Breeding Canola (Brassica napus L.) for Protein in Feed and Food
Abstract
1. Introduction
2. Transition from Rapeseed to Canola
3. Extracting Oil from Raw Seed
4. Effects of Processing on Meal Quality
5. Nutritional Value of Canola Meal Protein
6. Anti-Nutrients in Canola Meal
7. Seed Development in Canola
8. Seed Storage Proteins of the Brassicaceae Family
8.1. Cruciferin of B. napus
8.2. Napin of B. napus
9. Genetic Control of Seed Storage Proteins in Canola
10. Non-Genetic Control of Seed Storage Proteins in Canola
11. Canola Protein as a Novel Food Product
11.1. Allergenicity of Canola Seed Storage Proteins
11.2. Regulation of Novel Foods in the United States
11.3. Regulation of Novel Food in the European Union
11.4. Regulation of Novel Food in Canada
12. Canola Meal as a Protein Source in Animal Husbandry
12.1. Cattle
12.2. Swine
12.3. Poultry
12.4. Aquaculture
13. Seed Storage Proteins in Food Processing
14. Separation and Quantification of Seed Storage Proteins
15. Immunodetection and Quantification of Seed Storage Proteins
16. Manipulation of Seed Storage Proteins in Select Crop Species
16.1. Seed Storage Protein Manipulation through Conventional Breeding
16.2. Soybean
16.3. Wheat
16.4. Canola
17. Plant Breeding in the Omics Era
17.1. Genomics
17.1.1. Linkage Mapping
17.1.2. Association Mapping
18. Marking of a New Era of Crop Improvement with Genome Editing
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- United Nations, Department of Economic and Social Affairs, Population Division World Population Prospects 2019. Available online: https://population.un.org/wpp/ (accessed on 10 October 2021).
- Fedoroff, N.V.; Battisti, D.S.; Beachy, R.N.; Cooper, P.J.M.; Fischhoff, D.A.; Hodges, C.N.; Knauf, V.C.; Lobell, D.; Mazur, B.J.; Molden, D.; et al. Radically rethinking agriculture for the 21st century. Science 2010, 327, 833–834. [Google Scholar] [CrossRef]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef]
- Tamburino, L.; Bravo, G.; Clough, Y.; Nicholas, K.A. From population to production: 50 years of scientific literature on how to feed the world. Glob. Food Sec. 2020, 24. [Google Scholar] [CrossRef]
- Gerten, D.; Heck, V.; Jägermeyr, J.; Bodirsky, B.L.; Fetzer, I.; Jalava, M.; Kummu, M.; Lucht, W.; Rockström, J.; Schaphoff, S.; et al. Feeding ten billion people is possible within four terrestrial planetary boundaries. Nat. Sustain. 2020, 3, 200–208. [Google Scholar] [CrossRef]
- Röös, E.; Bajželj, B.; Smith, P.; Patel, M.; Little, D.; Garnett, T. Greedy or needy? Land use and climate impacts of food in 2050 under different livestock futures. Glob. Environ. Chang. 2017, 47, 1–12. [Google Scholar] [CrossRef]
- Springmann, M.; Clark, M.; Mason-D’Croz, D.; Wiebe, K.; Bodirsky, B.L.; Lassaletta, L.; de Vries, W.; Vermeulen, S.J.; Herrero, M.; Carlson, K.M.; et al. Options for keeping the food system within environmental limits. Nature 2018, 562, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Ramankutty, N.; Mehrabi, Z.; Waha, K.; Jarvis, L.; Kremen, C.; Herrero, M.; Rieseberg, L.H. Trends in Global Agricultural Land Use: Implications for Environmental Health and Food Security. Annu. Rev. Plant Biol. 2018, 69, 789–815. [Google Scholar] [CrossRef]
- Fitton, N.; Alexander, P.; Arnell, N.; Bajzelj, B.; Calvin, K.; Doelman, J.; Gerber, J.S.; Havlik, P.; Hasegawa, T.; Herrero, M.; et al. The vulnerabilities of agricultural land and food production to future water scarcity. Glob. Environ. Chang. 2019, 58, 101944. [Google Scholar] [CrossRef]
- Röös, E.; Bajželj, B.; Smith, P.; Patel, M.; Little, D.; Garnett, T. Protein futures for Western Europe: Potential land use and climate impacts in 2050. Reg. Environ. Chang. 2017, 17, 367–377. [Google Scholar] [CrossRef]
- Report of the World Food Conference1975. Available online: https://digitallibrary.un.org/record/701143?ln=en (accessed on 10 October 2021).
- Food Security Information Network Global report on food crises 2018. Available online: http://www.fao.org/3/a-br323e.pdf (accessed on 10 October 2021).
- Trade Reforms and Food Security: Conceptualizing the Linkages 2003. Available online: http://www.fao.org/docrep/005/y4671e/y4671e06.htm. (accessed on 10 October 2021).
- Semba, R.D. The rise and fall of protein malnutrition in global health. Ann. Nutr. Metab. 2016, 69, 79–88. [Google Scholar] [CrossRef]
- Hwalla, N.; El Labban, S.; Bahn, R.A. Nutrition security is an integral component of food security. Front. Life Sci. 2016, 9, 167–172. [Google Scholar] [CrossRef]
- Crichton, M.; Craven, D.; Mackay, H.; Marx, W.; De Van Der Schueren, M.; Marshall, S. A systematic review, meta-analysis and meta-regression of the prevalence of protein-energy malnutrition: Associations with geographical region and sex. Age Ageing 2019, 48, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Gerbens-Leenes, P.W.; Nonhebel, S.; Krol, M.S. Food consumption patterns and economic growth. Increasing affluence and the use of natural resources. Appetite 2010, 55, 597–608. [Google Scholar] [CrossRef]
- He, Y.; Yang, X.; Xia, J.; Zhao, L.; Yang, Y. Consumption of meat and dairy products in China: A review. Proc. Nutr. Soc. 2016, 75, 385–391. [Google Scholar] [CrossRef]
- Nelson, G.; Bogard, J.; Lividini, K.; Arsenault, J.; Riley, M.; Sulser, T.B.; Mason-D’Croz, D.; Power, B.; Gustafson, D.; Herrero, M.; et al. Income growth and climate change effects on global nutrition security to mid-century. Nat. Sustain. 2018, 1, 773–781. [Google Scholar] [CrossRef]
- Sabaté, J.; Sranacharoenpong, K.; Harwatt, H.; Wien, M.; Soret, S. The environmental cost of protein food choices. Public Health Nutr. 2015, 18, 2067–2073. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, D.; Pimentel, M. Sustainability of meat-based and plant-based diets and the environment. Am. J. Clin. Nutr. 2003, 78, 660–663. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Bazer, F.W.; Cross, H.R. Land-based production of animal protein: Impacts, efficiency, and sustainability. Ann. N. Y. Acad. Sci. 2014, 1328, 18–28. [Google Scholar] [CrossRef]
- Sabaté, J.; Soret, S. Sustainability of plant-based diets: Back to the future. Am. J. Clin. Nutr. 2014, 100, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Shepon, A.; Eshel, G.; Noor, E.; Milo, R. The opportunity cost of animal based diets exceeds all food losses. Proc. Natl. Acad. Sci. USA 2018, 115, 3804–3809. [Google Scholar] [CrossRef]
- Graham, F.; Russell, J.; Holdsworth, M.; Menon, M.; Barker, M. Exploring the relationship between environmental impact and nutrient content of sandwiches and beverages available in cafés in a UK university. Sustainability 2019, 11. [Google Scholar] [CrossRef]
- Nelson, M.E.; Hamm, M.W.; Hu, F.B.; Abrams, S.A.; Griffin, T.S. Alignment of healthy dietary patterns and environmental sustainability: A systematic review. Adv. Nutr. 2016, 7, 1005–1025. [Google Scholar] [CrossRef] [PubMed]
- Bentham, J.; Singh, G.M.; Danaei, G.; Green, R.; Lin, J.K.; Stevens, G.A.; Farzadfar, F.; Bennett, J.E.; Di Cesare, M.; Dangour, A.D.; et al. Multidimensional characterization of global food supply from 1961 to 2013. Nat. Food 2020, 1, 70–75. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization Pulses Nutritious Seeds for a Sustainable Future. Available online: https://www.fao.org/3/i5528e/i5528e.pdf (accessed on 10 October 2021).
- Blackstone, N.T.; El-Abbadi, N.H.; McCabe, M.S.; Griffin, T.S.; Nelson, M.E. Linking sustainability to the healthy eating patterns of the Dietary Guidelines for Americans: A modelling study. Lancet Planet. Health 2018, 2, e344–e352. [Google Scholar] [CrossRef]
- Tuomisto, H.L. Importance of considering environmental sustainability in dietary guidelines. Lancet Planet. Health 2018, 2, e331–e332. [Google Scholar] [CrossRef]
- Canada’s Dietary Guidelines for Health Professionals and Policy Makers 2019. Available online: https://food-guide.canada.ca/en/guidelines/ (accessed on 10 October 2021).
- Wanasundara, J.P.D.; McIntosh, T.C.; Perera, S.P.; Withana-Gamage, T.S.; Mitra, P. Canola/rapeseed protein-functionality and nutrition. OCL 2016, 23, D407. [Google Scholar] [CrossRef]
- Tan, S.H.; Mailer, R.J.; Blanchard, C.L.; Agboola, S.O. Canola Proteins for Human Consumption: Extraction, Profile, and Functional Properties. J. Food Sci. 2011, 76. [Google Scholar] [CrossRef] [PubMed]
- Book of Abstracts of 15th International Rapeseed Congress 2019. Available online: https://www.openagrar.de/receive/openagrar_mods_00049944 (accessed on 10 October 2021).
- Pudel, F. Rapeseed proteins recent results on extraction and application. In Canola and Rapeseed, 1st ed.; Thiyam-Holländer, U., Eskin, N.A.M., Matthäus, B., Eds.; CRC Press: Boca Raton, FL, USA, 2012; pp. 184–199. [Google Scholar]
- Jones, O.G. Recent advances in the functionality of non-animal-sourced proteins contributing to their use in meat analogs. Curr. Opin. Food Sci. 2016, 7, 7–13. [Google Scholar] [CrossRef]
- Chabanon, G.; Chevalot, I.; Framboisier, X.; Chenu, S.; Marc, I. Hydrolysis of rapeseed protein isolates: Kinetics, characterization and functional properties of hydrolysates. Process Biochem. 2007, 42, 1419–1428. [Google Scholar] [CrossRef]
- Yang, C.; Wang, Y.; Vasanthan, T.; Chen, L. Impacts of pH and heating temperature on formation mechanisms and properties of thermally induced canola protein gels. Food Hydrocoll. 2014, 40, 225–236. [Google Scholar] [CrossRef]
- Fetzer, A.; Herfellner, T.; Stäbler, A.; Menner, M.; Eisner, P. Influence of process conditions during aqueous protein extraction upon yield from pre-pressed and cold-pressed rapeseed press cake. Ind. Crops Prod. 2018, 112, 236–246. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Craig, B.M. Production and utilization of rapeseed in Canada. J. Am. Oil Chem. Soc. 1971, 48, 737–739. [Google Scholar] [CrossRef]
- Nieschlag, H.J.; Wolff, I.A. Industrial uses of high erucic oils. J. Am. Oil Chem. Soc. 1971, 48, 723–727. [Google Scholar] [CrossRef]
- Daun, J.K. Glucosinolate levels in western Canadian rapeseed and canola. J. Am. Oil Chem. Soc. 1986, 63, 639–643. [Google Scholar] [CrossRef]
- Stefansson, B.R.; Hougen, F.W.; Downey, R.K. Note on the isolation of rape plants with seed oil free from erucic acid. Can. J. Plant Sci. 1961, 41, 218–219. [Google Scholar] [CrossRef]
- de Wildt, D.J.; Speijers, G.J.A. Influence of dietary rapeseed oil and erucic acid upon myocardial performance and hemodynamics in rats. Toxicol. Appl. Pharmacol. 1984, 74, 99–108. [Google Scholar] [CrossRef]
- EFSA Panel on Contaminants in the Food Chain Erucic acid in feed and food. EFSA J. 2016, 14. [CrossRef]
- Jensen, S.K.; Liu, Y.-G.; Eggum, B.O. The effect of heat treatment on glucosinolates and nutritional value of rapeseed meal in rats. Anim. Feed Sci. Technol. 1995, 53, 17–28. [Google Scholar] [CrossRef]
- Vermorel, M.; Davicco, M.-J.J.; Evrard, J. Valorization of rapeseed meal. 3. Effects of glucosinolate content on food intake, weight gain, liver weight and plasma thyroid hormone levels in growing rats. Reprod. Nutr. Dev. 1987, 27, 57–66. [Google Scholar] [CrossRef]
- Lo, M.; Bell, J.M. Effects of various dietary glucosinolate on growth, feed intake, and thyroid function of rats. Can. J. Anim. Sci. 1972, 52, 295–302. [Google Scholar] [CrossRef]
- Tripathi, M.K.; Mishra, A.S. Glucosinolates in animal nutrition: A review. Anim. Feed Sci. Technol. 2007, 132, 1–27. [Google Scholar] [CrossRef]
- Stefansson, B.R.; Kondra, Z.P. Tower summer rape. Can. J. Plant Sci. 1975, 344, 343–344. [Google Scholar] [CrossRef]
- Rakow, G. Classical Genetics and Traditional Breeding; Edwards, D., Batley, J., Parkin, I., Kole, C., Eds.; CRC Press: Boca Raton, FL, USA, 2011; ISBN 9780429067310. [Google Scholar]
- Consensus Document on Key Nutrient and Key Toxicants in Low Erucic Acid Rapeseed (Canola) 2001. Available online: https://www.oecd.org/env/ehs/biotrack/46815125.pdf (accessed on 10 October 2021).
- Food and Agriculture Organization of the United Nations FAOSTAT Database. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 10 October 2021).
- The Economic Impact of Canola on the Canadian Econom: 2020 Update. Available online: https://www.canolacouncil.org/download/131/economic-impact/17818/economic-impact-report-canada_december-2020 (accessed on 10 October 2021).
- Goering, K.J.; Eslick, R.; Brelsford, D.L. A search for high erucic acid containing oils in the cruciferae. Econ. Bot. 1965, 19, 251–256. [Google Scholar] [CrossRef]
- Calhoun, W.; Crane, J.M.; Stamp, D.L. Development of a low glucosinolate, high erucic acid rapeseed breeding program. J. Am. Oil Chem. Soc. 1975, 52, 363–365. [Google Scholar] [CrossRef]
- Tang, M.; Zhang, Y.; Liu, Y.; Tong, C.; Cheng, X.; Zhu, W.; Li, Z.; Huang, J.; Liu, S. Mapping loci controlling fatty acid profiles, oil and protein content by genome-wide association study in Brassica napus. Crop J. 2019, 7, 217–226. [Google Scholar] [CrossRef]
- Chung, J.; Babka, H.L.; Graef, G.L.; Staswick, P.E.; Lee, D.J.; Cregan, P.B.; Shoemaker, R.C.; Specht, J.E. The seed protein, oil, and yield QTL on soybean linkage group I. (Cell Biology & Molecular Genetics).PDF. Crop Sci. 2003, 43, 1053–1067. [Google Scholar] [CrossRef]
- Hu, Z.Y.; Hua, W.; Zhang, L.; Deng, L.B.; Wang, X.F.; Liu, G.H.; Hao, W.J.; Wang, H.Z. Seed structure characteristics to form ultrahigh oil content in rapeseed. PLoS ONE 2013, 8, 2–11. [Google Scholar] [CrossRef]
- Ohlson, R. Modern processing of rapeseed. J. Am. Oil Chem. Soc. 1992, 69, 195–198. [Google Scholar] [CrossRef]
- McCurdy, S.M. Effects of processing on the functional properties of canola/rapeseed protein. J. Am. Oil Chem. Soc. 1990, 67, 281–284. [Google Scholar] [CrossRef]
- Mupondwa, E.; Li, X.; Wanasundara, J.P.D. Technoeconomic Prospects for Commercialization of Brassica (Cruciferous) Plant Proteins. JAOCS, J. Am. Oil Chem. Soc. 2018, 95, 903–922. [Google Scholar] [CrossRef]
- Unger, E.H. Crushing and Extraction. Canola Chem. Prod. Process. Util. 2011, 163–188. [Google Scholar]
- Mag, T.K. Canola oil processing in Canada. J. Am. Oil Chem. Soc. 1983, 60, 380–384. [Google Scholar] [CrossRef]
- Bailey, H.M.; Stein, H.H. Can the digestible indispensable amino acid score methodology decrease protein malnutrition. Anim. Front. 2019, 9, 18–23. [Google Scholar] [CrossRef]
- Le Thanh, B.V.; Beltranena, E.; Zhou, X.; Wang, L.F.; Zijlstra, R.T. Amino acid and energy digestibility of Brassica napus canola meal from different crushing plants fed to ileal-cannulated grower pigs. Anim. Feed Sci. Technol. 2019, 252, 83–91. [Google Scholar] [CrossRef]
- Foegeding, E.A.; Davis, J.P. Food protein functionality: A comprehensive approach. Food Hydrocoll. 2011, 25, 1853–1864. [Google Scholar] [CrossRef]
- Tandang-Silvas, M.R.G.; Tecson-Mendoza, E.M.; Mikami, B.; Utsumi, S.; Maruyama, N. Molecular design of seed storage proteins for enhanced food physiochemical properties. Annu. Rev. Food Sci. Technol. 2011, 2, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Perera, S.P.; McIntosh, T.C.; Wanasundara, J.P.D. Structural properties of cruciferin and napin of Brassica napus (canola) show distinct responses to changes in pH and temperature. Plants 2016, 5, 36. [Google Scholar] [CrossRef]
- Salazar-Villanea, S.; Bruininx, E.M.A.M.; Gruppen, H.; Carré, P.; Quinsac, A.; van der Poel, A.F.B. Effects of toasting time on digestive hydrolysis of soluble and insoluble 00-rapeseed meal proteins. JAOCS J. Am. Oil Chem. Soc. 2017, 94, 619–630. [Google Scholar] [CrossRef]
- Wanasundara, J.P.D.; Abeysekara, S.J.; McIntosh, T.C.; Falk, K.C. Solubility differences of major storage proteins of brassicaceae oilseeds. JAOCS, J. Am. Oil Chem. Soc. 2012, 89, 869–881. [Google Scholar] [CrossRef]
- Fukushima, D. Denaturation of Soybean proteins by organic solvents. Cereal Chem. 1969, 46, 156–163. [Google Scholar]
- Thacker, P.A.; Newkirk, R.W. Performance of growing-finishing pigs fed barley-based diets containing toasted or non-toasted canola meal. Can. J. Anim. Sci. 2011, 85, 53–59. [Google Scholar] [CrossRef]
- Mosenthin, R.; Messerschmidt, U.; Sauer, N.; Carré, P.; Quinsac, A.; Schöne, F. Effect of the desolventizing/toasting process on chemical composition and protein quality of rapeseed meal. J. Anim. Sci. Biotechnol. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Newkirk, R.W.; Classen, H.L. The effects of toasting canola meal on body weight, feed conversion efficiency, and mortality in broiler chickens. Poult. Sci. 2002, 81, 815–825. [Google Scholar] [CrossRef]
- Khattab, R.Y.; Arntfield, S.D. Functional properties of raw and processed canola meal. LWT Food Sci. Technol. 2009, 42, 1119–1124. [Google Scholar] [CrossRef]
- He, R.; He, H.Y.; Chao, D.; Ju, X.; Aluko, R. Effects of High Pressure and Heat Treatments on Physicochemical and Gelation Properties of Rapeseed Protein Isolate. Food Bioprocess Technol. 2014, 7, 1344–1353. [Google Scholar] [CrossRef]
- Lund, M.N.; Ray, C.A. Control of Maillard reactions in foods: Strategies and chemical mechanisms. J. Agric. Food Chem. 2017, 65, 4537–4552. [Google Scholar] [CrossRef] [PubMed]
- Quality of Western Canadian Canola 2018. Available online: https://www.grainscanada.gc.ca/en/grain-research/export-quality/oilseeds/canola/2018/pdf/canola-quality-report-18.pdf (accessed on 10 October 2021).
- Quality of Canadian Oilseed-Type Soybeans 2018. Available online: https://www.grainscanada.gc.ca/en/grain-research/export-quality/oilseeds/soybean-oil/2018/index.html (accessed on 10 October 2021).
- Adewole, D.I.; Rogiewicz, A.; Dyck, B.; Slominski, B.A. Effects of canola meal source on the standardized ileal digestible amino acids and apparent metabolizable energy contents for broiler chickens. Poult. Sci. 2017, 96, 4298–4306. [Google Scholar] [CrossRef]
- Kasprzak, M.M.; Houdijk, J.G.M.; Olukosi, O.A.; Appleyard, H.; Kightley, S.P.J.; Carré, P.; Sutton, T.; Wiseman, J. The content and standardized ileal digestibility of crude protein and amino acids in rapeseed co-products fed to pigs. Livest. Sci. 2018, 208, 22–27. [Google Scholar] [CrossRef]
- Millward, D.J. Amino acid scoring patterns for protein quality assessment. Br. J. Nutr. 2012, 108. [Google Scholar] [CrossRef] [PubMed]
- Schaafsma, G. The protein digestibility–corrected amino acid score. J. Nutr. 2000, 130, 1865S–1867S. [Google Scholar] [CrossRef]
- Hughes, G.J.; Ryan, D.J.; Mukherjea, R.; Schasteen, C.S. Protein digestibility-corrected amino acid scores (PDCAAS) for soy protein isolates and concentrate: Criteria for evaluation. J. Agric. Food Chem. 2011, 59, 12707–12712. [Google Scholar] [CrossRef]
- Bos, C.; Airinei, G.; Mariotti, F.; Benamouzig, R.; Bérot, S.; Evrard, J.; Fénart, E.; Tomé, D.; Gaudichon, C. The poor digestibility of rapeseed protein is balanced by its very high metabolic utilization in humans. J. Nutr. 2007, 137, 594–600. [Google Scholar] [CrossRef]
- Fleddermann, M.; Fechner, A.; Rößler, A.; Bähr, M.; Pastor, A.; Liebert, F.; Jahreis, G. Nutritional evaluation of rapeseed protein compared to soy protein for quality, plasma amino acids, and nitrogen balance—A randomized cross-over intervention study in humans. Clin. Nutr. 2013, 32, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Gilani, G.S.; Cockell, K.A.; Sepehr, E. Effects of antinutritional factors on protein digestibility and amino acid availability in foods. J. AOAC Int. 2005, 88, 967–987. [Google Scholar] [CrossRef]
- Bell, J.M. Factors affecting the nutritional value of canola meal: A review. Can. J. Anim. Sci. 1993, 73, 689–697. [Google Scholar] [CrossRef]
- Hannoufa, A.; Pillai, B.V.S.; Chellamma, S. Genetic enhancement of Brassica napus seed quality. Transgenic Res. 2014, 23, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Harloff, H.J.; Lemcke, S.; Mittasch, J.; Frolov, A.; Wu, J.G.; Dreyer, F.; Leckband, G.; Jung, C. A mutation screening platform for rapeseed (Brassica napus L.) and the detection of sinapine biosynthesis mutants. Theor. Appl. Genet. 2012, 124, 957–969. [Google Scholar] [CrossRef]
- Zhai, Y.; Yu, K.; Cai, S.; Hu, L.; Amoo, O.; Xu, L.; Yang, Y.; Ma, B.; Jiao, Y.; Zhang, C.; et al. Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 2019, 18, pbi.13281. [Google Scholar] [CrossRef]
- Sashidhar, N.; Harloff, H.; Jung, C. Identification of phytic acid mutants in oilseed rape ( Brassica napus) by large scale screening of mutant populations through amplicon sequencing. New Phytol. 2019, 225, 2022–2034. [Google Scholar] [CrossRef] [PubMed]
- Bérot, S.; Compoint, J.P.; Larré, C.; Malabat, C.; Guéguen, J. Large scale purification of rapeseed proteins (Brassica napus L.). J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2005, 818, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Steeves, T.A. The evolution and biological significance of seeds. Can. J. Bot. 1983, 61, 3550–3560. [Google Scholar] [CrossRef]
- Crouch, M.L.; Sussex, I.M. Development and storage-protein synthesis in Brassica napus L. embryos in vivo and in vitro. Planta 1981, 153, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Norton, G.; Harris, J.F. Compositional changes in developing rape seed (Brassica napus L.). Planta 1975, 123, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Lord, E.M.; Russell, S.D. The mechanisms of pollination and fertilization in plants. Annu. Rev. Cell Dev. Biol. 2002, 18, 81–105. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.; Slane, D.; Herud, O.; Kong, J.; Jürgens, G. Early embryogenesis in flowering plants: Setting up the basic body pattern. Annu. Rev. Plant Biol. 2012, 63, 483–506. [Google Scholar] [CrossRef]
- Capron, A.; Chatfield, S.; Provart, N.; Berleth, T. Embryogenesis: Pattern formation from a single cell. Arab. B. 2009, 7, e0126. [Google Scholar] [CrossRef]
- Tykarska, T. Rape embryogenesis I. The proembryo development. Acta Soc. Bot. Pol. 1976, 45, 3–16. [Google Scholar] [CrossRef]
- Tykarska, T. Rape embryogenesis. II. Development of embryo proper. Acta Soc. Bot. Pol. 1979, 48, 391–421. [Google Scholar] [CrossRef]
- Ilić-Grubor, K.; Attree, S.M.; Fowke, L.C. Comparative morphological study of zygotic and microspore-derived embryos of Brassica napus L. as revealed by scanning electron microscopy. Ann. Bot. 1998, 82, 157–165. [Google Scholar] [CrossRef]
- Fernandez, D.E.; Turner, F.R.; Crouch, M.L. In situ localization of storage protein mRNAs in developing meristems of Brassica napus embryos. Development 1991, 111, 299–313. [Google Scholar] [CrossRef]
- Tykarska, T. Rape embryogenesis. III. Embryo development in time. Acta Soc. Bot. Pol. 1980, 49, 369–385. [Google Scholar] [CrossRef]
- Yeung, E.C.; Rahman, M.H.; Thorpe, T.A. Comparative development of zygotic and microspore-derived embryos in Brassica napus L. cv Topas. I. Histodifferentiation. Int. J. Plant Sci. 1996, 157, 27–39. [Google Scholar] [CrossRef]
- Shewry, P.R.; Halford, N.G. Cereal seed storage proteins: Structures, properties and role in grain utilization. J. Exp. Bot. 2002, 53, 947–958. [Google Scholar] [CrossRef]
- Wanasundara, J.P.D. Proteins of brassicaceae oilseeds and their potential as a plant protein source. Crit. Rev. Food Sci. Nutr. 2011, 51, 635–677. [Google Scholar] [CrossRef]
- Galland, M.; Huguet, R.; Arc, E.; Cueff, G.; Job, D.; Rajjou, L. Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol. Cell. Proteom. 2014, 13, 252–268. [Google Scholar] [CrossRef] [PubMed]
- Herman, E.M.; Larkins, B.A. Protein storage bodies and vacoules. Plant Cell 1999, 11, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Job, C.; Rajjou, L.; Lovigny, Y.; Belghazi, M.; Job, D.; Loïc, R.; Yoann, L.; Maya, B.; Dominique, J.; Lovigny, Y.; et al. Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol. 2005, 138, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Bieker, S.; Riester, L.; Doll, J.; Franzaring, J.; Fangmeier, A.; Zentgraf, U. Nitrogen supply drives senescence-related seed storage protein expression in rapeseed leaves. Genes 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Osborne, T.B.; Osbornb, T.B. The Vegetable Proteins, 2nd ed.; Plimmer, R.H.A., Hopkins, F.G., Eds.; Longmans, Green and Co.: London, UK, 1924. [Google Scholar]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 2008, 9, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Jolivet, P.; Boulard, C.; Bellamy, A.; Larré, C.; Barre, M.; Rogniaux, H.; D’Andréa, S.; Chardot, T.; Nesi, N. Protein composition of oil bodies from mature Brassica napus seeds. Proteomics 2009, 9, 3268–3284. [Google Scholar] [CrossRef]
- Kubala, S.; Garnczarska, M.; Wojtyla, Ł.; Clippe, A.; Kosmala, A.; Żmieńko, A.; Lutts, S.; Quinet, M. Deciphering priming-induced improvement of rapeseed (Brassica napus L.) germination through an integrated transcriptomic and proteomic approach. Plant Sci. 2015, 231, 94–113. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; He, D.; Gupta, R.; Yang, P. Physiological and proteomic analyses on artificially aged Brassica napus seed. Front. Plant Sci. 2015, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hajduch, M.; Casteel, J.E.; Hurrelmeyer, K.E.; Song, Z.; Agrawal, G.K.; Thelen, J.J. Proteomic analysis of seed filling in Brassica napus. Developmental characterization of metabolic isozymes using high-resolution two-dimensional gel electrophoresis. Plant Physiol. 2006, 141, 32–46. [Google Scholar] [CrossRef]
- Borisjuk, L.; Neuberger, T.; Schwender, J.; Heinzel, N.; Sunderhaus, S.; Fuchs, J.; Hay, J.O.; Tschiersch, H.; Braun, H.-P.; Denolf, P.; et al. Seed architecture shapes embryo metabolism in oilseed rape. Plant Cell 2013, 25, 1625–1640. [Google Scholar] [CrossRef] [PubMed]
- Hoglund, A.-S.; Rodin, J.; Larsson, E.; Rask, L.; Höglund, A.S.; Rödin, J.; Larsson, E.; Rask, L. Distribution of napin and cruciferin in developing rape seed embryos. Plant Physiol. 1992, 98, 509–515. [Google Scholar] [CrossRef]
- Sjödahl, S.; Gustavsson, H.O.; Rödin, J.; Lenman, M.; Höglund, A.S.; Rask, L. Cruciferin gene families are expressed coordinately but with tissue-specific differences during Brassica napus seed development. Plant Mol. Biol. 1993, 23, 1165–1176. [Google Scholar] [CrossRef]
- Tandang-Silvas, M.R.G.; Fukuda, T.; Fukuda, C.; Prak, K.; Cabanos, C.; Kimura, A.; Itoh, T.; Mikami, B.; Utsumi, S.; Maruyama, N. Conservation and divergence on plant seed 11S globulins based on crystal structures. Biochim. Biophys. Acta Proteins Proteomics 2010, 1804, 1432–1442. [Google Scholar] [CrossRef]
- Nietzel, T.; Dudkina, N.V.; Haase, C.; Denolf, P.; Semchonok, D.A.; Boekema, E.J.; Braun, H.P.; Sunderhaus, S. The native structure and composition of the cruciferin complex in Brassica napus. J. Biol. Chem. 2013, 288, 2238–2245. [Google Scholar] [CrossRef]
- Müntz, K. Deposition of storage proteins. Plant Mol. Biol. 1998, 38, 77–99. [Google Scholar] [CrossRef] [PubMed]
- Jung, R.; Paul Scott, M.; Nam, Y.W.; Beaman, T.W.; Bassüner, R.; Saalbach, I.; Müntz, K.; Nielsen, N.C. The role of proteolysis in the processing and assembly of 11S seed globulins. Plant Cell 1998, 10, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Aider, M.; Barbana, C. Canola proteins: Composition, extraction, functional properties, bioactivity, applications as a food ingredient and allergenicity—A practical and critical review. Trends Food Sci. Technol. 2011, 22, 21–39. [Google Scholar] [CrossRef]
- Kim, J.H.; Varankovich, N.V.; Stone, A.K.; Nickerson, M.T. Nature of protein-protein interactions during the gelation of canola protein isolate networks. Food Res. Int. 2016, 89, 408–414. [Google Scholar] [CrossRef]
- Pudel, F.; Tressel, R.P.; Düring, K. Production and properties of rapeseed albumin. Lipid Technol. 2015, 27, 112–114. [Google Scholar] [CrossRef]
- Byczyńska, A.; Barciszewski, J. The biosynthesis, structure and properties of napin—The storage protein from rape seeds. J. Plant Physiol. 1999, 154, 417–425. [Google Scholar] [CrossRef]
- Lönnerdal, B.; Janson, J.-C. Studies on Brassica seed proteins. Biochim. Biophys. Acta Protein Struct. 1972, 278, 175–183. [Google Scholar] [CrossRef]
- Ericson, M.L.; Rodin, J.; Lenman, M.; Glimelius, K.; Josefsson, L.G.; Rask, L.; Rödin, J.; Lenman, M.; Glimelius, K.; Josefsson, L.G.; et al. Structure of the rapeseed 1.7 S storage protein, napin, and its precursor. J. Biol. Chem. 1986, 261, 14576–14581. [Google Scholar] [CrossRef]
- Mylne, J.S.; Hara-Nishimura, I.; Rosengren, K.J. Seed storage albumins: Biosynthesis, trafficking and structures. Funct. Plant Biol. 2014, 41, 671–677. [Google Scholar] [CrossRef]
- Boutilier, K.; Hattori, J.; Baum, B.R.; Miki, B.L. Evolution of 2S albumin seed storage protein genes in the Brassicaceae. Biochem. Syst. Ecol. 1999, 27, 223–234. [Google Scholar] [CrossRef]
- Josefsson, L.G.; Lenman, M.; Ericson, M.L.; Rask, L. Structure of a gene encoding the 1.7 S storage protein, napin, from Brassica napus. J. Biol. Chem. 1987, 262, 12196–12201. [Google Scholar] [CrossRef]
- Scofield, S.R.; Crouch, M.L. Nucleotide sequence of a member of the napin storage protein family from Brassica napus. J. Biol. Chem. 1987, 262, 12202–12208. [Google Scholar] [CrossRef]
- José-Estanyol, M.; Gomis-Rüth, F.X.; Puigdomènech, P. The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol. Biochem. 2004, 42, 355–365. [Google Scholar] [CrossRef]
- Malabat, C.; nchez-Vioque, R.I.S.; Rabiller, C.; Gu guen, J. Emulsifying and foaming properties of native and chemically modified peptides from the 2S and 12S proteins of rapeseed (Brassica napus L.). J. Am. Oil Chem. Soc. 2001, 78, 235–242. [Google Scholar] [CrossRef]
- Schwartz, J.M.; Solé, V.; Guéguen, J.; Ropers, M.H.; Riaublanc, A.; Anton, M. Partial replacement of β-casein by napin, a rapeseed protein, as ingredient for processed foods: Thermoreversible aggregation. LWT Food Sci. Technol. 2015, 63, 562–568. [Google Scholar] [CrossRef]
- Malabat, C.; Atterby, H.; Chaudhry, Q.; Renard, M.; Guéguen, J.; De Recherche, U.; Végétales, P.; Kingdom, U.; Rheu, L. Genetic variability of rapeseed protein composition. In Proceedings of 11th International Rapeseed Congress, Royal Veterinary and Agricultural University, Frederiksberg, Denmark, 6 July 2003. [Google Scholar]
- Kawakatsu, T.; Takaiwa, F. Cereal seed storage protein synthesis: Fundamental processes for recombinant protein production in cereal grains. Plant Biotechnol. J. 2010, 8, 939–953. [Google Scholar] [CrossRef]
- Le Signor, C.; Aimé, D.; Bordat, A.; Belghazi, M.; Labas, V.; Gouzy, J.; Young, N.D.; Prosperi, J.M.; Leprince, O.; Thompson, R.D.; et al. Genome-wide association studies with proteomics data reveal genes important for synthesis, transport and packaging of globulins in legume seeds. New Phytol. 2017, 214, 1597–1613. [Google Scholar] [CrossRef] [PubMed]
- Members of the Complex Trait Consortium The nature and identification of quantitative trait loci: A community’s view. Nat. Rev. Genet. 2003, 4, 911–916. [CrossRef]
- Zhao, J.; Becker, H.C.; Zhang, D.; Zhang, Y.; Ecke, W. Conditional QTL mapping of oil content in rapeseed with respect to protein content and traits related to plant development and grain yield. Theor. Appl. Genet. 2006, 113, 33–38. [Google Scholar] [CrossRef]
- Würschum, T.; Liu, W.; Maurer, H.P.; Abel, S.; Reif, J.C. Dissecting the genetic architecture of agronomic traits in multiple segregating populations in rapeseed (Brassica napus L.). Theor. Appl. Genet. 2012, 124, 153–161. [Google Scholar] [CrossRef]
- Teh, L.; Möllers, C. Genetic variation and inheritance of phytosterol and oil content in a doubled haploid population derived from the winter oilseed rape Sansibar × Oase cross. Theor. Appl. Genet. 2016, 129, 181–199. [Google Scholar] [CrossRef] [PubMed]
- Behnke, N.; Suprianto, E.; Möllers, C. A major QTL on chromosome C05 significantly reduces acid detergent lignin (ADL) content and increases seed oil and protein content in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 2018, 131, 2477–2492. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.; Wang, H.; Wang, X.; Guo, L.; Gu, J.; Zhao, W.; Li, B.; Chen, D.; Raboanatahiry, N.; Li, M. Genetic dissection of seed oil and protein content and identification of networks associated with oil content in Brassica napus. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, W.; Yu, K.; Sun, L.; Gao, J.; Zhou, X.; Peng, Q.; Fu, S.; Hu, M.; Long, W.; et al. Unconditional and conditional QTL analyses of seed fatty acid composition in Brassica napus L. BMC Plant Biol. 2018, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Chao, H.; Wang, H.; Zhao, W.; Zhang, L.; Raboanatahiry, N.; Wang, X.; Wang, B.; Jia, H.; Li, M. Stable, environmental specific and novel QTL identification as well as genetic dissection of fatty acid metabolism in Brassica napus. Front. Plant Sci. 2018, 9, 1–25. [Google Scholar] [CrossRef]
- Toroser, D.; Thormann, C.E.; Osborn, T.C.; Mithen, R. RFLP mapping of quantitative trait loci controlling seed aliphatic-glucosinolate content in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 1995, 91, 802–808. [Google Scholar] [CrossRef]
- Uzunova, M.; Ecke, W.; Weissleder, K.; Röbbelen, G. Mapping the genome of rapeseed (Brassica napus L.). I. Construction of an RFLP linkage map and localization of QTLs for seed glucosinolate content. Theor. Appl. Genet. 1995, 90, 194–204. [Google Scholar] [CrossRef]
- Howell, P.M.; Sharpe, A.G.; Lydiate, D.J. Homoeologous loci control the accumulation of seed glucosinolates in oilseed rape (Brassica napus). Genome 2003, 46, 454–460. [Google Scholar] [CrossRef]
- Schatzki, J.; Ecke, W.; Becker, H.C.; Möllers, C. Mapping of QTL for the seed storage proteins cruciferin and napin in a winter oilseed rape doubled haploid population and their inheritance in relation to other seed traits. Theor. Appl. Genet. 2014, 127, 1213–1222. [Google Scholar] [CrossRef]
- Wen, J.; Xu, J.F.; Long, Y.; Wu, J.G.; Xu, H.M.; Meng, J.L.; Shi, C.H. QTL mapping based on the embryo and maternal genetic systems for non-essential amino acids in rapeseed (Brassica napus L.) meal. J. Sci. Food Agric. 2016, 96, 465–473. [Google Scholar] [CrossRef]
- Fauteux, F.; Strömvik, M.V. Seed storage protein gene promoters contain conserved DNA motifs in Brassicaceae, Fabaceae and Poaceae. BMC Plant Biol. 2009, 9, 1–11. [Google Scholar] [CrossRef]
- Sjödahl, S.; Gustavsson, H.O.; Rödin, J.; Rask, L. Deletion analysis of the Brassica napus cruciferin gene cru 1 promoter in transformed tobacco: Promoter activity during early and late stages of embryogenesis is influenced by cis-acting elements in partially separate regions. Planta 1995, 197, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Stålberg, K.; Ellerstöm, M.; Ezcurra, I.; Ablov, S.; Rask, L. Disruption of an overlapping E-box/ABRE motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta 1996, 199, 515–519. [Google Scholar] [CrossRef]
- Parcy, F.; Valon, C.; Raynal, M.; Gaubier-Comella, P.; Delseny, M.; Giraudat, J. Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 1994, 6, 1567–1582. [Google Scholar] [CrossRef]
- Kroj, T.; Savino, G.; Valon, C.; Giraudat, J.; Parcy, F. Regulation of storage protein gene expression in Arabidopsis. Development 2003, 130, 6065–6073. [Google Scholar] [CrossRef]
- Lara, P.; Oñate-Sánchez, L.; Abraham, Z.; Ferrándiz, C.; Díaz, I.; Carbonero, P.; Vicente-Carbajosa, J. Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J. Biol. Chem. 2003, 278, 21003–21011. [Google Scholar] [CrossRef] [PubMed]
- Ezcurra, I.; Wycliffe, P.; Nehlin, L.; Ellerström, M.; Rask, L. Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 2000, 24, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Vicient, C.M.; Bies-Etheve, N.; Delseny, M. Changes in gene expression in the leafy cotyledon1 (lec1) and fusca3 (fus3) mutants of Arabidopsis thaliana L. J. Exp. Bot. 2000, 51, 995–1003. [Google Scholar] [CrossRef]
- Kagaya, Y.; Toyoshima, R.; Okuda, R.; Usui, H.; Yamamoto, A.; Hattori, T. LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 2005, 46, 399–406. [Google Scholar] [CrossRef]
- Pelletier, J.M.; Kwong, R.W.; Park, S.; Le, B.H.; Baden, R.; Cagliari, A.; Hashimoto, M.; Munoz, M.D.; Fischer, R.L.; Goldberg, R.B.; et al. LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. USA 2017, 114, E6710–E6719. [Google Scholar] [CrossRef]
- Gao, C.; Qi, S.; Liu, K.; Li, D.; Jin, C.; Li, Z.; Huang, G.; Hai, J.; Zhang, M.; Chen, M. MYC2, MYC3, and MYC4 function redundantly in seed storage protein accumulation in Arabidopsis. Plant Physiol. Biochem. 2016, 108, 63–70. [Google Scholar] [CrossRef]
- Verdier, J.; Thompson, R.D. Transcriptional regulation of storage protein synthesis during dicotyledon seed filling. Plant Cell Physiol. 2008, 49, 1263–1271. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 1–26. [Google Scholar] [CrossRef] [PubMed]
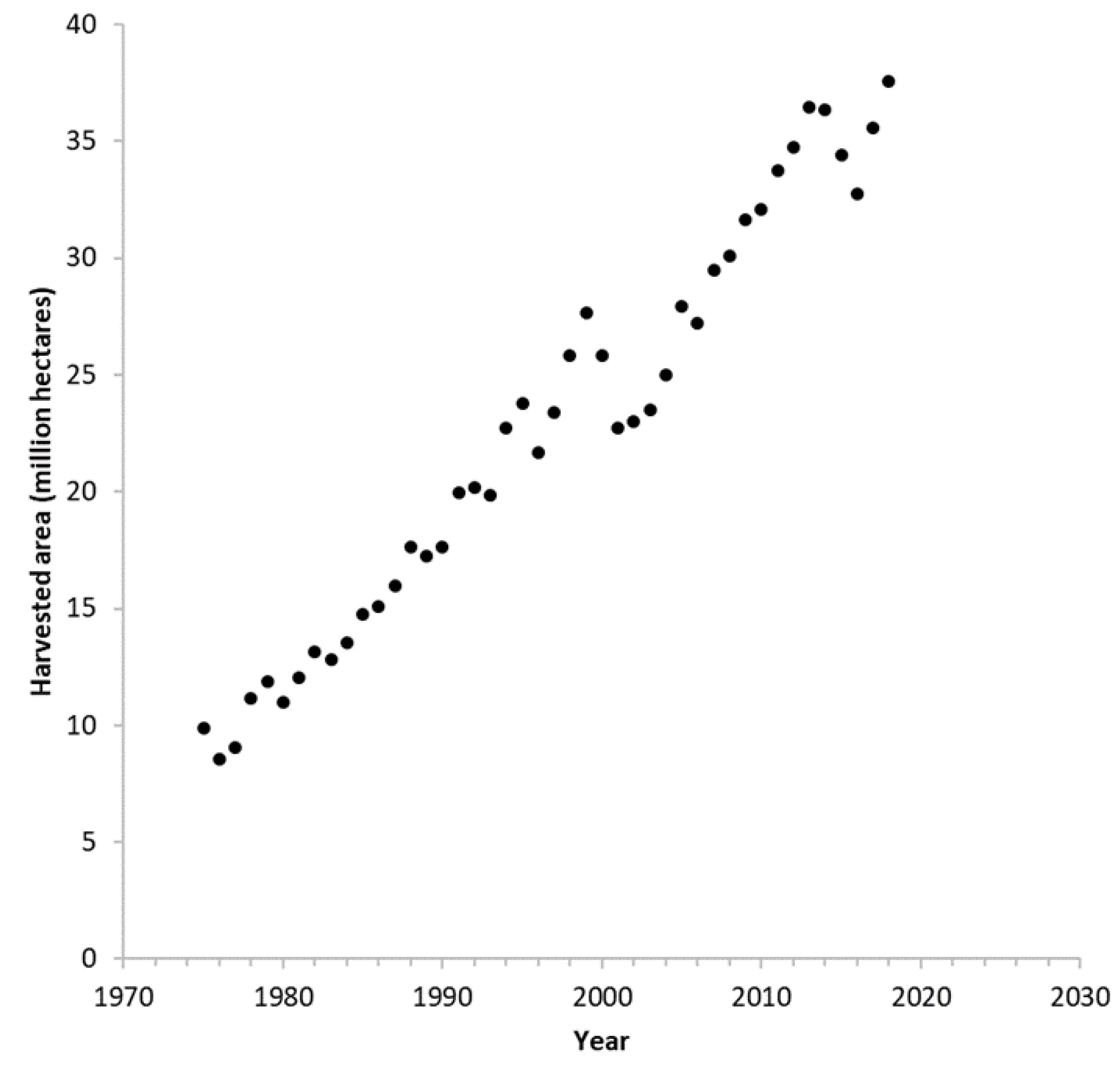
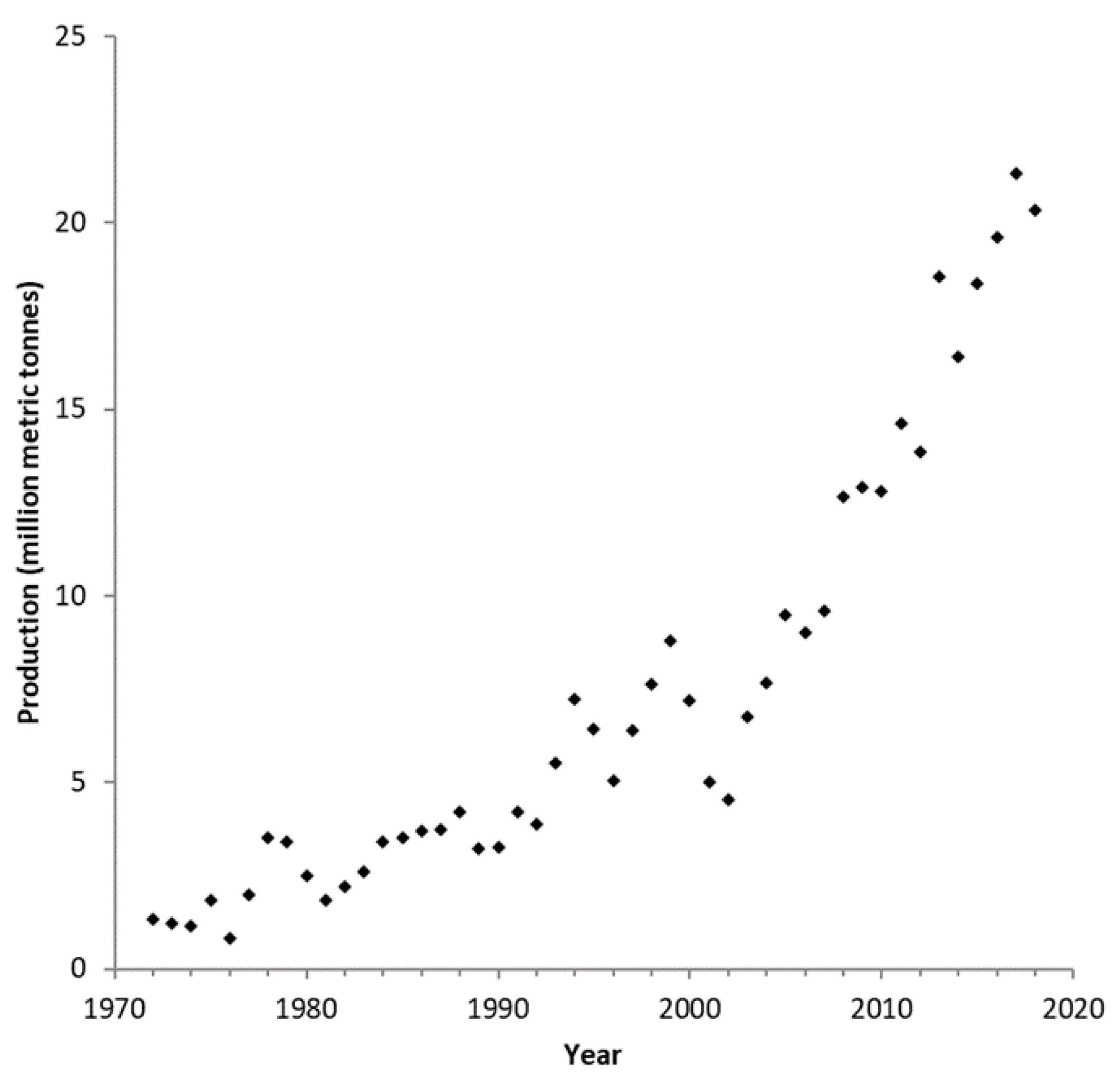
- Barthet, V.J. Comparison between Canadian canola harvest and export surveys. Plants 2016, 5, 1209–1214. [Google Scholar] [CrossRef]
- Triboï, E.; Martre, P.; Triboï-Blondel, A.M.; Croue, S. De Environmentally-induced changes in protein composition in developing grains of wheat are related to changes in total protein content. J. Exp. Bot. 2003, 54, 1731–1742. [Google Scholar] [CrossRef]
- Staswick, P.E.; Huang, J.F.; Rhee, Y. Nitrogen and methyl jasmonate induction of soybean vegetative storage protein genes. Plant Physiol. 1991, 96, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Paek, N.C.; Sexton, P.J.; Naeve, S.L.; Shibles, R. Differential accumulation of soybean seed storage protein subunits in response to sulfur and nitrogen nutritional sources. Plant Prod. Sci. 2000, 3, 268–274. [Google Scholar] [CrossRef]
- Borpatragohain, P.; Rose, T.J.; King, G.J. Fire and brimstone: Molecular interactions between sulfur and glucosinolate biosynthesis in model and crop brassicaceae. Front. Plant Sci. 2016, 7, 1–18. [Google Scholar] [CrossRef]
- Wang, Z.-H.; Li, S.-X.; Malhi, S. Effects of fertilization and other agronomic measures on nutritional quality of crops. J. Sci. Food Agric. 2008, 88, 7–23. [Google Scholar] [CrossRef]
- Grant, C.A.; Clayton, G.W.; Johnston, A.M. Sulphur fertilizer and tillage effects on canola seed quality in the Black soil zone of western Canada. Can. J. Plant Sci. 2003, 83, 745–758. [Google Scholar] [CrossRef]
- Poisson, E.; Trouverie, J.; Brunel-Muguet, S.; Akmouche, Y.; Pontet, C.; Pinochet, X.; Avice, J.C. Seed yield components and seed quality of oilseed rape are impacted by sulfur fertilization and its interactions with nitrogen fertilization. Front. Plant Sci. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Chardigny, J.-M.M.; Walrand, S. Plant protein for food: Opportunities and bottlenecks. OCL Oilseeds fats, Crop. Lipids 2016, 23, D404. [Google Scholar] [CrossRef]
- Mejia, L.A.; Korgaonkar, C.K.; Schweizer, M.; Chengelis, C.; Marit, G.; Ziemer, E.; Grabiel, R.; Empie, M. A 13-week sub-chronic dietary toxicity study of a cruciferin-rich canola protein isolate in rats. Food Chem. Toxicol. 2009, 47, 2645–2654. [Google Scholar] [CrossRef] [PubMed]
- Mejia, L.A.; Korgaonkar, C.K.; Schweizer, M.; Chengelis, C.; Novilla, M.; Ziemer, E.; Williamson-Hughes, P.S.; Grabiel, R.; Empie, M. A 13-week dietary toxicity study in rats of a napin-rich canola protein isolate. Regul. Toxicol. Pharmacol. 2009, 55, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Masilamani, M.; Commins, S.; Shreffler, W. Determinants of food allergy. Immunol. Allergy Clin. North Am. 2012, 32, 11–33. [Google Scholar] [CrossRef]
- Moreno, F.J.; Clemente, A. 2S Albumin Storage Proteins: What Makes them Food Allergens? Open Biochem. J. 2008, 2, 16–28. [Google Scholar] [CrossRef]
- Sicherer, S.H.; Sampson, H.A. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J. Allergy Clin. Immunol. 2018, 141, 41–58. [Google Scholar] [CrossRef]
- Waserman, S.; Bégin, P.; Watson, W. IgE-mediated food allergy. Clin. Rev. Allergy Immunol. 2019, 57, 244–260. [Google Scholar] [CrossRef]
- Gould, H.J.; Sutton, B.J. IgE in allergy and asthma today. Nat. Rev. Immunol. 2008, 8, 205–217. [Google Scholar] [CrossRef]
- Monsalve, R.I.; Gonzalez de la Pena, M.A.; Lopez-Otiin, C.; Fiandor, A.; Fernandez, C.; Villalba, M.; Rodriguez, R. Detection, isolation and complete amino acid sequence of an aeroallergenic protein from rapeseed flour. Clin Exp Allergy 1997, 27, 833–841. [Google Scholar] [CrossRef]
- Puumalainen, T.J.; Puustinen, A.; Poikonen, S.; Turjanmaa, K.; Palosuo, T.; Vaali, K. Proteomic identification of allergenic seed proteins, napin and cruciferin, from cold-pressed rapeseed oils. Food Chem. 2015, 175, 381–385. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Dietetic Products Nutrition and Allergies Scientific Opinion on the safety of “rapeseed protein isolate” as a Novel Food ingredient. EFSA J. 2013, 11, 1–23. [CrossRef]
- Fiocchi, A.; Dahdah, L.; Riccardi, C.; Mazzina, O.; Fierro, V. Preacutionary labelling of cross-reactive foods: The case of rapeseed. Asthma Res. Pract. 2016, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bonds, R.S.; Midoro-Horiuti, T.; Goldblum, R. A structural basis for food allergy: The role of cross-reactivity. Curr. Opin. Allergy Clin. Immunol. 2008, 8, 82–86. [Google Scholar] [CrossRef] [PubMed]
- L’Hocine, L.; Pitre, A.; Pitre, M. Detection and identification of allergens from Canadian mustard varieties of Sinapis alba and Brassica juncea. Biomolecules 2019, 9, 489. [Google Scholar] [CrossRef] [PubMed]
- Government of Canada Regulations amending the food and drug regulations (1220—Enhanced labeling for food allergen and gluten sources and added sulphites). Canada Gaz. Part II 2011, 145, 325.
- Mustard: A Priority Food Allergen in Canada 2009 . Available online: https://www.canada.ca/en/health-canada/services/food-nutrition/reports-publications/food-labelling/mustard-priority-food-allergen-canada-systematic-review.html (accessed on 10 October 2021).
- Marambe, H.K.; McIntosh, T.C.; Cheng, B.; Wanasundara, J.P.D.D. Quantification of major 2S allergen protein of yellow mustard using anti-Sin a 1 epitope antibody. Food Control 2014, 44, 233–241. [Google Scholar] [CrossRef]
- Gylling, H. Rapeseed oil does not cause allergic reactions. Allergy Eur. J. Allergy Clin. Immunol. 2006, 61, 895. [Google Scholar] [CrossRef]
- Paul, R.H.; Frestedt, J.; Magurany, K. GRAS from the ground up: Review of the Interim Pilot Program for GRAS notification. Food Chem. Toxicol. 2017, 105, 140–150. [Google Scholar] [CrossRef] [PubMed]
- van der Spiegel, M.; Noordam, M.Y.; van der Fels-Klerx, H.J. Safety of novel protein sources (insects, microalgae, seaweed, duckweed, and rapeseed) and legislative aspects for their application in food and feed production. Compr. Rev. Food Sci. Food Saf. 2013, 12, 662–678. [Google Scholar] [CrossRef]
- Day, L. Proteins from land plants—Potential resources for human nutrition and food security. Trends Food Sci. Technol. 2013, 32, 25–42. [Google Scholar] [CrossRef]
- Kim, S.W.; Less, J.F.; Wang, L.; Yan, T.; Kiron, V.; Kaushik, S.J.; Lei, X.G. Meetin global feed protein demand: Challenge, opportunity, and strategy. Annu. Rev. Anim. Biosci. 2019, 7, 221–243. [Google Scholar] [CrossRef]
- Lomascolo, A.; Uzan-Boukhris, E.; Sigoillot, J.C.; Fine, F. Rapeseed and sunflower meal: A review on biotechnology status and challenges. Appl. Microbiol. Biotechnol. 2012, 95, 1105–1114. [Google Scholar] [CrossRef]
- Thomke, S. Review of rapeseed meal in animal nutrition: Ruminant animals. J. Am. Oil Chem. Soc. 1981, 58, 805–810. [Google Scholar] [CrossRef]
- Swanepoel, N.; Robinson, P.H.; Erasmus, L.J. Determining the optimal ratio of canola meal and high protein dried distillers grain protein in diets of high producing Holstein dairy cows. Anim. Feed Sci. Technol. 2014, 189, 41–53. [Google Scholar] [CrossRef][Green Version]
- Huhtanen, P.; Hetta, M.; Swensson, C. Evaluation of canola meal as a protein supplement for dairy cows: A review and a meta-analysis. Can. J. Anim. Sci. 2012, 91, 529–543. [Google Scholar] [CrossRef]
- He, M.L.; Gibb, D.; McKinnon, J.J.; McAllister, T.A. Effect of high dietary levels of canola meal on growth performance, carcass quality and meat fatty acid profiles of feedlot cattle. Can. J. Anim. Sci. 2013, 93, 269–280. [Google Scholar] [CrossRef]
- Mejicanos, G.; Sanjayan, N.; Kim, I.H.; Nyachoti, C.M. Recent advances in canola meal utilization in swine nutrition. J. Anim. Sci. Technol. 2016, 58, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Landero, J.L.; Beltranena, E.; Zijlstra, R.T. Growth performance and preference studies to evaluate solvent-extracted Brassica napus or Brassica juncea canola meal fed to weaned pigs. J. Anim. Sci. 2012, 90, 406–408. [Google Scholar] [CrossRef] [PubMed]
- Chai, S.J.; Cole, D.; Nisler, A.; Mahon, B.E. Poultry: The most common food in outbreaks with known pathogens, United States, 1998-2012. Epidemiol. Infect. 2017, 145, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.A.; Islam, A.F.F.; Iji, P.A.A. Energy utilization and performance of broiler chickens raised on diets with vegetable proteins or conventional feeds. Asian J. Poult. Sci. 2012, 6, 117–128. [Google Scholar] [CrossRef]
- Ravindran, V.; Abdollahi, M.R.; Bootwalla, S.M. Nutrient analysis, metabolizable energy, and digestible amino acids of soybean meals of different origins for broilers. Poult. Sci. 2014, 93, 2567–2577. [Google Scholar] [CrossRef] [PubMed]
- Vieira, S.L.; Lima, I.L. Live performance, water intake and excreta characteristics of broilers fed all vegetable diets based on corn and soybean meal. Int. J. Poult. Sci. 2005, 4, 365–368. [Google Scholar] [CrossRef]
- Kasprzak, M.M.; Houdijk, J.G.M.; Liddell, S.; Davis, K.; Olukosi, O.A.; Kightley, S.; White, G.A.; Wiseman, J. Rapeseed napin and cruciferin are readily digested by poultry. J. Anim. Physiol. Anim. Nutr. (Berl). 2017, 101, 658–666. [Google Scholar] [CrossRef]
- Proudfoot, F.C.; Hulan, H.W.; McRae, K.B. Effect of feeding poultry diets supplemented with rapeseed meal as a primary protein source to juvenile and adult meat breeder genotypes. Can. J. Anim. Sci. 1983, 63, 957–965. [Google Scholar] [CrossRef]
- Bryan, D.D.S.L.; Abbott, D.A.; Van Kessel, A.G.; Classen, H.L. In vivo digestion characteristics of protein sources fed to broilers. Poult. Sci. 2019, 98, 3313–3325. [Google Scholar] [CrossRef] [PubMed]
- The State of World Fisheries and Aquaculture 2018 . Available online: https://www.fao.org/policy-support/tools-and-publications/resources-details/en/c/1269332/ (accessed on 10 October 2021).
- Turchini, G.M.; Trushenski, J.T.; Glencross, B.D. Throughts for the future of aquaculture nutrition: REaligning perspectives to reflect contemporary issues related to judicious use of marine resources in aquafeeds. N. Am. J. Aquac. 2019, 81, 13–39. [Google Scholar] [CrossRef]
- Adem, H.N.; Tressel, R.P.; Pudel, F.; Slawski, H.; Schulz, C. Rapeseed use in aquaculture. OCL Oilseeds fats 2014, 21, 1–9. [Google Scholar] [CrossRef]
- Nagel, F.; Slawski, H.; Adem, H.; Tressel, R.P.; Wysujack, K.; Schulz, C. Albumin and globulin rapeseed protein fractions as fish meal alternative in diets fed to rainbow trout (Oncorhynchus mykiss W.). Aquaculture 2012, 354–355, 121–127. [Google Scholar] [CrossRef]
- Slawski, H.; Adem, H.; Tressel, R.P.; Wysujack, K.; Koops, U.; Kotzamanis, Y.; Wuertz, S.; Schulz, C. Total fish meal replacement with rapeseed protein concentrate in diets fed to rainbow trout (Oncorhynchus mykiss Walbaum). Aquac. Int. 2012, 20, 443–453. [Google Scholar] [CrossRef]
- Luo, Z.; Li, X.D.; Wang, W.M.; Tan, X.Y.; Liu, X. Partial replacement of fish meal by a mixture of soybean meal and rapeseed meal in practical diets for juvenile Chinese mitten crab Eriocheir sinensis: Effects on growth performance and in vivo digestibility. Aquac. Res. 2011, 42, 1615–1622. [Google Scholar] [CrossRef]
- Suárez, J.A.; Gaxiola, G.; Mendoza, R.; Cadavid, S.; Garcia, G.; Alanis, G.; Suárez, A.; Faillace, J.; Cuzon, G. Substitution of fish meal with plant protein sources and energy budget for white shrimp Litopenaeus vannamei (Boone, 1931). Aquaculture 2009, 289, 118–123. [Google Scholar] [CrossRef]
- Cruz-Suarez, L.E.; Ricque-Marie, D.; Tapia-Salazar, M.; McCallum, I.M.; Hickling, D. Assessment of differently processed feed pea (Pisum sativum) meals and canola meal (Brassica sp.) in diets for blue shrimp (Litopenaeus stylirostris). Aquaculture 2001, 196, 87–104. [Google Scholar] [CrossRef]
- Kumar, P.; Chatli, M.K.; Mehta, N.; Singh, P.; Malav, O.P.; Verma, A.K. Meat analogues: Health promising sustainable meat substitutes. Crit. Rev. Food Sci. Nutr. 2017, 57, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Hoek, A.C.; Luning, P.A.; Weijzen, P.; Engels, W.; Kok, F.J.; de Graaf, C. Replacement of meat by meat substitutes. A survey on person- and product-related factors in consumer acceptance. Appetite 2011, 56, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, B.L.; Boom, R.M.; van der Goot, A.J. Structuring processes for meat analogues. Trends Food Sci. Technol. 2018, 81, 25–36. [Google Scholar] [CrossRef]
- Wu, J.; Muir, A.D. Comparative structural, emulsifying, and biological properties of 2 major canola proteins, cruciferin and napin. J. Food Sci. 2008, 73. [Google Scholar] [CrossRef]
- Kohno-Murase, J.; Murase, M.; Ichikawa, H.; Imamura, J. Improvement in the quality of seed storage protein by transformation of Brassica napus with an antisense gene for cruciferin. Theor. Appl. Genet. 1995, 91, 627–631. [Google Scholar] [CrossRef]
- Falco, S.C.; Guida, T.; Locke, M.; Mauvais, J.; Sanders, C.; Ward, R.T.; Webber, P. Transgenic Canola and Soyban Seeds with Increased Lysine. Nat. Biotechnol. 1995, 13, 577–582. [Google Scholar] [CrossRef]
- Cheung, L.; Wanasundara, J.; Nickerson, M.T. The Effect of pH and NaCl Levels on the Physicochemical and Emulsifying Properties of a Cruciferin Protein Isolate. Food Biophys. 2014, 9, 105–113. [Google Scholar] [CrossRef]
- Cheung, L.; Wanasundara, J.; Nickerson, M.T. Effect of pH and NaCl on the Emulsifying Properties of a Napin Protein Isolate. Food Biophys. 2014, 10, 30–38. [Google Scholar] [CrossRef]
- Gerzhova, A.; Mondor, M.; Benali, M.; Aider, M. Study of total dry matter and protein extraction from canola meal as affected by the pH, salt addition and use of zeta-potential/turbidimetry analysis to optimize the extraction conditions. Food Chem. 2016, 201, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Congdon, R.W.; Muth, G.W.; Splittgerber, A.G. The binding interaction of Coomassie blue with proteins. Anal. Biochem. 1993, 213, 407–413. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- Moritz, C.P. 40 years Western blotting: A scientific birthday toast. J. Proteomics 2020, 212. [Google Scholar] [CrossRef]
- Janes, K.A. An analysis of critical factors for quantitative immunoblotting. Sci. Signal. 2015, 8, rs2. [Google Scholar] [CrossRef] [PubMed]
- Rödin, J.; Rask, L. Characterization of the 12S storage protein of Brassica napus (cruciferin): Disulfide bonding between subunits. Physiol. Plant. 1990, 79, 421–426. [Google Scholar] [CrossRef]
- Kawagoe, Y.; Suzuki, K.; Tasaki, M.; Yasuda, H.; Akagi, K.; Katoh, E.; Nishizawa, N.K.; Ogawa, M.; Takaiwa, F. The critical role of disulfide bond formation in protein sorting in the endosperm of rice. Plant Cell 2005, 17, 1141–1153. [Google Scholar] [CrossRef]
- Wan, L.; Ross, A.R.S.; Yang, J.; Hegedus, D.D.; Kermode, A.R. Phosphorylation of the 12 S globulin cruciferin in wild-type and abi1-1 mutant Arabidopsis thaliana (thale cress) seeds. Biochem. J. 2007, 404, 247–256. [Google Scholar] [CrossRef]
- Murphy, D.J.; Cummins, I.; Kang, A.S. Synthesis of the major oil-body membrane protein in developing rapeseed (Brassica napus) embryos. Integration with storage-lipid and storage-protein synthesis and implications for the mechanism of oil-body formation. Biochem. J. 1989, 258, 285–293. [Google Scholar] [CrossRef]
- Murphy, D.J.; Cummins, I. Biosynthesis of Seed Storage Products during Embryogenesis in Rapeseed, Brassica napus. J. Plant Physiol. 1989, 135, 63–69. [Google Scholar] [CrossRef]
- Sikdar, M.S.I.; Bowra, S.; Schmidt, D.; Dionisio, G.; Holm, P.B.; Vincze, E. Targeted modification of storage protein content resulting in improved amino acid composition of barley grain. Transgenic Res. 2016, 25, 19–31. [Google Scholar] [CrossRef]
- Gil-Humanes, J.; Pistón, F.; Giménez, M.J.; Martín, A.; Barro, F. The Introgression of RNAi Silencing of γ-Gliadins into Commercial Lines of Bread Wheat Changes the Mixing and Technological Properties of the Dough. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Kim, H.J.; Lee, J.Y.; Yoon, U.H.; Lim, S.H.; Kim, Y.M. Effects of reduced prolamin on seed storage protein composition and the nutritional quality of rice. Int. J. Mol. Sci. 2013, 14, 17073–17084. [Google Scholar] [CrossRef]
- Wu, Y.; Messing, J. Proteome balancing of the maize seed for higher nutritional value. Front. Plant Sci. 2014, 5, 1–5. [Google Scholar] [CrossRef]
- Henderson, I.R.; Salt, D.E. Natural genetic variation and hybridization in plants. J. Exp. Bot. 2017, 68, 5415–5417. [Google Scholar] [CrossRef] [PubMed]
- Carmona, S.; Alvarez, J.B.; Caballero, L. Genetic diversity for morphological traits and seed storage proteins in Spanish rivet wheat. Biol. Plant. 2010, 54, 69–75. [Google Scholar] [CrossRef]
- Liu, S.; Ohta, K.; Dong, C.; Thanh, V.C.; Ishimoto, M.; Qin, Z.; Hirata, Y. Genetic diversity of soybean (Glycine max (L.) Merrill) 7S globulin protein subunits. Genet. Resour. Crop Evol. 2006, 53, 1209–1219. [Google Scholar] [CrossRef]
- Kim, W.S.; Gillman, J.D.; Krishnan, H.B. Identification of a plant introduction soybean line with genetic lesions affecting two distinct glycinin subunits and evaluation of impacts on protein content and composition. Mol. Breed. 2013, 32, 291–298. [Google Scholar] [CrossRef]
- Hu, Y.; Wu, G.; Cao, Y.; Wu, Y.; Xiao, L.; Li, X.; Lu, C. Breeding response of transcript profiling in developing seeds of Brassica napus. BMC Mol. Biol. 2009, 10. [Google Scholar] [CrossRef]
- Rizzo, G.; Baroni, L. Soy, Soy Foods and Their Role in Vegetarian Diets. Nutrients 2018, 10, 43. [Google Scholar] [CrossRef]
- Nishinari, K.; Fang, Y.; Guo, S.; Phillips, G.O. Soy proteins: A review on composition, aggregation and emulsification. Food Hydrocoll. 2014, 39, 301–318. [Google Scholar] [CrossRef]
- Nishinari, K.; Fang, Y.; Nagano, T.; Guo, S.; Wang, R. Soy as a Food Ingredient, 2nd ed.; Yada, R.Y., Ed.; Elsevier Science and Technology: Cambridge, UK, 2018; ISBN 9780081007297. [Google Scholar]
- Singh, P.; Kumar, R.; Sabapathy, S.N.; Bawa, A.S. Functional and edible uses of soy protein products. Compr. Rev. Food Sci. Food Saf. 2008, 7, 14–28. [Google Scholar] [CrossRef]
- Saio, K.; Kamiya, M.; Watanabe, T.; Koyama, E.; Yamazaki, S.; Chem, A.B.; Wolf, W.J.; Babcock, T.E.; Smith, A.K.; Teramachi, Y.; et al. Food processing characteristics of soybean 11s and 7s proteins: Part I. Effect of difference of protein components among soybean varieties on formation of tofu-gel. Agric. Biol. Chem. 1969, 33, 1301–1308. [Google Scholar] [CrossRef]
- Poysa, V.; Woodrow, L.; Yu, K. Effect of soy protein subunit composition on tofu quality. Food Res. Int. 2006, 39, 309–317. [Google Scholar] [CrossRef]
- Zarkadas, C.G.; Gagnon, C.; Poysa, V.; Khanizadeh, S.; Cober, E.R.; Chang, V.; Gleddie, S. Protein quality and identification of the storage protein subunits of tofu and null soybean genotypes, using amino acid analysis, one- and two-dimensional gel electrophoresis, and tandem mass spectrometry. Food Res. Int. 2007, 40, 111–128. [Google Scholar] [CrossRef]
- Takahashi, M.; Uematsu, Y.; Kashiwaba, K.; Yagasaki, K.; Hajika, M.; Matsunaga, R.; Komatsu, K.; Ishimoto, M. Accumulation of high levels of free amino acids in soybean seeds through integration of mutations conferring seed protein deficiency. Planta 2003, 217, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.S.; Krishnan, H.B. Expression of an 11 kDa methionine-rich delta-zein in transgenic soybean results in the formation of two types of novel protein bodies in transitional cells situated between the vascular tissue and storage parenchyma cells. Plant Biotechnol. J. 2004, 2, 199–210. [Google Scholar] [CrossRef]
- Kim, W.-S.; Jez, J.M.; Krishnan, H.B. Effects of proteome rebalancing and sulfur nutrition on the accumulation of methionine rich δ-zein in transgenic soybeans. Front. Plant Sci. 2014, 5, 1–12. [Google Scholar] [CrossRef]
- Schmidt, M.A.; Herman, E.M. Proteome rebalancing in soybean seeds can be exploited to enhance foreign protein accumulation. Plant Biotechnol. J. 2008, 6, 832–842. [Google Scholar] [CrossRef]
- Wieser, H. Chemistry of gluten proteins. Food Microbiol. 2007, 24, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Altenbach, S.B.; Tanaka, C.K.; Pineau, F.; Lupi, R.; Drouet, M.; Beaudouin, E.; Morisset, M.; Denery-Papini, S. Assessment of the allergenic potential of transgenic wheat (Triticum aestivum) with reduced levels of ω5-gliadins, the major sensitizing allergen in wheat-dependent exercise-induced anaphylaxis. J. Agric. Food Chem. 2015, 63, 9323–9332. [Google Scholar] [CrossRef] [PubMed]
- Gil-Humanes, J.; Piston, F.; Tollefsen, S.; Sollid, L.M.; Barro, F. Effective shutdown in the expression of celiac disease-related wheat gliadin T-cell epitopes by RNA interference. Proc. Natl. Acad. Sci. USA 2010, 107, 17023–17028. [Google Scholar] [CrossRef] [PubMed]
- Barro, F.; Rooke, L.; Bekes, F.; Gras, P.; Tatham, A.S.; Fido, R.; Lazzeri, P.A.; Shrewy, P.R.; Barcelo, P.; Tatha, A.S.; et al. Transformation of wheat with high molecular weight subunit genes results in improved functional properties. Nat. Biotechnol. 1997, 15, 871–875. [Google Scholar] [CrossRef] [PubMed]
- Altpeter, F.; Vasil, V.; Srivastava, V.; Vasil, I.K. Integration and expression of the high-molecular-weight glutenin subunit 1Ax1 gene in wheat. Nat. Biotechnol. 1996, 14, 1246–1251. [Google Scholar] [CrossRef] [PubMed]
- Blechl, A.E.; Anderson, O.D. Expression of a novel high-molecular-weight glutenin subunit gene in transgenic wheat. Nat. Biotechnol. 1996, 14, 875–879. [Google Scholar] [CrossRef]
- Altenbach, S.B.; Tanaka, C.K.; Seabourn, B.W. Silencing of omega-5 gliadins in transgenic wheat eliminates a major source of environmental variability and improves dough mixing properties of flour. BMC Plant Biol. 2014, 14. [Google Scholar] [CrossRef]
- Pistón, F.; Gil-Humanes, J.; Rodríguez-Quijano, M.; Barro, F. Down-Regulating γ-Gliadins in bread wheat leads to Non-Specific increases in other gluten proteins and has no major effect on dough gluten strength. PLoS ONE 2011, 6, 1–10. [Google Scholar] [CrossRef]
- Troise, A.D.; Wilkin, J.D.; Fiore, A. Impact of rapeseed press-cake on Maillard reaction in a cookie model system. Food Chem. 2018, 243, 365–372. [Google Scholar] [CrossRef]
- Altenbach, S.B.; Kuo, C.C.; Staraci, L.C.; Pearson, K.W.; Wainwright, C.; Georgescu, A.; Townsend, J. Accumulation of a Brazil nut albumin in seeds of transgenic canola results in enhanced levels of seed protein methionine. Plant Mol. Biol. 1992, 18, 235–245. [Google Scholar] [CrossRef]
- Guerche, P.; De Almeida, E.R.P.; Schwarztein, M.A.; Gander, E.; Krebbers, E.; Pelletier, G. Expression of the 2S albumin from Bertholletia excelsa in Brassica napus. MGG Mol. Gen. Genet. 1990, 221, 306–314. [Google Scholar] [CrossRef]
- Kohno-Murase, J.; Murase, M.; Ichikawa, H.; Imamura, J. Effects of an antisense napin gene on seed storage compounds in transgenic Brassica napus seeds. Plant Mol. Biol. 1994, 26, 1115–1124. [Google Scholar] [CrossRef] [PubMed]
- Rolletschek, H.; Schwender, J.; König, C.; Chapman, K.D.; Romsdahl, T.; Lorenz, C.; Braun, H.P.; Denolf, P.; van Audenhove, K.; Munz, E.; et al. Cellular plasticity in response to suppression of storage proteins in the Brassica napus embryo. Plant Cell 2020, 32, 2383–2401. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.; Batley, J.; Snowdon, R.J. Accessing complex crop genomes with next-generation sequencing. Theor. Appl. Genet. 2013, 126, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and Prospects of Association Mapping in Plants. Plant Genome 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Mason, A.S.; Higgins, E.E.; Snowdon, R.J.; Batley, J.; Stein, A.; Werner, C.; Parkin, I.A.P. A user guide to the Brassica 60K Illumina InfiniumTM SNP genotyping array. Theor. Appl. Genet. 2017, 130, 621–633. [Google Scholar] [CrossRef]
- Clarke, W.E.; Higgins, E.E.; Plieske, J.; Wieseke, R.; Sidebottom, C.; Khedikar, Y.; Batley, J.; Edwards, D.; Meng, J.; Li, R.; et al. A high-density SNP genotyping array for Brassica napus and its ancestral diploid species based on optimised selection of single-locus markers in the allotetraploid genome. Theor. Appl. Genet. 2016, 129, 1887–1899. [Google Scholar] [CrossRef]
- You, Q.; Yang, X.; Peng, Z.; Xu, L.; Wang, J. Development and applications of a high throughput genotyping tool for polyploid crops: Single nucleotide polymorphism (SNP) array. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 2008, 3, 1–7. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, 1–10. [Google Scholar] [CrossRef]
- Scheben, A.; Batley, J.; Edwards, D. Genotyping-by-sequencing approaches to characterize crop genomes: Choosing the right tool for the right application. Plant Biotechnol. J. 2017, 15, 149–161. [Google Scholar] [CrossRef]
- Myles, S.; Peiffer, J.; Brown, P.J.; Ersoz, E.S.; Zhang, Z.; Costich, D.E.; Buckler, E. Association mapping: Critical considerations shift from genotyping to experimental design. Plant Cell 2009, 21, 2194–2202. [Google Scholar] [CrossRef]
- Xu, Y.; Li, P.; Yang, Z.; Xu, C. Genetic mapping of quantitative trait loci in crops. Crop J. 2017, 5, 175–184. [Google Scholar] [CrossRef]
- Cockram, J.; Mackay, I. Genetic Mapping Populations for Conducting High-Resolution Trait Mapping in Plants. Adv. Biochem. Eng. Biotechnol. 2018, 164, 109–138. [Google Scholar]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop Breeding Chips and Genotyping Platforms: Progress, Challenges, and Perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef]
- Lorieux, M. MapDisto: Fast and efficient computation of genetic linkage maps. Mol. Breed. 2012, 30, 1231–1235. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Soto-Cerda, B.J. Association Mapping in Plant Genomes. In Genetic Diversity in Plants; InTech: London, UK, 2012. [Google Scholar]
- Yu, J.; Buckler, E.S. Genetic association mapping and genome organization of maize. Curr. Opin. Biotechnol. 2006, 17, 155–160. [Google Scholar] [CrossRef]
- Risch, N.; Merikangas, K. The future of genetic studies of complex human diseases. Science 1996, 273, 1516–1517. [Google Scholar] [CrossRef]
- Thornsberry, J.M.; Goodman, M.M.; Doebley, J.; Kresovich, S.; Nielsen, D.; Buckler, E.S. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 2001, 28, 286–289. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.J.; Yan, J. Crop genome-wide association study: A harvest of biological relevance. Plant J. 2019, 97, 8–18. [Google Scholar] [CrossRef]
- Tian, D.; Wang, P.; Tang, B.; Teng, X.; Li, C.; Liu, X.; Zou, D.; Song, S.; Zhang, Z. GWAS Atlas: A curated resource of genome-wide variant-trait associations in plants and animals. Nucleic Acids Res. 2020, 48, D927–D932. [Google Scholar] [CrossRef] [PubMed]
- Ersoz, E.S.; Yu, J.; Buckler, E.S. Applications of Linkage Disequilibrium and Association Mapping in Maize. In Molecular Genetic Approaches to Maize Improvement; Springer Berlin Heidelberg: Berlin, Heidelberg, 2009; pp. 173–195. [Google Scholar]
- Li, J.; Bus, A.; Spamer, V.; Stich, B. Comparison of statistical models for nested association mapping in rapeseed (Brassica napus L.) through computer simulations. BMC Plant Biol. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Han, B. Natural Variations and Genome-Wide Association Studies in Crop Plants. Annu. Rev. Plant Biol. 2014, 65, 531–551. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, X.; Wang, J.; Li, M.; Wang, Q.; Tian, F.; Su, Z.; Pan, Y.; Liu, D.; Lipka, A.E.; et al. GAPIT version 2: An enhanced integrated tool for genomic association and prediction. Plant Genome 2016, 9. [Google Scholar] [CrossRef]
- Chen, P.; Shen, Z.; Ming, L.; Li, Y.; Dan, W.; Lou, G.; Peng, B.; Wu, B.; Li, Y.; Zhao, D.; et al. Genetic basis of variation in rice seed storage protein (Albumin, globulin, prolamin, and glutelin) content revealed by genome-wide association analysis. Front. Plant Sci. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Deng, M.; Li, D.; Luo, J.; Xiao, Y.; Liu, H.; Pan, Q.; Zhang, X.; Jin, M.; Zhao, M.; Yan, J. The genetic architecture of amino acids dissection by association and linkage analysis in maize. Plant Biotechnol. J. 2017, 15, 1250–1263. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Liu, H.; Chen, J.; Shi, T.; Zhang, C.; Sun, D.; He, Z.; Hao, Y.; Chen, W. Genome-wide association studies of free amino acid levels by six multi-locus models in bread wheat. Front. Plant Sci. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Lee, S.; Van, K.; Sung, M.; Nelson, R.; LaMantia, J.; McHale, L.K.; Mian, M.A.R. Genome-wide association study of seed protein, oil and amino acid contents in soybean from maturity groups I to IV. Theor. Appl. Genet. 2019, 132, 1639–1659. [Google Scholar] [CrossRef]
- Angelovici, R.; Lipka, A.E.; Deason, N.; Gonzalez-Jorge, S.; Lin, H.; Cepela, J.; Buell, R.; Gore, M.A.; DellaPenna, D. Genome-wide analysis of branched-chain amino acid levels in Arabidopsis Seeds. Plant Cell 2013, 25, 4827–4843. [Google Scholar] [CrossRef]
- Hatzig, S.; Breuer, F.; Nesi, N.; Ducournau, S.; Wagner, M.H.; Leckband, G.; Abbadi, A.; Snowdon, R.J. Hidden effects of seed quality breeding on germination in oilseed rape (Brassica napus L.). Front. Plant Sci. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Qian, L.; Qian, W.; Snowdon, R.J. Sub-genomic selection patterns as a signature of breeding in the allopolyploid Brassica napus genome. BMC Genomics 2014, 15, 1–17. [Google Scholar] [CrossRef]
- Li, F.; Chen, B.; Xu, K.; Wu, J.; Song, W.; Bancroft, I.; Harper, A.L.; Trick, M.; Liu, S.; Gao, G.; et al. Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA Res. 2014, 21, 355–367. [Google Scholar] [CrossRef]
- Qu, C.; Jia, L.; Fu, F.; Zhao, H.; Lu, K.; Wei, L.; Xu, X.; Liang, Y.; Li, S.; Wang, R.; et al. Genome-wide association mapping and Identification of candidate genes for fatty acid composition in Brassica napus L. using SNP markers. BMC Genom. 2017, 18, 1–17. [Google Scholar] [CrossRef]
- Guan, M.; Huang, X.; Xiao, Z.; Jia, L.; Wang, S.; Zhu, M.; Qiao, C.; Wei, L.; Xu, X.; Liang, Y.; et al. Association mapping analysis of fatty acid content in different ecotypic rapeseed using mrMLM. Front. Plant Sci. 2019, 9. [Google Scholar] [CrossRef]
- Wang, J.; Jian, H.; Wei, L.; Qu, C.; Xu, X.; Lu, K.; Qian, W.; Li, J.; Li, M.; Liu, L. Genome-Wide Analysis of Seed Acid Detergent Lignin (ADL) and hull content in rapeseed (Brassica napus L.). PLoS ONE 2015, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Huang, H.; Yi, X.; Zhang, Y.; Yang, Q.; Zhang, C.; Fan, C.; Zhou, Y. Dissection of genetic architecture for glucosinolate accumulations in leaves and seeds of Brassica napus by genome-wide association study. Plant Biotechnol. J. 2020, 18, 1472–1484. [Google Scholar] [CrossRef]
- Qu, C.M.; Li, S.M.; Duan, X.J.; Fan, J.H.; Jia, L.D.; Zhao, H.Y.; Lu, K.; Li, J.N.; Xu, X.F.; Wang, R. Identification of candidate genes for seed glucosinolate content using association mapping in brassica napus L. Genes 2015, 6, 1215–1229. [Google Scholar] [CrossRef] [PubMed]
- Jan, H.U.; Guan, M.; Yao, M.; Liu, W.; Wei, D.; Abbadi, A.; Zheng, M.; He, X.; Chen, H.; Guan, C.; et al. Genome-wide haplotype analysis improves trait predictions in Brassica napus hybrids. Plant Sci. 2019, 283, 157–164. [Google Scholar] [CrossRef]
- Werner, C.R.; Voss-Fels, K.P.; Miller, C.N.; Qian, W.; Hua, W.; Guan, C.-Y.; Snowdon, R.J.; Qian, L. Effective Genomic Selection in a Narrow-Genepool Crop with Low-Density Markers: Asian Rapeseed as an Example. Plant Genome 2018, 11, 170084. [Google Scholar] [CrossRef]
- Qian, L.; Qian, W.; Snowdon, R.J. Haplotype hitchhiking promotes trait coselection in Brassica napus. Plant Biotechnol. J. 2016, 14, 1578–1588. [Google Scholar] [CrossRef]
- Liu, S.; Fan, C.; Li, J.; Cai, G.; Yang, Q.; Wu, J.; Yi, X.; Zhang, C.; Zhou, Y. A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor. Appl. Genet. 2016, 129, 1203–1215. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Liu, J.; Hua, W.; Sun, X.; Wang, X.; Wang, H. Identification of stable QTLs for seed oil content by combined linkage and association mapping in Brassica napus. Plant Sci. 2016, 252, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Voss-Fels, K.; Cui, Y.; Jan, H.U.; Samans, B.; Obermeier, C.; Qian, W.; Snowdon, R.J. Deletion of a Stay-Green Gene Associates with Adaptive Selection in Brassica napus. Mol. Plant 2016, 9, 1559–1569. [Google Scholar] [CrossRef]
- Lin, Y.; Pajak, A.; Marsolais, F.; McCourt, P.; Riggs, C.D. Characterization of a Cruciferin deficient mutant of Arabidopsis and its utility for overexpression of foreign proteins in plants. PLoS ONE 2013, 8, 1–8. [Google Scholar] [CrossRef]
- Huang, S.; Adams, W.R.; Zhou, Q.; Malloy, K.P.; Voyles, D.A.; Anthony, J.; Kriz, A.L.; Luethy, M.H. Improving Nutritional Quality of Maize Proteins by Expressing Sense and Antisense Zein Genes. J. Agric. Food Chem. 2004, 52, 1958–1964. [Google Scholar] [CrossRef]
- Lange, M.; Vincze, E.; Wieser, H.; Schjoerring, J.K.; Holm, P.B. Suppression of C-hordein synthesis in barley by antisense constructs results in a more balanced amino acid composition. J. Agric. Food Chem. 2007, 55, 6074–6081. [Google Scholar] [CrossRef]
- Goossens, A.; Van Montagu, M.; Angenon, G. Co-introduction of an antisense gene for an endogenous seed storage protein can increase expression of a transgene in Arabidopsis thaliana seeds. FEBS Lett. 1999, 456, 160–164. [Google Scholar] [CrossRef]
- Withana-Gamage, T.S.; Hegedus, D.D.; Qiu, X.; Yu, P.; May, T.; Lydiate, D.; Wanasundara, J.P.D. Characterization of Arabidopsis thaliana lines with altered seed storage protein profiles using synchrotron-powered FT-IR spectromicroscopy. J. Agric. Food Chem. 2013, 61, 901–912. [Google Scholar] [CrossRef]
- Hegedus, D.D.; Baron, M.; Labbe, N.; Coutu, C.; Lydiate, D.; Lui, H.; Rozwadowski, K. A strategy for targeting recombinant proteins to protein storage vacuoles by fusion to Brassica napus napin in napin-depleted seeds. Protein Expr. Purif. 2014, 95, 162–168. [Google Scholar] [CrossRef]
- Langner, T.; Kamoun, S.; Belhaj, K. CRISPR crops: Plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 2018, 56, 479–512. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA—Guided DNA Endonuclease in Adaptive BActerial Immunity. Science 2012, 337, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wu, J.J.; Tang, T.; De Liu, K.; Dai, C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Braatz, J.; Harloff, H.-J.J.; Mascher, M.; Stein, N.; Himmelbach, A.; Jung, C. CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 2017, 174, 935–942. [Google Scholar] [CrossRef]
- Sun, Q.; Lin, L.; Liu, D.; Wu, D.; Fang, Y.; Wu, J.; Wang, Y. CRISPR/Cas9-mediated multiplex genome editing of the BnWRKY11 and BnWRKY70 genes in Brassica napus L. Int. J. Mol. Sci. 2018, 19, 1–19. [Google Scholar] [CrossRef]
- Okuzaki, A.; Ogawa, T.; Koizuka, C.; Kaneko, K.; Inaba, M.; Imamura, J.; Koizuka, N. CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 2018, 131, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Hao, M.; Wang, W.; Wang, H.; Chen, F.; Chu, W.; Zhang, B.; Mei, D.; Cheng, H.; Hu, Q. An Efficient CRISPR/Cas9 Platform for Rapidly Generating Simultaneous Mutagenesis of Multiple Gene Homoeologs in Allotetraploid Oilseed Rape. Front. Plant Sci. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Zhang, H.; Yang, Q.; Meng, Q.; Han, S.; Nwafor, C.C.; Khan, M.H.U.; Fan, C.; Zhou, Y. Promoter variations in a homeobox gene, BnA10.LMI1, determine lobed leaves in rapeseed (Brassica napus L.). Theor. Appl. Genet. 2018, 131, 2699–2708. [Google Scholar] [CrossRef] [PubMed]


| Soybean | Canola | |
|---|---|---|
| Protein content (%) 1 | 34.50 | 20.20 |
| Oil content (%) 1 | 18.40 | 44.50 |
| Oil-free protein of the meal (%) 2 | 43.80 | 37.80 |
| Oleic acid (% in oil) | 21.90 | 62.90 |
| Linoleic acid (% in oil) | 53.60 | 18.70 |
| α-Linolenic acid (% in oil) | 9.00 | 9.40 |
| Total saturated fatty acids (% in oil) | 15.20 | 6.70 |
| Erucic acid (% in oil) | n.a. | 0.01 |
| Total seed glucosinolates (μmol/g, 8.5% moisture) | n.a. | 10.00 |
| Total glucosinolates of the meal (µmol/g, oil-free, dry basis) | n.a. | 21.00 |
| Soybean | Canola | |
|---|---|---|
| Quality (%) | ||
| Moisture | 9.65 | 8.89 ± 0.43 |
| Crude protein | 46.10 | 37.82 ± 2.09 |
| Amino acids (%) | ||
| Alanine | 1.97 | 1.64 ± 0.08 |
| Arginine | 3.27 | 2.15 ± 0.11 |
| Aspartic acid | 5.04 | 2.51 ± 0.12 |
| Cysteine | 0.61 | 0.81 ± 0.04 |
| Glutamic acid | 8.34 | 6.2 ± 0.41 |
| Glycine | 1.98 | 1.87 ± 0.10 |
| Histidine | 1.11 | 0.91 ± 0.05 |
| Isoleucine | 1.81 | 1.26 ± 0.09 |
| Leucine | 3.45 | 2.54 ± 0.14 |
| Lysine | 2.90 | 2.09 ± 0.08 |
| Methionine | 0.58 | 0.69 ± 0.03 |
| Phenylalanine | 2.25 | 1.44 ± 0.07 |
| Proline | 2.39 | 2.21 ± 0.11 |
| Serine | 2.31 | 1.47 ± 0.09 |
| Threonine | 1.73 | 1.52 ± 0.06 |
| Tryptophan | 0.69 | 0.52 ± 0.03 |
| Tyrosine | 1.74 | 1.06 ± 0.04 |
| Valine | 1.88 | 1.63 ± 0.09 |
| Weeks after Pollination | 1 | 2 | 3 | 4 | 5 | 6 | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Development | ||||||||||||||||||||||||||||||||||||
| Crouch and Sussex (1981) Planta 153: 64–74 | H | T | C | |||||||||||||||||||||||||||||||||
| Custers et al. (1999) Protoplasma 208: 257–264 | G | H | T | C | ||||||||||||||||||||||||||||||||
| Fernandez et al. (1991) Development 111: 299–313 | H | T | C | |||||||||||||||||||||||||||||||||
| Ilic-Grubor et al. (1998) Ann. Bot. 82: 157–165 | H | T | C | |||||||||||||||||||||||||||||||||
| Yeung et al. (1996) Int. J. Plant Sci. 157(1): 27–39 | H | T | C | |||||||||||||||||||||||||||||||||
| CRU mRNA | ||||||||||||||||||||||||||||||||||||
| DeLisle and Crouch (1989) Plant Physiol. 91: 617–623 | * | *** | ||||||||||||||||||||||||||||||||||
| Finkelstein et al. (1985) Plant Physiol. 78: 630–636 | * | *** | ||||||||||||||||||||||||||||||||||
| Sjodahl et al. (1993) Plant Mol. Biol. 23: 1165–1176 | * | *** | ||||||||||||||||||||||||||||||||||
| CRU protein | ||||||||||||||||||||||||||||||||||||
| Crouch and Sussex (1981) Planta 153: 64–74 | * | *** | ||||||||||||||||||||||||||||||||||
| NAPIN mRNA | ||||||||||||||||||||||||||||||||||||
| DeLisle and Crouch (1989) Plant Physiol. 91: 617–623 | * | *** | ||||||||||||||||||||||||||||||||||
| Finkelstein et al. (1985) Plant Physiol. 78: 630–636 | * | *** | ||||||||||||||||||||||||||||||||||
| NAPIN protein | ||||||||||||||||||||||||||||||||||||
| Crouch and Sussex (1981) Planta 153: 64–74 | * | *** | ||||||||||||||||||||||||||||||||||
| Entry | Entry Name | Protein Names | Length |
|---|---|---|---|
| P33522 | CRU4_BRANA | Cruciferin CRU4 (11S globulin) (12S storage protein) [Cleaved into: Cruciferin CRU4 alpha chain; Cruciferin CRU4 beta chain] | 465 |
| P11090 | CRUA_BRANA | Cruciferin (11S globulin) (12S storage protein) [Cleaved into: Cruciferin subunit alpha; Cruciferin subunit beta] | 488 |
| P33524 | CRU2_BRANA | Cruciferin BnC2 (11S globulin) (12S storage protein) [Cleaved into: Cruciferin BnC2 subunit alpha; Cruciferin BnC2 subunit beta] | 496 |
| P33525 | CRU3_BRANA | Cruciferin CRU1 (11S globulin) (12S storage protein) [Cleaved into: Cruciferin CRU1 alpha chain; Cruciferin CRU1 beta chain] | 509 |
| P33523 | CRU1_BRANA | Cruciferin BnC1 (11S globulin) (12S storage protein) [Cleaved into: Cruciferin BnC1 subunit alpha; Cruciferin BnC1 subunit beta] | 490 |
| Entry | Entry Name | Protein Names | Length |
|---|---|---|---|
| P24565 | 2SSI_BRANA | Napin-1A (Napin BnIa) [Cleaved into: Napin-1A small chain; Napin-1A large chain] | 110 |
| P09893 | 2SSE_BRANA | Napin embryo-specific (1.7S seed storage protein) [Cleaved into: Napin embryo-specific small chain; Napin embryo-specific large chain] | 186 |
| P17333 | 2SS4_BRANA | Napin (1.7S seed storage protein) [Cleaved into: Napin small chain; Napin large chain] | 180 |
| P27740 | 2SSB_BRANA | Napin-B (1.7S seed storage protein) [Cleaved into: Napin-B small chain; Napin-B large chain] | 178 |
| P01090 | 2SS2_BRANA | Napin-2 (1.7S seed storage protein) [Cleaved into: Napin-2 small chain; Napin-2 large chain] | 178 |
| P01091 | 2SS1_BRANA | Napin-1 (1.7S seed storage protein) [Cleaved into: Napin-1 small chain; Napin-1 large chain] (Fragment) | 133 |
| P80208 | 2SS3_BRANA | Napin-3 (1.7S seed storage protein) (Napin BnIII) (Napin nIII) [Cleaved into: Napin-3 small chain; Napin-3 large chain] | 125 |
| GRAS Number | Notifier | Substance | Intended Use | Date of Closure | FDA Response |
|---|---|---|---|---|---|
| 683 | DSM Innovation Company | Canola protein isolate | Dietary protein Food processing | 2017 | no questions |
| 682 | Cargill Inc. | Lecithin from canola | Food processing Dietary fat Dietary choline | 2017 | no questions |
| 533 | American Lecithin Company | Lecithin from canola | Food processing Dietary fat Dietary choline | 2015 | no questions |
| 425 | Danone Trading B.V. | Canola oil (low-erucic-acid rapeseed oil) | Dietary fat (infant formula) | 2012 | no questions |
| 386 | BioExx Specialty Proteins, Ltd. | Canola protein isolate and hydrolyzed canola protein isolate | Food processing Dietary protein | 2011 | no questions |
| 327 | Archer Daniels Midland Company | Cruciferin-rich canola/rapeseed protein isolate and napin-rich canola/rapeseed protein isolate | Dietary protein | 2010 | no questions |
| Trait | Population Size | Reference |
|---|---|---|
| Erucic acid content | 215 | [302] |
| Erucic acid content | 203 | [303] |
| Erucic acid content | 472 | [304] |
| Fatty acid composition | 435 | [305] |
| Fatty acid composition | 520 | [59] |
| Fatty acid content | 370 | [306] |
| Fibre content | 520 | [307] |
| Glucosinolate | 521 | [308] |
| Glucosinolate | 203 | [303] |
| Glucosinolate | 520 | [309] |
| Glucosinolate | 1425 | [310] |
| Glucosinolate | 203 | [311] |
| Glucosinolate | 203 | [312] |
| Glucosinolates | 215 | [302] |
| Oil content | 472 | [304] |
| Oil content | 521 | [313] |
| Oil content | 105 | [314] |
| Oil content | 370 | [59] |
| Oil content | 521 | [313] |
| Oil content | 203 | [311] |
| Oil content | 203 | [315] |
| Protein content | 370 | [59] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
So, K.K.Y.; Duncan, R.W. Breeding Canola (Brassica napus L.) for Protein in Feed and Food. Plants 2021, 10, 2220. https://doi.org/10.3390/plants10102220
So KKY, Duncan RW. Breeding Canola (Brassica napus L.) for Protein in Feed and Food. Plants. 2021; 10(10):2220. https://doi.org/10.3390/plants10102220
Chicago/Turabian StyleSo, Kenny K. Y., and Robert W. Duncan. 2021. "Breeding Canola (Brassica napus L.) for Protein in Feed and Food" Plants 10, no. 10: 2220. https://doi.org/10.3390/plants10102220
APA StyleSo, K. K. Y., & Duncan, R. W. (2021). Breeding Canola (Brassica napus L.) for Protein in Feed and Food. Plants, 10(10), 2220. https://doi.org/10.3390/plants10102220





