidermatum [83]. Instead, P. aphanidermatum reduced important lipoproteins that help P. fluorescens In5 control pathogens [41]. Therefore, the presence of a particular microbial community member or co-infection can cause shifts in microbial communities and, in turn, may affect the establishment and persistence of or protection conferred by a biocontrol agent.
Resource competition within bacterial communities can also affect pathogen invasion success [43,84], suggesting that community architecture can have a large impact on plant–pathogen interactions. Plants hosting microbial communities with greater niche overlap showed stronger pathogen resistance, which corresponds to a decrease in susceptibility to pathogen invasions in plants harboring more diverse microbial communities [53,54,55,56]. By utilizing concepts about niche theory, exploring the niche breadth of candidate microbial agents can also contribute to a better understanding of the success of their introduction. For example, the lifestyle of the oomycete—whether it regularly colonizes the rhizosphere or phyllosphere of the plant—may influence the strategy for plant protection. A subset of phyllosphere and root microbiota is transferable between the two habitats while others are niche-specific [85], so the design of treatments should consider the target area of inoculation (i.e., seed or foliar application) and the success of colonization. An analysis of genomes from plant-associated bacteria shows that their genomes include more carbohydrate metabolism functions [86], possibly indicating that bacteria derived from plants are better suited to establish and persist on a plant than non-plant associated microorganisms.
Available niche space and the ability to outperform pre-established microbes may influence success of the introduction of biocontrol agents. Therefore, the persistence and stability of an introduced biocontrol agent should be monitored. Even if an ideal community to suppress disease is achieved, a main ecological question remains regarding how it is maintained. Within ecological communities, complex interactions arise that make it difficult to control subsequent processes. For example, intraspecific oomycete and bacterial–oomycete interactions can facilitate infections through virulence signaling or the facilitation of opportunistic infections [81,87,88]. Factors such as plant age, resident microbial community, and other environmental aspects can shape the microbiome of a plant [71,89] and may potentially affect the establishment, persistence, and influence of transplanted microbes. However, even if some introduced microorganisms do not persist, they can have lasting effects on plants. For example, after 100 days, willows inoculated with different starting rhizosphere soil showed differences in growth according to their initial microbial inoculation despite the convergence of the microbiome [90]. Therefore, the context of these intricate interactions could change the disease outcome.
2.1.3. Phenotypic Plasticity
Phenotypic plasticity can cause context-specific outcomes, which generally arise from the effect of genotype by environment (G×E) interactions on a trait’s expression. When a trait exhibits plasticity, its expression changes depending on environmental conditions. The microbiome can be considered an “extended genotype” due to the genetic repertoire included in the plants own genome as well as its microbiota [91], and it can also exhibit plasticity in the expression of its traits [89]. Once an MBCA is established, it is also subject to G×E interactions, which can affect its effectiveness to control plant diseases [92]. In terms of environmental effects, certain functions or characteristics of MBCAs can be turned on or off depending on various environmental factors [48,49]. Genotypic effects from the plant may influence microbiome traits involved in microbial recognition or triggering an immune response, which could in turn, affect the MBCAs’ disease suppressiveness [92]. Together, G×E effects and the phenotypic plasticity that can influence MBCAs’ efficacies make it imperative to test a variety of environmental and genotype combinations before deploying MBCA solutions. Environmental-dependent expression of biocontrol agents can give rise to non-parallel reaction norms (Figure 1), meaning that G×E interactions are apparent, and potentially, some G×E combinations are much more effective than others. Context dependency may also be due to the influence of the environment serving as the source of the plant’s microbiota [89,93]. This means that the location of the plant and correspondingly available microbes, including their genetic material that is incorporated into the microbiome, can feedback and influence an MBCA’s success.
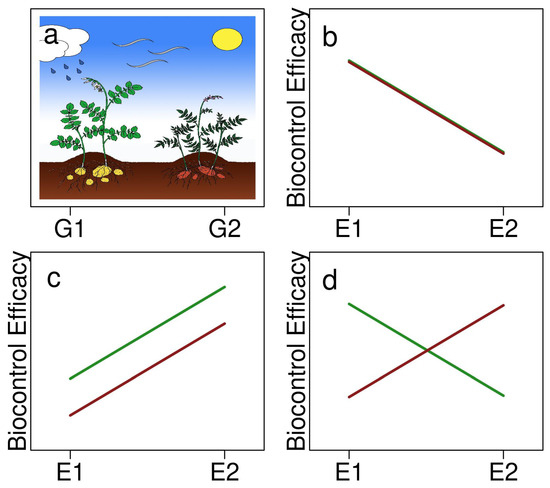
Figure 1.
A representation of two potato genotypes, (a) G1 (green) and G2 (red), and (b–d) three possible outcomes of an MBCA’s biocontrol efficacy (trait phenotype) across two different environments (E1 and E2) as illustrated by reaction norms showing: (b) the loss of efficacy in E2 but no difference between genotypes, (c) reduced efficacy in E1 and G2 but parallel responses across genotypes, and (d) G×E interaction, where G2 is more effective in E2 and G1 more effective in E1.
2.2. The Effect of Spatial Dynamics on Microbial Biocontrol Agents: Metapopulation and Metacommunity Perspectives
Spatial dynamics can have a role in the persistence and success of microbiome manipulations as well as the occurrence of plant pathogens. Epidemiological models and studies have long accounted for spatial scales to study disease pressure in human diseases, including work on ‘landscape epidemiology’ dating back to the 1930s [94]. Landscape and spatial lens have also been used to investigate crop pests [95,96], and the advancement of drone technology will likely assist such investigations in the future [97,98,99,100]. A number of oomycete pathogens have demonstrated spatially- and density-dependent patterns of disease occurrence, including Pythium and Phytophthora species [101,102,103]. More recently, building on population spatial dynamics, ecological frameworks including metapopulation and metacommunity perspectives have been described as a useful tool through which to analyze the interaction of multiple populations or species across a landscape [104]. Such perspectives may contribute to a better understanding of the dynamics within a pathosystem and help to determine how to decrease disease pressure [105,106].
Spatial analyses of metapopulations show that increasing distance between host plant fields and using a mixture of resistant and susceptible host plants can reduce disease severity [103]. Although a metapopulation perspective can be informative to analyze plant–pathogen interactions and their evolution [107,108], it focuses on the distribution and dynamics of single species in landscapes. However, within that same scale, multiple pairs of interacting species comprise a metacommunity [104,109], which can help offer a framework to examine not only host–pathogen interactions, but also those of biocontrol organisms and multiple members of the plant microbiome.
The metacommunity framework, which incorporates interacting species at multiple ecological, spatial, and evolutionary scales [104,110], can be useful to examine plant–pathogen dynamics [111]. Within a metacommunity lens, stochastic processes such as drift and dispersal are considered, which can greatly affect pattern of biodiversity in small communities [112]. Metacommunity dynamics can also affect the evolution of pathogens due to horizontal gene transfer with fungal cohabitants, which has been documented in the following oomycetes Phytophthora ramorum, Phytophthora sojae, Phytophthora infestans, and Hyaloperonospora parasitica [113]. Spatial dynamics of metacommunities that include pathogenic organisms should not only be considered on a macro (between field and plant scales), but also on a microscale, including within the rhizosphere [114] or plant.
A metacommunity perspective can help inform biocontrol applications as the dynamics of an MBCA’s population is likely to be influenced by multiple interactions among other members of the local and regional pool of microorganisms (Figure 2). Recent work has described the utility of using metacommunity theory to study symbiont evolutionary ecology [115], and such interactions among symbionts, such as the timing and sequence of multiple infections, can influence parasitic epidemics within plants [67]. In the case of agricultural settings, the regional species pool is influenced by the previous crop, which in turn, affects oomycete and bacterial communities [116,117].
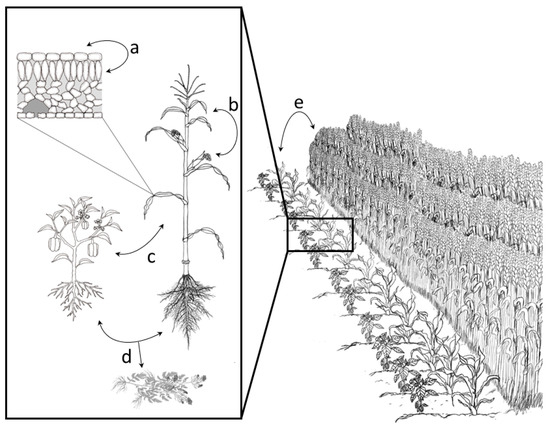
Figure 2.
Illustration demonstrating interacting spatial scales within agricultural systems that can be considered through a metacommunity framework. Exchange of microorganisms among these spatial scales can impact the success of microbial biological controls. These spatial scales include dispersal of microorganisms (a) within leaf microhabitats, including within the leaf and on the leaf surface; (b) between leaves on the same plant; (c) between neighboring plant hosts of different species, shown here within an intercropping system; (d) from pre-cropping material (either due to legacy effects in the soil or due to remaining plant debris); and (e) from neighboring fields.
Intercropping can be one way to expand the regional species pool and increase patch heterogeneity, which can help to decrease disease pressure through additional species’ interactions. The prevention of infection from potential pathogens may depend on having a more diverse metacommunity from which mutualistic microbial members can arise [118]. Spatial crop configurations that produce more diverse microbial communities [119,120] may help disease suppression through potential niche overlap between endophytes and pathogens [121]. Additionally, intercropping may contribute to reduced disease incidence through induced resistance from other plants or the production of allelopathic compounds [122]. Phytophthora infestans can be reduced with intercrops of cereal or grass-clover [123], and maize-pepper systems show reduced Phytophthora capsici severity through antimicrobial compounds secreted by maize [124].
3. Challenges and Limitations of Microbial Manipulations in Agricultural Systems
In the last decade, more research papers consider the microbiome in the control of oomycete pathogens (Table 1). Many of the aforementioned ecological concepts, including G×E interactions, community context, and spatial dynamics have been discussed in recent literature. These research avenues can help broaden the understanding of when MBCAs can work and in which context, but there are still many challenges with implementing effective MBCAs against oomycete pathogens.

Table 1.
Primary research papers with the search terms ‘microbiome’, ‘oomycete’, and ‘control’ were assessed to determine main research areas focusing on microbiome for biological control. The articles were categorized according to their incorporation of ecological concepts or discussion of control mechanisms in relationship to the microbiome. Search was conducted using Elsevier’s Scopus database and Web of Science and includes literature from 2013 to 2020 (see Supplementary Materials for more information).
For example, microbial communities are attributed to suppress diseases (i.e., within disease-suppressive soils), however, they are difficult to recreate in the field due to the involvement of multiple species [74]. The vast array of microbial diversity may make it challenging to create persistent alterations to a plant’s microbiome in the field. Weinhold and colleagues [151] genetically modified the wild tobacco plant (Nicotiana attenuate) to constitutively express antimicrobial peptide 1 (Mc-AMP1) of the common ice plant (Mesembryanthemum crystallinum), which selects against Gram-positive bacteria. Despite differences in microbial communities in greenhouse experiments—mainly through a reduction in beneficial bacterial, field trials showed no major alterations between microbial communities in engineered plants and normal plants. Rather than seeing systematic differences in modified plants in the field, it was found that root associated bacterial communities were altered only in subtle ways, which the authors attributed to the resilience of diverse microbial communities in nature. Such studies may help explain why MBCAs that look promising in more controlled environments, such as greenhouses and labs, fail in the field or yield inconsistent results [152]. These disparate effects may also be due to differences in plant physiology when grown in controlled environments compared to the field [153].
Because genotype and environment have an influence on the leaf microbiome [89], it is possible that colonization and persistence of MCBAs may vary based on these attributes and their interactions. Therefore, it would be unlikely to find a ‘silver bullet’ that would provide protection against oomycete pathogens under varying host genetic background and ambient environmental conditions. Due to these constraints, the application of MBCAs for a disease can benefit from a systems approach [79] to try to better select optimum conditions and genotype combinations for the use of an MBCA.
Evolution of pathogen strains and adaptation to biocontrol agents is another factor that may change dynamics of host–pathogen interactions [154], and many questions remain about evolution of resistance to biocontrol agents. Multiple infections of Albugo candida may have enabled host range expansion through genomic introgressions [88]. In this case, the primary infection reduces the defense mechanisms of the host, allowing for a secondary infection of A. candida and genetic exchange and the production of new and more challenging pathogens. Horizontal gene transfer (HGT) among oomycete and fungi may have enabled oomycetes to become successful plant pathogens [113], and HGT among co-occurring fungi and oomycetes may serve as an additional genetic reservoirs to provide resistance to biocontrol agents. Another frontier of the microbiome is its viral constituents. Oomycete plant pathogens have been found to harbor viruses [155] that, in some cases, may also amplify their virulence by increasing sporulation [156] and possibly interfering with host–pathogen interactions.
4. Outlook
Microbiome manipulations can help plant hosts resist diseases [157,158] and may provide a powerful tool to help protect plants from oomycete pathogens. In co-evolving, natural systems, the microbiome may provide additional genetic tools to help its host adapt to disease. A challenge in agricultural systems is that they are not representative of natural host–pathogen systems because of the alteration of co-evolutionary dynamics. Typically, arms races can ensue in natural systems with sexually reproducing hosts, whereby the host acquires adaptations to enable it to contend with a particular pathogen and subsequently the pathogen adapts to overcome these new arms [159]. In natural systems, the microbiome may contribute to the genetic arsenal to combat oomycete pathogens. In agricultural systems, cultivars of the host plant are artificially selected for particular characteristics usually related to commercial desirability. As every generation is reset with artificially selected cultivars, the evolutionary capabilities of host–microbial partnerships may be limited due to reduced opportunities for vertical transmission and selection for pathogen suppressive traits. More work is incorporating the microbiome in breeding research including for disease resistance [6,125,160].
Recent studies have accounted for the phylogenetic distance of host plants on yield outcomes and soil microbiome community [161], and although effects on yield have not been shown, changes in soil microbiome community are apparent. Such studies help unite concepts of evolutionary ecology and microbial ecology to better understand how microbes can be utilized in agricultural practices against oomycete pathogens. Selection on soil microbiomes was shown to influence flowering traits [162], suggesting that the engineering of the microbiome for disease resistance may also be possible.
Applications of microbial consortia rather than just one species could help lead to resilient communities that have disease-fighting capacity—either through induced resistance or through direct antagonism [40,163]. In a host-mutualist context, pathogenic microorganisms can otherwise be kept in check by interactions with other members of the microbiome [118]. Although it has been argued that most combined use of biocontrol agents results in antagonistic relationships between MBCAs and only one of the MCBAs is responsible for most of the suppression [164], a number of microbial consortia have shown improved results against oomycete pathogens [42,55,165]. These conflicting results suggest that a community approach or the application of multiple microbes could provide more protection rather than a single strain in certain circumstances. Although the dynamics of a community of MBCAs, rather than just a few strains or species, would be more intricate to study, an additional obstacle is the registration procedure of MBCAs. Registration of single strains is already challenging, and due to the time and cost associated in registering MBCAs, most are often on the market as biofertilizers [166]. Therefore, the adaptation of registration procedures to also account for communities would be needed before such control strategies could be implemented.
The success of biocontrol solutions is more likely to occur if an MBCA persists after its application, so identifying factors or assembling communities that facilitate persistence is an important goal. According to Kinnunen and colleagues [167], a successful establishment of a new microbial community member is defined as the maintenance of a metabolically active population for a significant period of time, and it can depend on niche space, stochastic processes such as drift, and community diversity. Including members that aid in the formation of biofilms—structured microbial communities embedded in an extracellular matrix—may facilitate the biocontrol of oomycete pathogens [168,169,170]. Biofilms are thought to contribute to the control of pathogens through diverse means such as provoking induced systemic resistance, producing antimicrobial compounds, and excluding pathogens from niche space [171]. However, biofilm formation can also assist Phytophthora parasitica infections [172], and their assistance or inhibition of infection may be situational and depend on the characteristics of the biofilm and its inhabitants. Additional work is needed to help elucidate and differentiate biofilm characteristics that aid in protection and in infection. Aside from the impact of biofilm formation, the identification of other biotic and abiotic factors to enable the success of MBCAs in different environment could help direct the successful establishment of an MBCA. Due to environmental effects, more focus on functional rather than taxonomic attributes of the microbiota is needed [173].
Biocontrol solutions need individualized solutions for plant genotype, local environment, and local microbial constituents. Such customized methods for pathogen control take more work to understand effective treatments under different situations, and an “one size fits all” model for biocontrol solutions is not realistic. Additionally, given the ability for pathogenic organisms to adapt, solutions must be pliable. Although there are a number of obstacles to implementing microbial biocontrol solutions on a larger scale, the societal desire to decrease pesticide use can help derive the necessary activation energy to find microbiome-based biocontrol solutions.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/plants10122697/s1.
Author Contributions
Conceptualization, K.E.S. with input from T.M.; writing—original draft preparation, K.E.S.; visualization, K.E.S. with input from T.M.; writing—review and editing, T.M. and K.E.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
We would like to thank Cecilia Panzetti and Jonas Lehner for the contribution of their illustrations to the figures and Blake Matthews for constructive feedback. We would also like to acknowledge the support of Susanne Vogelgsang and thank the two anonymous reviewers.
Conflicts of Interest
The authors declare no conflict of interest.
References
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C.G.; Carey, H.V.; Domazet-Lošo, T.; Douglas, A.E.; Dubilier, N.; Eberl, G.; Fukami, T.; Gilbert, S.F.; et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA 2013, 110, 3229. [Google Scholar] [CrossRef] [Green Version]
- Hacquard, S.; Garrido-Oter, R.; González, A.; Spaepen, S.; Ackermann, G.; Lebeis, S.; McHardy, A.; Dangl, J.; Knight, R.; Ley, R.; et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 2015, 17, 603–616. [Google Scholar] [CrossRef] [Green Version]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Ravel, J. The vocabulary of microbiome research: A proposal. Microbiome 2015, 3, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313. [Google Scholar] [CrossRef]
- Bakker, M.G.; Manter, D.K.; Sheflin, A.M.; Weir, T.L.; Vivanco, J.M. Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil 2012, 360, 1–13. [Google Scholar] [CrossRef]
- Lozupone, C.A. Unraveling interactions between the microbiome and the host immune system to decipher mechanisms of disease. mSystems 2018, 3, e00183-17. [Google Scholar] [CrossRef] [Green Version]
- Zilber-Rosenberg, I.; Rosenberg, E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Christian, N.; Whitaker, B.; Clay, K. Microbiomes: Unifying animal and plant systems through the lens of community ecology theory. Front. Microbiol. 2015, 6, 869. [Google Scholar] [CrossRef] [Green Version]
- Hird, S.M. Evolutionary biology needs wild microbiomes. Front. Microbiol. 2017, 8, 725. [Google Scholar] [CrossRef]
- Busby, P.E.; Soman, C.; Wagner, M.R.; Friesen, M.L.; Kremer, J.; Bennett, A.; Morsy, M.; Eisen, J.A.; Leach, J.E.; Dangl, J.L. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017, 15, e2001793. [Google Scholar] [CrossRef] [PubMed]
- Baldrian, P. Forest microbiome: Diversity, complexity and dynamics. FEMS Microbiol. Rev. 2016, 41, 109–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendes, R.; Raaijmakers, J.M. Cross-kingdom similarities in microbiome functions. ISME J. 2015, 9, 1905–1907. [Google Scholar] [CrossRef] [Green Version]
- Flandroy, L.; Poutahidis, T.; Berg, G.; Clarke, G.; Dao, M.C.; Decaestecker, E.; Furman, E.; Haahtela, T.; Massart, S.; Plovier, H.; et al. The impact of human activities and lifestyles on the interlinked microbiota and health of humans and of ecosystems. Sci. Total Environ. 2018, 627, 1018–1038. [Google Scholar] [CrossRef] [PubMed]
- Rook, G.A. Regulation of the immune system by biodiversity from the natural environment: An ecosystem service essential to health. Proc. Natl. Acad. Sci. USA 2013, 110, 18360. [Google Scholar] [CrossRef] [Green Version]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.A.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330. [Google Scholar] [CrossRef] [Green Version]
- Costello, E.K.; Stagaman, K.; Dethlefsen, L.; Bohannan, B.J.M.; Relman, D.A. The application of ecological theory toward an understanding of the human microbiome. Science 2012, 336, 1255. [Google Scholar] [CrossRef] [Green Version]
- Relman, D.A. How to build healthy growth-promoting gut communities. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 379. [Google Scholar] [CrossRef]
- Walter, J.; Maldonado-Gómez, M.X.; Martínez, I. To engraft or not to engraft: An ecological framework for gut microbiome modulation with live microbes. Curr. Opin. Biotechnol. 2018, 49, 129–139. [Google Scholar] [CrossRef]
- van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; de Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.; Tijssen, J.G.; et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Shahinas, D.; Silverman, M.; Sittler, T.; Chiu, C.; Kim, P.; Allen-Vercoe, E.; Weese, S.; Wong, A.; Low, D.E.; Pillai, D.R. Toward an understanding of changes in diversity associated with fecal microbiome transplantation based on 16S rRNA gene deep sequencing. mBio 2012, 3, e00338-12. [Google Scholar] [CrossRef] [Green Version]
- Clemente, J.; Ursell, L.; Parfrey, L.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [Green Version]
- Dash, S.; Clarke, G.; Berk, M.; Jacka, F.N. The gut microbiome and diet in psychiatry: Focus on depression. Curr. Opin. Psychiatry 2015, 28, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Grube, M.; Schloter, M.; Smalla, K. Unraveling the plant microbiome: Looking back and future perspectives. Front. Microbiol. 2014, 5, e00338-12. [Google Scholar] [CrossRef] [Green Version]
- Thines, M.; Kamoun, S. Oomycete–plant coevolution: Recent advances and future prospects. Curr. Opin. Plant Biol. 2010, 13, 427–433. [Google Scholar] [CrossRef]
- Judelson, H.S. Dynamics and innovations within oomycete genomes: Insights into biology, pathology, and evolution. Eukaryot. Cell 2012, 11, 1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, A.J.; Anderson, V.L.; Robertson, E.J.; Secombes, C.J.; van West, P. New insights into animal pathogenic oomycetes. Trends Microbiol. 2008, 16, 13–19. [Google Scholar] [CrossRef]
- Mendoza, L.; Hernandez, F.; Ajello, L. Life cycle of the human and animal oomycete pathogen Pythium insidiosum. J. Clin. Microbiol. 1993, 31, 2967. [Google Scholar] [CrossRef] [Green Version]
- Ploch, S.; Thines, M. Obligate biotrophic pathogens of the genus Albugo are widespread as asymptomatic endophytes in natural populations of Brassicaceae. Mol. Ecol. 2011, 20, 3692–3699. [Google Scholar] [CrossRef] [PubMed]
- Spring, O.; Gomez-Zeledon, J.; Hadziabdic, D.; Trigiano, R.N.; Thines, M.; Lebeda, A. Biological characteristics and assessment of virulence diversity in pathosystems of economically important biotrophic oomycetes. Crit. Rev. Plant Sci. 2018, 37, 439–495. [Google Scholar] [CrossRef]
- Derevnina, L.; Petre, B.; Kellner, R.; Dagdas, Y.F.; Sarowar, M.N.; Giannakopoulou, A.; De la Concepcion, J.C.; Chaparro-Garcia, A.; Pennington, H.G.; van West, P.; et al. Emerging oomycete threats to plants and animals. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2016, 371, 20150459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bebber, D.P.; Gurr, S.J. Crop-destroying fungal and oomycete pathogens challenge food security. Fungal Genet. Biol. 2015, 74, 62–64. [Google Scholar] [CrossRef]
- Tyler, B.M. Genetics and genomics of the oomycete–host interface. Trends Genet. 2001, 17, 611–614. [Google Scholar] [CrossRef]
- Klinter, S.; Bulone, V.; Arvestad, L. Diversity and evolution of chitin synthases in oomycetes (Straminipila: Oomycota). Mol. Phylogenet. Evol. 2019, 139, 106558. [Google Scholar] [CrossRef]
- Mélida, H.; Sandoval-Sierra, J.V.; Diéguez-Uribeondo, J.; Bulone, V. Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryot. Cell 2012, 12, 194–203. [Google Scholar] [CrossRef] [Green Version]
- Gaulin, E.; Bottin, A.; Dumas, B. Sterol biosynthesis in oomycete pathogens. Plant Signal. Behav. 2010, 5, 258–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahlin, P.; Srivastava, V.; Ekengren, S.; McKee, L.S.; Bulone, V. Comparative analysis of sterol acquisition in the oomycetes Saprolegnia parasitica and Phytophthora infestans. PLoS ONE 2017, 12, e0170873. [Google Scholar] [CrossRef]
- Madoui, M.A.; Bertrand-Michel, J.; Gaulin, E.; Dumas, B. Sterol metabolism in the oomycete Aphanomyces euteiches, a legume root pathogen. New Phytol. 2009, 183, 291–300. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Chen, P.C.; Tsay, T.T. The biocontrol efficacy and antibiotic activity of Streptomyces plicatus on the oomycete Phytophthora capsici. Biol. Control 2016, 98, 34–42. [Google Scholar] [CrossRef]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance Beyond Efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [Green Version]
- Michelsen, C.F.; Watrous, J.; Glaring, M.A.; Kersten, R.; Koyama, N.; Dorrestein, P.C.; Stougaard, P. Nonribosomal peptides, key biocontrol components for Pseudomonas fluorescens In5, isolated from a Greenlandic suppressive soil. mBio 2015, 6, e00079-15. [Google Scholar] [CrossRef] [Green Version]
- Ezziyyani, M.; Requena, M.E.; Egea-Gilabert, C.; Candela, M.E. Biological control of Phytophthora root rot of pepper Using Trichoderma harzianum and Streptomyces rochei in combination. J. Phytopathol. 2007, 155, 342–349. [Google Scholar] [CrossRef]
- van Dijk, K.; Nelson, E.B. Fatty acid competition as a mechanism by which Enterobacter cloacae suppresses Pythium ultimum sporangium germination and damping-off. Appl. Environ. Microbiol. 2000, 66, 5340–5347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parnell, J.J.; Berka, R.; Young, H.A.; Sturino, J.M.; Kang, Y.; Barnhart, D.M.; DiLeo, M.V. From the lab to the farm: An industrial perspective of plant beneficial microorganisms. Front. Plant Sci. 2016, 7, 1110. [Google Scholar] [CrossRef]
- Dorn, B.; Musa, T.; Krebs, H.; Fried, P.M.; Forrer, H.R. Control of late blight in organic potato production: Evaluation of copper-free preparations under field, growth chamber and laboratory conditions. Eur. J. Plant Pathol. 2007, 119, 217–240. [Google Scholar] [CrossRef]
- Cunniffe, N.J.; Gilligan, C.A. A theoretical framework for biological control of soil-borne plant pathogens: Identifying effective strategies. J. Theor. Biol. 2011, 278, 32–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köhl, J.; Postma, J.; Nicot, P.; Ruocco, M.; Blum, B. Stepwise screening of microorganisms for commercial use in biological control of plant–pathogenic fungi and bacteria. Biol. Control 2011, 57, 1–12. [Google Scholar] [CrossRef]
- Smits, W.K.; Kuipers, O.P.; Veening, J.W. Phenotypic variation in bacteria: The role of feedback regulation. Nat. Rev. Microbiol. 2006, 4, 259–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Kim, Y.S.; Balaraju, K.; Seo, Y.S.; Park, J.; Ryu, C.M.; Park, S.H.; Kim, J.F.; Kang, S.; Jeon, Y. Molecular changes associated with spontaneous phenotypic variation of Paenibacillus polymyxa, a commonly used biocontrol agent, and temperature-dependent control of variation. Sci. Rep. 2020, 10, 16586. [Google Scholar] [CrossRef] [PubMed]
- Ambrico, A.; Trupo, M.; Magarelli, R.A. Influence of phenotypic dissociation in Bacillus subtilis strain ET-1 on Iturin A production. Curr. Microbiol. 2019, 76, 1487–1494. [Google Scholar] [CrossRef]
- Stone, B.W.G.; Weingarten, E.A.; Jackson, C.R. The role of the phyllosphere microbiome in plant health and function. Annu. Plant Rev. Online 2018, 1, 533–556. [Google Scholar] [CrossRef]
- Griggs, R.G.; Steenwerth, K.L.; Mills, D.A.; Cantu, D.; Bokulich, N.A. Sources and assembly of microbial communities in vineyards as a functional component of winegrowing. Front. Microbiol. 2021, 12, 673810. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wei, Z.; Kowalchuk, G.A.; Xu, Y.; Shen, Q.; Jousset, A. Rhizosphere microbiome functional diversity and pathogen invasion resistance build up during plant development. Environ. Microbiol. 2020, 22, 5005–5018. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Tao, J.; Liu, T.; Liu, Y.; Xiao, N.; Li, T.; Gu, Y.; Yin, H.; Meng, D. Responses of phyllosphere microbiota and plant health to application of two different biocontrol agents. AMB Express 2019, 9, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jousset, A.; Becker, J.; Chatterjee, S.; Karlovsky, P.; Scheu, S.; Eisenhauer, N. Biodiversity and species identity shape the antifungal activity of bacterial communities. Ecology 2014, 95, 1184–1190. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Köberl, M.; Rybakova, D.; Müller, H.; Grosch, R.; Smalla, K. Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 2017, 93, fix050. [Google Scholar] [CrossRef] [PubMed]
- Arif, I.; Batool, M.; Schenk, P.M. Plant microbiome engineering: Expected benefits for improved crop growth and resilience. Trends Biotechnol. 2020, 38, 1385–1396. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Jin, K.; Raaijmakers, J.M. Designing a home for beneficial plant microbiomes. Curr. Opin. Plant Biol. 2021, 62, 102025. [Google Scholar] [CrossRef] [PubMed]
- Adam, E.; Groenenboom, A.E.; Kurm, V.; Rajewska, M.; Schmidt, R.; Tyc, O.; Weidner, S.; Berg, G.; de Boer, W.; Falcão Salles, J. Controlling the microbiome: Microhabitat adjustments for successful biocontrol strategies in soil and human gut. Front. Microbiol. 2016, 7, 1079. [Google Scholar] [CrossRef] [PubMed]
- Vassileva, M.; Flor-Peregrin, E.; Malusá, E.; Vassilev, N. Towards better understanding of the interactions and efficient application of plant beneficial prebiotics, probiotics, postbiotics and synbiotics. Front. Plant Sci. 2020, 11, 1068. [Google Scholar] [CrossRef] [PubMed]
- Kehe, J.; Kulesa, A.; Ortiz, A.; Ackerman, C.M.; Thakku, S.G.; Sellers, D.; Kuehn, S.; Gore, J.; Friedman, J.; Blainey, P.C. Massively parallel screening of synthetic microbial communities. Proc. Natl. Acad. Sci. USA 2019, 116, 12804–12809. [Google Scholar] [CrossRef] [Green Version]
- Johns, N.I.; Blazejewski, T.; Gomes, A.L.C.; Wang, H.H. Principles for designing synthetic microbial communities. Curr. Opin. Microbiol. 2016, 31, 146–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Yang, T.; Friman, V.P.; Kowalchuk, G.A.; Hautier, Y.; Li, M.; Wei, Z.; Xu, Y.; Shen, Q.; Jousset, A. Introduction of probiotic bacterial consortia promotes plant growth via impacts on the resident rhizosphere microbiome. Proc. R. Soc. B Biol. Sci. 2021, 288, 20211396. [Google Scholar] [CrossRef]
- Hubbell, S.P. The Unified Neutral Theory of Biodiversity and Biogeography, Monographs in Population Biology; Princeton Univ. Press: Princeton, NJ, USA, 2001. [Google Scholar]
- Carlström, C.I.; Field, C.M.; Bortfeld-Miller, M.; Müller, B.; Sunagawa, S.; Vorholt, J.A. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nat. Ecol. Evol. 2019, 3, 1445–1454. [Google Scholar] [CrossRef]
- Leopold, D.R.; Busby, P.E. Host genotype and colonist arrival order jointly govern plant microbiome composition and function. Curr. Biol. 2020, 30, 3260–3266.e5. [Google Scholar] [CrossRef] [PubMed]
- Halliday, F.W.; Umbanhowar, J.; Mitchell, C.E. Interactions among symbionts operate across scales to influence parasite epidemics. Ecol. Lett. 2017, 20, 1285–1294. [Google Scholar] [CrossRef]
- Tkacz, A.; Bestion, E.; Bo, Z.; Hortala, M.; Poole, P.S. Influence of plant fraction, soil, and plant species on microbiota: A multikingdom comparison. mBio 2020, 11, e02785-19. [Google Scholar] [CrossRef] [Green Version]
- Wei, Z.; Gu, Y.; Friman, V.P.; Kowalchuk, G.A.; Xu, Y.; Shen, Q.; Jousset, A. Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 2019, 5, eaaw0759. [Google Scholar] [CrossRef] [Green Version]
- Walsh, C.M.; Becker-Uncapher, I.; Carlson, M.; Fierer, N. Variable influences of soil and seed-associated bacterial communities on the assembly of seedling microbiomes. ISME J. 2021, 15, 2748–2762. [Google Scholar] [CrossRef] [PubMed]
- Amend, A.S.; Cobian, G.M.; Laruson, A.J.; Remple, K.; Tucker, S.J.; Poff, K.E.; Antaky, C.; Boraks, A.; Jones, C.A.; Kuehu, D.; et al. Phytobiomes are compositionally nested from the ground up. PeerJ 2019, 7, e6609. [Google Scholar] [CrossRef] [Green Version]
- Jack, A.L.H.; Nelson, E.B. A seed-recruited microbiome protects developing seedlings from disease by altering homing responses of Pythium aphanidermatum zoospores. Plant Soil 2018, 422, 209–222. [Google Scholar] [CrossRef]
- Löbmann, M.T.; Vetukuri, R.R.; de Zinger, L.; Alsanius, B.W.; Grenville-Briggs, L.J.; Walter, A.J. The occurrence of pathogen suppressive soils in Sweden in relation to soil biota, soil properties, and farming practices. Appl. Soil Ecol. 2016, 107, 57–65. [Google Scholar] [CrossRef]
- Gómez Expósito, R.; de Bruijn, I.; Postma, J.; Raaijmakers, J.M. Current insights into the role of rhizosphere bacteria in disease suppressive soils. Front. Microbiol. 2017, 8, 2529. [Google Scholar] [CrossRef] [PubMed]
- Bonanomi, G.; Antignani, V.; Capodilupo, M.; Scala, F. Identifying the characteristics of organic soil amendments that suppress soilborne plant diseases. Soil Biol. Biochem. 2010, 42, 136–144. [Google Scholar] [CrossRef]
- Fukami, T. Historical contingency in community assembly: Integrating niches, species pools, and priority effects. Annu. Rev. Ecol. Evol. Syst. 2015, 46, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Bazghaleh, N.; Prashar, P.; Woo, S.; Vandenberg, A. Effects of lentil genotype on the colonization of beneficial Trichoderma species and biocontrol of Aphanomyces root rot. Microorganisms 2020, 8, 1290. [Google Scholar] [CrossRef]
- Singh, P.; Santoni, S.; This, P.; Péros, J.P. Genotype-environment interaction shapes the microbial assemblage in grapevine’s phyllosphere and carposphere: An NGS aproach. Microorganisms 2018, 6, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrett, K.; Alcalá-Briseño, R.; Andersen, K.; Buddenhagen, C.; Choudhury, R.; Fulton, J.; Nopsa, J.H.; Poudel, R.; Xing, Y. Network analysis: A systems framework to address grand challenges in plant pathology. Annu. Rev. Phytopathol. 2018, 56, 559–580. [Google Scholar] [CrossRef]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.T.; Weigel, D.; Kemen, E.M. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef] [Green Version]
- Larousse, M.; Rancurel, C.; Syska, C.; Palero, F.; Etienne, C.; Industri, B.; Nesme, X.; Bardin, M.; Galiana, E. Tomato root microbiota and Phytophthora parasitica-associated disease. Microbiome 2017, 5, 56. [Google Scholar] [CrossRef]
- Hol, W.H.G.; Garbeva, P.; Hordijk, C.; Hundscheid, M.P.J.; Gunnewiek, P.J.A.K.; van Agtmaal, M.; Kuramae, E.E.; de Boer, W. Non-random species loss in bacterial communities reduces antifungal volatile production. Ecology 2015, 96, 2042–2048. [Google Scholar] [CrossRef] [Green Version]
- Hennessy, R.C.; Glaring, M.A.; Olsson, S.; Stougaard, P. Transcriptomic profiling of microbe–microbe interactions reveals the specific response of the biocontrol strain P. fluorescens In5 to the phytopathogen Rhizoctonia solani. BMC Res. Notes 2017, 10, 376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Z.; Yang, T.; Friman, V.P.; Xu, Y.; Shen, Q.; Jousset, A. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 2015, 6, 8413. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.; Salas Gonzalez, I.; Mittelviefhaus, M.; Clingenpeel, S.; Herrera Paredes, S.; Miao, J.; Wang, K.; Devescovi, G.; Stillman, K.; Monteiro, F.; et al. Genomic features of bacterial adaptation to plants. Nat. Genet. 2018, 50, 138–150. [Google Scholar] [CrossRef] [Green Version]
- Kong, P.; Hong, C. Soil bacteria as sources of virulence signal providers promoting plant infection by Phytophthora pathogens. Sci. Rep. 2016, 6, 33239. [Google Scholar] [CrossRef] [Green Version]
- McMullan, M.; Gardiner, A.; Bailey, K.; Kemen, E.; Ward, B.J.; Cevik, V.; Robert-Seilaniantz, A.; Schultz-Larsen, T.; Balmuth, A.; Holub, E.; et al. Evidence for suppression of immunity as a driver for genomic introgressions and host range expansion in races of Albugo candida, a generalist parasite. eLife 2015, 4, e04550. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; Del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef]
- Yergeau, E.; Bell, T.H.; Champagne, J.; Maynard, C.; Tardif, S.; Tremblay, J.; Greer, C.W. Transplanting soil microbiomes leads to lasting effects on willow growth, but not on the rhizosphere microbiome. Front. Microbiol. 2015, 6, 1436. [Google Scholar] [CrossRef] [Green Version]
- Henry, L.P.; Bruijning, M.; Forsberg, S.K.G.; Ayroles, J.F. The microbiome extends host evolutionary potential. Nat. Commun. 2021, 12, 5141. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.P.; Handelsman, J.; Goodman, R.M. Genetic basis in plants for interactions with disease-suppressive bacteria. Proc. Natl. Acad. Sci. USA 1999, 96, 4786–4790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ottesen, A.R.; Gorham, S.; Reed, E.; Newell, M.J.; Ramachandran, P.; Canida, T.; Allard, M.; Evans, P.; Brown, E.; White, J.R. Using a control to better understand phyllosphere microbiota. PLoS ONE 2016, 11, e0163482. [Google Scholar] [CrossRef] [Green Version]
- Ostfeld, R.S.; Glass, G.E.; Keesing, F. Spatial epidemiology: An emerging (or re-emerging) discipline. Trends Ecol. Evol. 2005, 20, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Otten, W.; Gilligan, C.A. Soil structure and soil-borne diseases: Using epidemiological concepts to scale from fungal spread to plant epidemics. Eur. J. Soil Sci. 2006, 57, 26–37. [Google Scholar] [CrossRef]
- Campbell, C.; Madden, L. Introduction to Plant Disease Epidemiology; Wiley: New York, NY, USA, 1990. [Google Scholar]
- Neupane, K.; Baysal-Gurel, F. Automatic identification and monitoring of plant diseases using unmanned aerial vehicles: A review. Remote Sens. 2021, 13, 3841. [Google Scholar] [CrossRef]
- Rodríguez, J.; Lizarazo, I.; Prieto, F.; Angulo-Morales, V. Assessment of potato late blight from UAV-based multispectral imagery. Comput. Electron. Agric. 2021, 184, 106061. [Google Scholar] [CrossRef]
- Maes, W.H.; Steppe, K. Perspectives for remote sensing with unmanned aerial vehicles in precision agriculture. Trends Plant Sci. 2019, 24, 152–164. [Google Scholar] [CrossRef]
- Calderón, R.; Montes-Borrego, M.; Landa, B.B.; Navas-Cortés, J.A.; Zarco-Tejada, P.J. Detection of downy mildew of opium poppy using high-resolution multi-spectral and thermal imagery acquired with an unmanned aerial vehicle. Precis. Agric. 2014, 15, 639–661. [Google Scholar] [CrossRef]
- Packer, A.; Clay, K. Soil pathogens and spatial patterns of seedling mortality in a temperate tree. Nature 2000, 404, 278–281. [Google Scholar] [CrossRef]
- Ristaino, J.B.; Larkin, R.P.; Campbell, C.L. Spatial and temporal dynamics of Phytophthora epidemics in commercial bell pepper fields. Phytopathology 1993, 83, 1312–1320. [Google Scholar] [CrossRef]
- Skelsey, P.; Rossing, W.A.H.; Kessel, G.J.T.; van der Werf, W. Invasion of Phytophthora infestans at the landscape level: How do spatial scale and weather modulate the consequences of spatial heterogeneity in host resistance? Phytopathology 2010, 100, 1146–1161. [Google Scholar] [CrossRef] [Green Version]
- Leibold, M.; Holyoak, M.; Mouquet, N. The metacommunity concept: A framework for multi-scale community ecology. Ecol. Lett. 2004, 7, 601–613. [Google Scholar] [CrossRef]
- Plantegenest, M.; May, C.L.; Fabre, F. Landscape epidemiology of plant diseases. J. R. Soc. Interface 2007, 4, 963–972. [Google Scholar] [CrossRef] [Green Version]
- Park, A.W.; Gubbins, S.; Gilligan, C.A. Invasion and persistence of plant parasites in a spatially structured host population. Oikos 2001, 94, 162–174. [Google Scholar] [CrossRef]
- Thrall, P.H.; Burdon, J.J. Host-pathogen dynamics in a metapopulation context: The ecological and evolutionary consequences of being spatial. J. Ecol. 1997, 85, 743–753. [Google Scholar] [CrossRef]
- Papaïx, J.; Burdon, J.J.; Lannou, C.; Thrall, P.H. Evolution of pathogen specialisation in a host metapopulation: Joint effects of host and pathogen dispersal. PLoS Comput. Biol. 2014, 10, e1003633. [Google Scholar] [CrossRef] [PubMed]
- Holyoak, M.; Leibold, M.; Holt, R. Metacommunities: Spatial Dynamics and Ecological Communities; University of Chicago Press: Chicago, IL, USA, 2005. [Google Scholar]
- Urban, M.C.; Skelly, D.K. Evolving metacommunities: Toward an evolutionary perspective on metacommunities. Ecology 2006, 87, 1616–1626. [Google Scholar] [CrossRef] [Green Version]
- Borer, E.T.; Laine, A.L.; Seabloom, E.W. A multiscale approach to plant disease using the metacommunity concept. Annu. Rev. Phytopathol. 2016, 54, 397–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, B.; Levine, J.M. Ecological drift and the distribution of species diversity. Proc. R. Soc. B Biol. Sci. 2017, 284, 20170507. [Google Scholar] [CrossRef] [PubMed]
- Richards, T.A.; Soanes, D.M.; Jones, M.D.M.; Vasieva, O.; Leonard, G.; Paszkiewicz, K.; Foster, P.G.; Hall, N.; Talbot, N.J. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc. Natl. Acad. Sci. USA 2011, 108, 15258–15263. [Google Scholar] [CrossRef] [Green Version]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2009, 321, 341–361. [Google Scholar] [CrossRef] [Green Version]
- Mihaljevic, J.R. Linking metacommunity theory and symbiont evolutionary ecology. Trends Ecol. Evol. 2012, 27, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, R.; Nicolaisen, M. Cropping history shapes fungal, oomycete and nematode communities in arable soils and affects cavity spot in carrot. Agric. Ecosyst. Environ. 2018, 257, 120–131. [Google Scholar] [CrossRef]
- Li, X.; Jousset, A.; de Boer, W.; Carrión, V.J.; Zhang, T.; Wang, X.; Kuramae, E.E. Legacy of land use history determines reprogramming of plant physiology by soil microbiome. ISME J. 2019, 13, 738–751. [Google Scholar] [CrossRef] [Green Version]
- Pillai, P.; Gouhier, T.C.; Vollmer, S.V. The cryptic role of biodiversity in the emergence of host–microbial mutualisms. Ecol. Lett. 2014, 17, 1437–1446. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Gao, D.; Zhou, X.; Chen, S.; Li, C.; Wu, F. Intercropping with potato-onion enhanced the soil microbial diversity of tomato. Microorganisms 2020, 8, 834. [Google Scholar] [CrossRef]
- Wang, G.; Bei, S.; Li, J.; Bao, X.; Zhang, J.; Schultz, P.A.; Li, H.; Li, L.; Zhang, F.; Bever, J.D.; et al. Soil microbial legacy drives crop diversity advantage: Linking ecological plant–soil feedback with agricultural intercropping. J. Appl. Ecol. 2021, 58, 496–506. [Google Scholar] [CrossRef]
- Blumenstein, K.; Albrectsen, B.R.; Martín, J.A.; Hultberg, M.; Sieber, T.N.; Helander, M.; Witzell, J. Nutritional niche overlap potentiates the use of endophytes in biocontrol of a tree disease. BioControl 2015, 60, 655–667. [Google Scholar] [CrossRef]
- Zhu, S.; Morel, J.B. Molecular mechanisms underlying microbial disease control in intercropping. Mol. Plant-Microbe Interact. 2018, 32, 20–24. [Google Scholar] [CrossRef] [Green Version]
- Bouws, H.; Finckh, M.R. Effects of strip intercropping of potatoes with non-hosts on late blight severity and tuber yield in organic production. Plant Pathol. 2008, 57, 916–927. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, Y.; Qi, L.; Mei, X.; Liao, J.; Ding, X.; Deng, W.; Fan, L.; He, X.; Vivanco, J.M.; et al. Plant-plant-microbe mechanisms involved in soil-borne disease suppression on a maize and pepper intercropping system. PLoS ONE 2015, 9, e115052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wille, L.; Messmer, M.M.; Bodenhausen, N.; Studer, B.; Hohmann, P. Heritable variation in pea for resistance against a root rot complex and its characterization by amplicon sequencing. Front. Plant Sci. 2020, 11, 542153. [Google Scholar] [CrossRef]
- Noel, Z.A.; Chang, H.X.; Chilvers, M.I. Variation in soybean rhizosphere oomycete communities from Michigan fields with contrasting disease pressures. Appl. Soil Ecol. 2020, 150, 103435. [Google Scholar] [CrossRef]
- Somera, T.S.; Freilich, S.; Mazzola, M. Comprehensive analysis of the apple rhizobiome as influenced by different Brassica seed meals and rootstocks in the same soil/plant system. Appl. Soil Ecol. 2021, 157, 103766. [Google Scholar] [CrossRef]
- Lakkis, S.; Trotel-Aziz, P.; Rabenoelina, F.; Schwarzenberg, A.; Nguema-Ona, E.; Clément, C.; Aziz, A. Strengthening grapevine resistance by Pseudomonas fluorescens PTA-CT2 relies on distinct defense pathways in susceptible and partially resistant genotypes to downy mildew and gray mold diseases. Front. Plant Sci. 2019, 10, 1112. [Google Scholar] [CrossRef] [PubMed]
- Bellini, A.; Ferrocino, I.; Cucu, M.A.; Pugliese, M.; Garibaldi, A.; Gullino, M.L. A compost treatment acts as a suppressive agent in Phytophthora capsici–Cucurbita pepo pathosystem by modifying the rhizosphere microbiota. Front. Plant Sci. 2020, 11, 885. [Google Scholar] [CrossRef]
- Wang, L.; Mazzola, M. Effect of soil physical conditions on emission of allyl isothiocyanate and subsequent microbial inhibition in response to Brassicaceae seed meal amendment. Plant Dis. 2018, 103, 846–852. [Google Scholar] [CrossRef]
- Soonvald, L.; Loit, K.; Runno-Paurson, E.; Astover, A.; Tedersoo, L. The role of long-term mineral and organic fertilisation treatment in changing pathogen and symbiont community composition in soil. Appl. Soil Ecol. 2019, 141, 45–53. [Google Scholar] [CrossRef]
- Asiloglu, R.; Samuel, S.O.; Sevilir, B.; Akca, M.O.; Acar Bozkurt, P.; Suzuki, K.; Murase, J.; Turgay, O.C.; Harada, N. Biochar affects taxonomic and functional community composition of protists. Biol. Fertil. Soils 2021, 57, 15–29. [Google Scholar] [CrossRef]
- Lee, S.; An, R.; Grewal, P.; Yu, Z.; Borherova, Z.; Lee, J. High-performing windowfarm hydroponic system: Transcriptomes of fresh produce and microbial communities in response to beneficial bacterial treatment. Mol. Plant-Microbe Interact. 2016, 29, 965–976. [Google Scholar] [CrossRef]
- McGehee, C.S.; Raudales, R.E.; Elmer, W.H.; McAvoy, R.J. Efficacy of biofungicides against root rot and damping-off of microgreens caused by Pythium spp. Crop Prot. 2019, 121, 96–102. [Google Scholar] [CrossRef]
- De Vrieze, M.; Pandey, P.; Bucheli, T.D.; Varadarajan, A.R.; Ahrens, C.H.; Weisskopf, L.; Bailly, A. Volatile organic compounds from native potato-associated Pseudomonas as potential anti-oomycete agents. Front. Microbiol. 2015, 6, 1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinchilla, D.; Bruisson, S.; Meyer, S.; Zühlke, D.; Hirschfeld, C.; Joller, C.; L’Haridon, F.; Mène-Saffrané, L.; Riedel, K.; Weisskopf, L. A sulfur-containing volatile emitted by potato-associated bacteria confers protection against late blight through direct anti-oomycete activity. Sci. Rep. 2019, 9, 18778. [Google Scholar] [CrossRef] [Green Version]
- Bruisson, S.; Zufferey, M.; L’Haridon, F.; Trutmann, E.; Anand, A.; Dutartre, A.; De Vrieze, M.; Weisskopf, L. Endophytes and epiphytes from the grapevine leaf microbiome as potential biocontrol agents against phytopathogens. Front. Microbiol. 2019, 10, 2726. [Google Scholar] [CrossRef] [Green Version]
- Agisha, V.N.; Kumar, A.; Eapen, S.J.; Sheoran, N.; Suseelabhai, R. Broad-spectrum antimicrobial activity of volatile organic compounds from endophytic Pseudomonas putida BP25 against diverse plant pathogens. Biocontrol Sci. Technol. 2019, 29, 1069–1089. [Google Scholar] [CrossRef]
- Khalaf, E.M.; Raizada, M.N. Bacterial seed endophytes of domesticated cucurbits antagonize Fungal and Oomycete pathogens including powdery mildew. Front. Microbiol. 2018, 9, 42. [Google Scholar] [CrossRef]
- Cucu, M.A.; Gilardi, G.; Pugliese, M.; Ferrocino, I.; Gullino, M.L. Effects of biocontrol agents and compost against the Phytophthora capsici of zucchini and their impact on the rhizosphere microbiota. Appl. Soil Ecol. 2020, 154, 103659. [Google Scholar] [CrossRef]
- Aleandri, M.P.; Chilosi, G.; Bruni, N.; Tomassini, A.; Vettraino, A.M.; Vannini, A. Use of nursery potting mixes amended with local Trichoderma strains with multiple complementary mechanisms to control soil-borne diseases. Crop Prot. 2015, 67, 269–278. [Google Scholar] [CrossRef]
- Granada, S.D.; Ramírez-Restrepo, S.; López-Luján, L.; Peláez-Jaramillo, C.A.; Bedoya-Pérez, J.C. Screening of a biological control bacterium to fight avocado diseases: From agroecosystem to bioreactor. Biocatal. Agric. Biotechnol. 2018, 14, 109–115. [Google Scholar] [CrossRef]
- Velázquez-Becerra, C.; Macías-Rodríguez, L.I.; López-Bucio, J.; Flores-Cortez, I.; Santoyo, G.; Hernández-Soberano, C.; Valencia-Cantero, E. The rhizobacterium Arthrobacter agilis produces dimethylhexadecylamine, a compound that inhibits growth of phytopathogenic fungi in vitro. Protoplasma 2013, 250, 1251–1262. [Google Scholar] [CrossRef]
- Méndez-Bravo, A.; Cortazar-Murillo, E.M.; Guevara-Avendaño, E.; Ceballos-Luna, O.; Rodríguez-Haas, B.; Kiel-Martínez, A.L.; Hernández-Cristóbal, O.; Guerrero-Analco, J.A.; Reverchon, F. Plant growth-promoting rhizobacteria associated with avocado display antagonistic activity against Phytophthora cinnamomi through volatile emissions. PLoS ONE 2018, 13, e0194665. [Google Scholar] [CrossRef] [Green Version]
- Cheffi, M.; Bouket, A.C.; Alenezi, F.N.; Luptakova, L.; Belka, M.; Vallat, A.; Rateb, M.E.; Tounsi, S.; Triki, M.A.; Belbahri, L. Olea europaea L. root endophyte Bacillus velezensis OEE1 counteracts oomycete and fungal harmful pathogens and harbours a large repertoire of secreted and volatile metabolites and beneficial functional genes. Microorganisms 2019, 7, 314. [Google Scholar] [CrossRef] [Green Version]
- Nandhini, M.; Rajini, S.B.; Udayashankar, A.C.; Niranjana, S.R.; Lund, O.S.; Shetty, H.S.; Prakash, H.S. Diversity, plant growth promoting and downy mildew disease suppression potential of cultivable endophytic fungal communities associated with pearl millet. Biol. Control 2018, 127, 127–138. [Google Scholar] [CrossRef]
- Vetukuri, R.R.; Masini, L.; McDougal, R.; Panda, P.; de Zinger, L.; Brus-Szkalej, M.; Lankinen, A.; Grenville-Briggs, L.J. The presence of Phytophthora infestans in the rhizosphere of a wild Solanum species may contribute to off-season survival and pathogenicity. Appl. Soil Ecol. 2020, 148, 103475. [Google Scholar] [CrossRef]
- Fiore-Donno, A.M.; Bonkowski, M. Different community compositions between obligate and facultative oomycete plant parasites in a landscape-scale metabarcoding survey. Biol. Fertil. Soils 2021, 57, 245–256. [Google Scholar] [CrossRef]
- Bakker, M.G.; Moorman, T.B.; Kaspar, T.C.; Manter, D.K. Isolation of cultivation-resistant oomycetes, first detected as amplicon sequences, from roots of herbicide-terminated winter rye. Phytobiomes J. 2017, 1, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Ebadzadsahrai, G.; Higgins Keppler, E.A.; Soby, S.D.; Bean, H.D. Inhibition of fungal growth and induction of a novel volatilome in response to Chromobacterium vaccinii volatile organic compounds. Front. Microbiol. 2020, 11, 1035. [Google Scholar] [CrossRef]
- Weinhold, A.; Karimi Dorcheh, E.; Li, R.; Rameshkumar, N.; Baldwin, I.T. Antimicrobial peptide expression in a wild tobacco plant reveals the limits of host-microbe-manipulations in the field. eLife 2018, 7, e28715. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, I.M.B.; Hockenhull, J.; Jensen, D.F.; Gerhardson, B.; Hökeberg, M.; Tahvonen, R.; Teperi, E.; Sundheim, L.; Henriksen, B. Selection of biological control agents for controlling soil and seed-borne diseases in the field. Eur. J. Plant Pathol. 1997, 103, 775–784. [Google Scholar] [CrossRef]
- Poorter, H.; Fiorani, F.; Pieruschka, R.; Wojciechowski, T.; van der Putten, W.H.; Kleyer, M.; Schurr, U.; Postma, J. Pampered inside, pestered outside? Differences and similarities between plants growing in controlled conditions and in the field. New Phytol. 2016, 212, 838–855. [Google Scholar] [CrossRef]
- Bardin, M.; Ajouz, S.; Comby, M.; Lopez-Ferber, M.; Graillot, B.; Siegwart, M.; Nicot, P.C. Is the efficacy of biological control against plant diseases likely to be more durable than that of chemical pesticides? Front. Plant Sci. 2015, 6, 566. [Google Scholar] [CrossRef]
- Cai, G.; Myers, K.; Fry, W.E.; Hillman, B.I. Phytophthora infestans RNA virus 2, a novel RNA virus from Phytophthora infestans, does not belong to any known virus group. Arch. Virol. 2019, 164, 567–572. [Google Scholar] [CrossRef]
- Cai, G.; Fry, W.E.; Hillman, B.I. PiRV-2 stimulates sporulation in Phytophthora infestans. Virus Res. 2019, 271, 197674. [Google Scholar] [CrossRef]
- Kwak, M.J.; Kong, H.G.; Choi, K.; Kwon, S.K.; Song, J.Y.; Lee, J.; Lee, P.A.; Choi, S.Y.; Seo, M.; Lee, H.J.; et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 2018, 36, 1100. [Google Scholar] [CrossRef] [PubMed]
- Zahn, G.; Amend, A.S. Foliar microbiome transplants confer disease resistance in a critically-endangered plant. PeerJ 2017, 5, e4020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clay, K.; Kover, P.X. The red queen hypothesis and plant/pathogen interactions. Annu. Rev. Phytopathol. 1996, 34, 29–50. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Jousset, A. Plant breeding goes microbial. Trends Plant Sci. 2017, 22, 555–558. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, I.; Bokulich, N.A.; Caporaso, J.G.; Enders, L.S.; Ghanem, W.; Ingerslew, K.S. Phylogenetic farming: Can evolutionary history predict crop rotation via the soil microbiome? Evol. Appl. 2020, 13, 1984–1999. [Google Scholar] [CrossRef] [Green Version]
- Panke-Buisse, K.; Poole, A.C.; Goodrich, J.K.; Ley, R.E.; Kao-Kniffin, J. Selection on soil microbiomes reveals reproducible impacts on plant function. Isme J. 2014, 9, 980. [Google Scholar] [CrossRef]
- Syed Ab Rahman, S.F.; Singh, E.; Pieterse, C.M.J.; Schenk, P.M. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018, 267, 102–111. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.M.; Jeffries, P.; Pautasso, M.; Jeger, M.J. Combined use of biocontrol agents to manage plant diseases in theory and practice. Phytopathology 2011, 101, 1024–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunne, C.; Moënne-Loccoz, Y.; McCarthy, J.; Higgins, P.; Powell, J.; Dowling, D.N.; O’Gara, F. Combining proteolytic and phloroglucinol-producing bacteria for improved biocontrol of Pythium-mediated damping-off of sugar beet. Plant Pathol. 1998, 47, 299–307. [Google Scholar] [CrossRef]
- Velivelli, S.L.S.; De Vos, P.; Kromann, P.; Declerck, S.; Prestwich, B.D. Biological control agents: From field to market, problems, and challenges. Trends Biotechnol. 2014, 32, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Kinnunen, M.; Dechesne, A.; Proctor, C.; Hammes, F.; Johnson, D.; Quintela-Baluja, M.; Graham, D.; Daffonchio, D.; Fodelianakis, S.; Hahn, N.; et al. A conceptual framework for invasion in microbial communities. ISME J. 2016, 10, 2773–2779. [Google Scholar] [CrossRef]
- Hultberg, M.; Alsberg, T.; Khalil, S.; Alsanius, B. Suppression of disease in tomato infected by Pythium ultimum with a biosurfactant produced by Pseudomonas koreensis. BioControl 2010, 55, 435–444. [Google Scholar] [CrossRef]
- D’aes, J.; De Maeyer, K.; Pauwelyn, E.; Höfte, M. Biosurfactants in plant–Pseudomonas interactions and their importance to biocontrol. Environ. Microbiol. Rep. 2010, 2, 359–372. [Google Scholar] [CrossRef]
- Sang, M.K.; Kim, K.D. Biocontrol activity and root colonization by Pseudomonas corrugata strains CCR04 and CCR80 against Phytophthora blight of pepper. BioControl 2014, 59, 437–448. [Google Scholar] [CrossRef]
- Pandin, C.; Le Coq, D.; Canette, A.; Aymerich, S.; Briandet, R. Should the biofilm mode of life be taken into consideration for microbial biocontrol agents? Microb. Biotechnol. 2017, 10, 719–734. [Google Scholar] [CrossRef] [Green Version]
- Larousse, M.; Galiana, E. Microbial partnerships of pathogenic oomycetes. PLoS Pathog. 2017, 13, e1006028. [Google Scholar] [CrossRef]
- Lemanceau, P.; Blouin, M.; Muller, D.; Moënne-Loccoz, Y. Let the core microbiota be functional. Trends Plant Sci. 2017, 22, 583–595. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).