Phytochemical Investigation of Bioactive Compounds from White Kidney Beans (Fruits of Phaseolus multiflorus var. Albus): Identification of Denatonium with Osteogenesis-Inducing Effect
Abstract
:1. Introduction
2. Results and Discussion
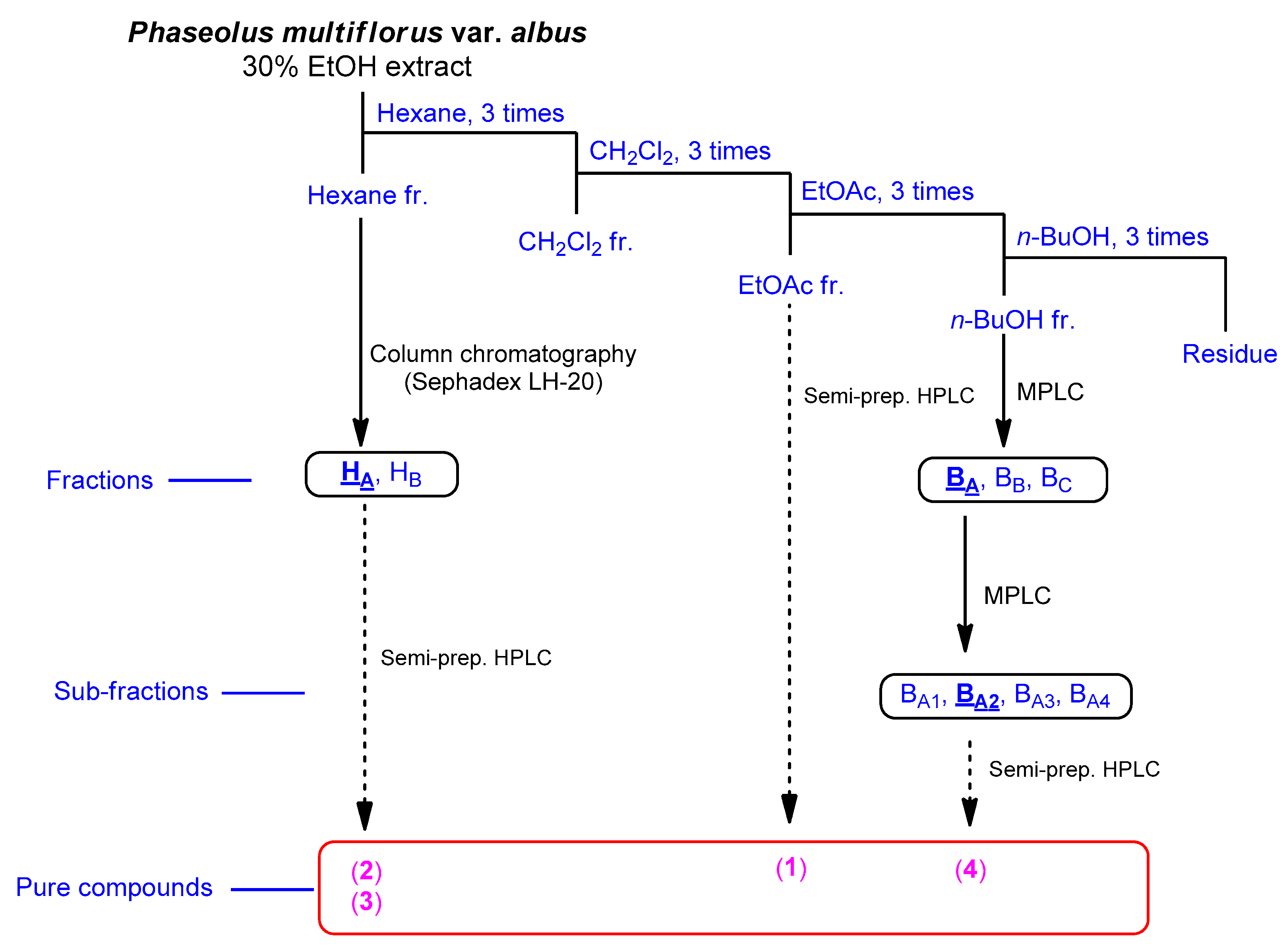
2.1. Isolation of Compounds
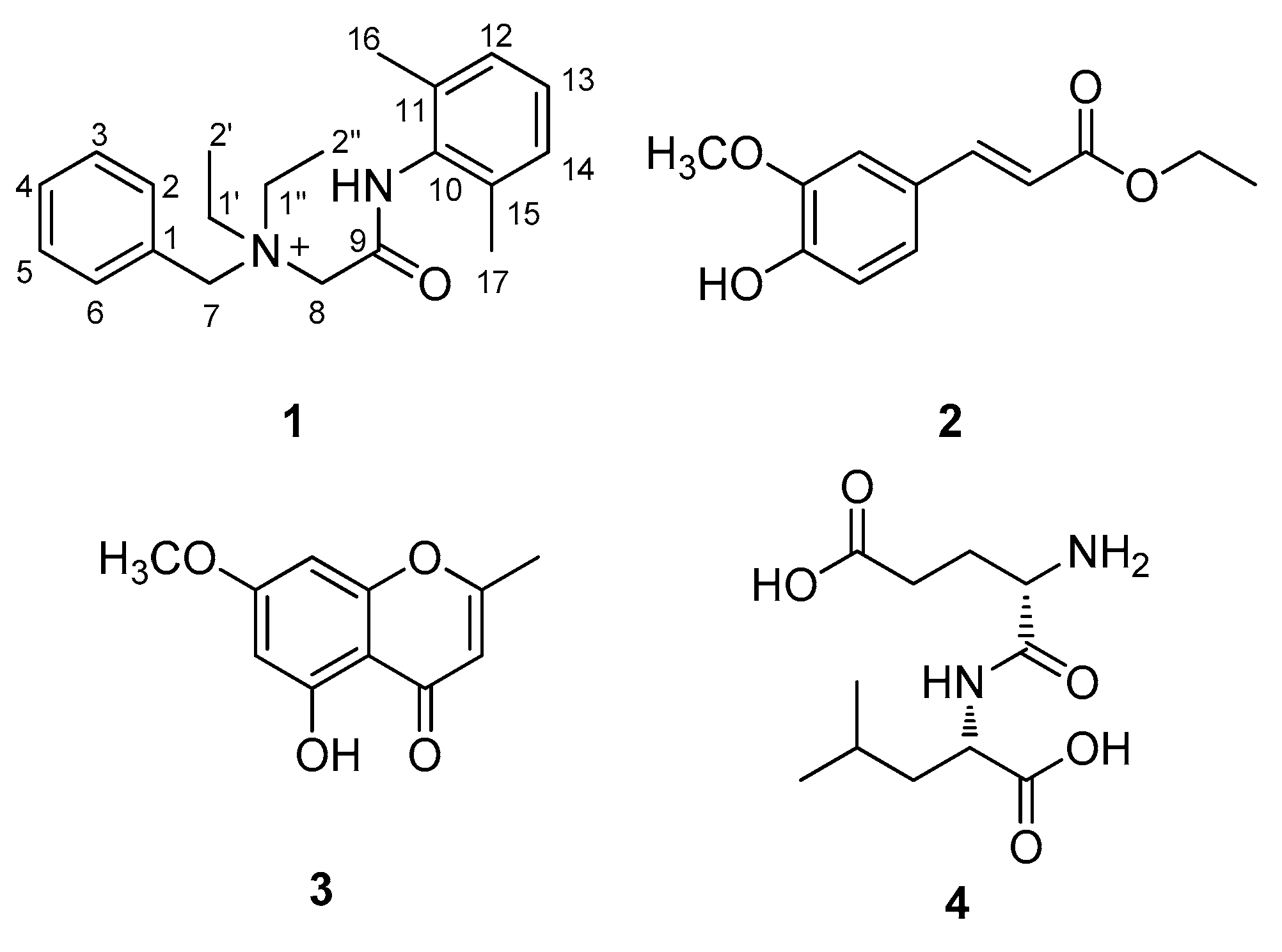
2.2. Elucidation of Compound Structures
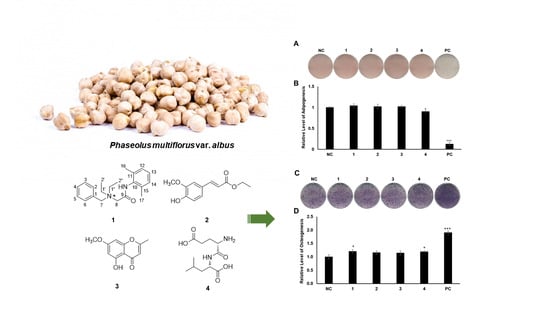
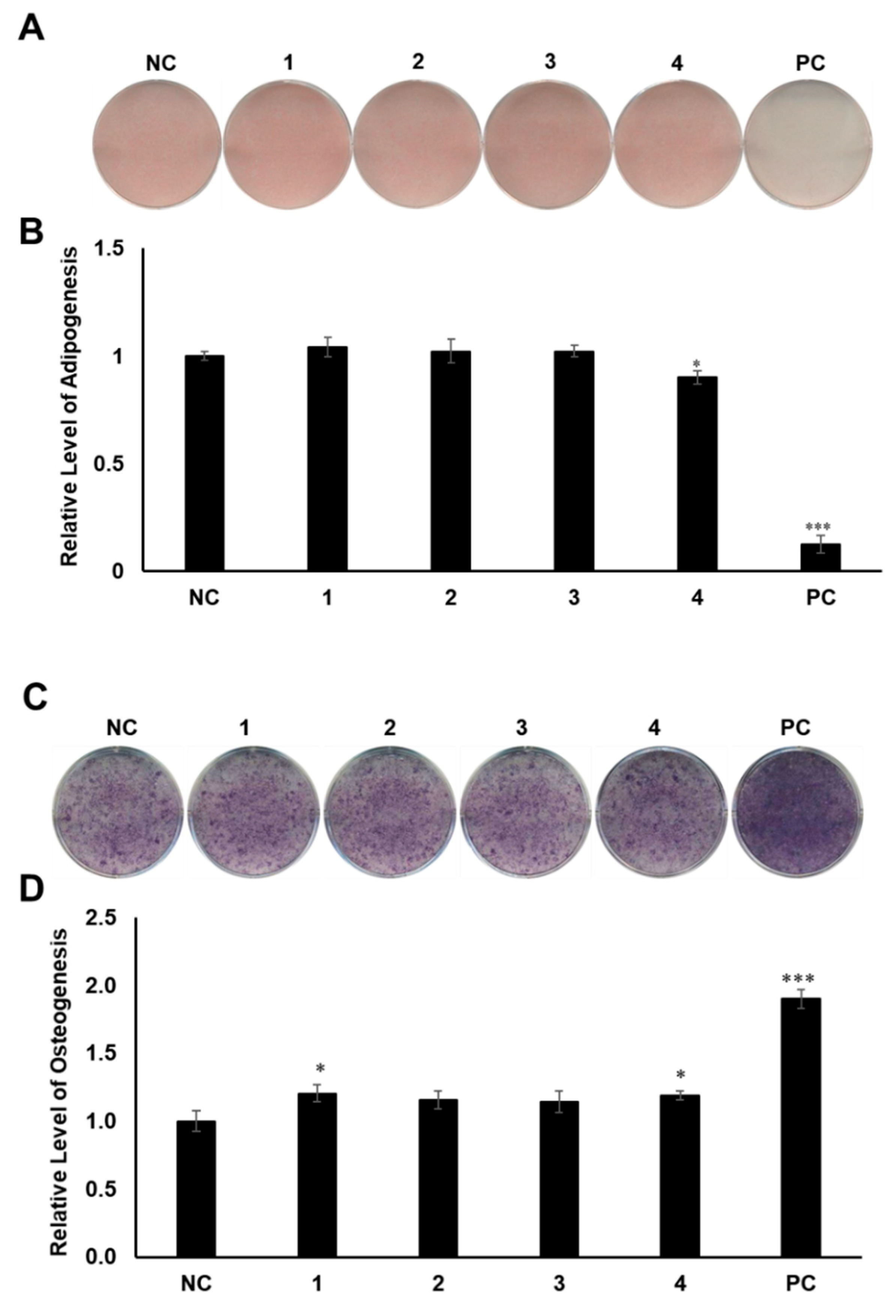
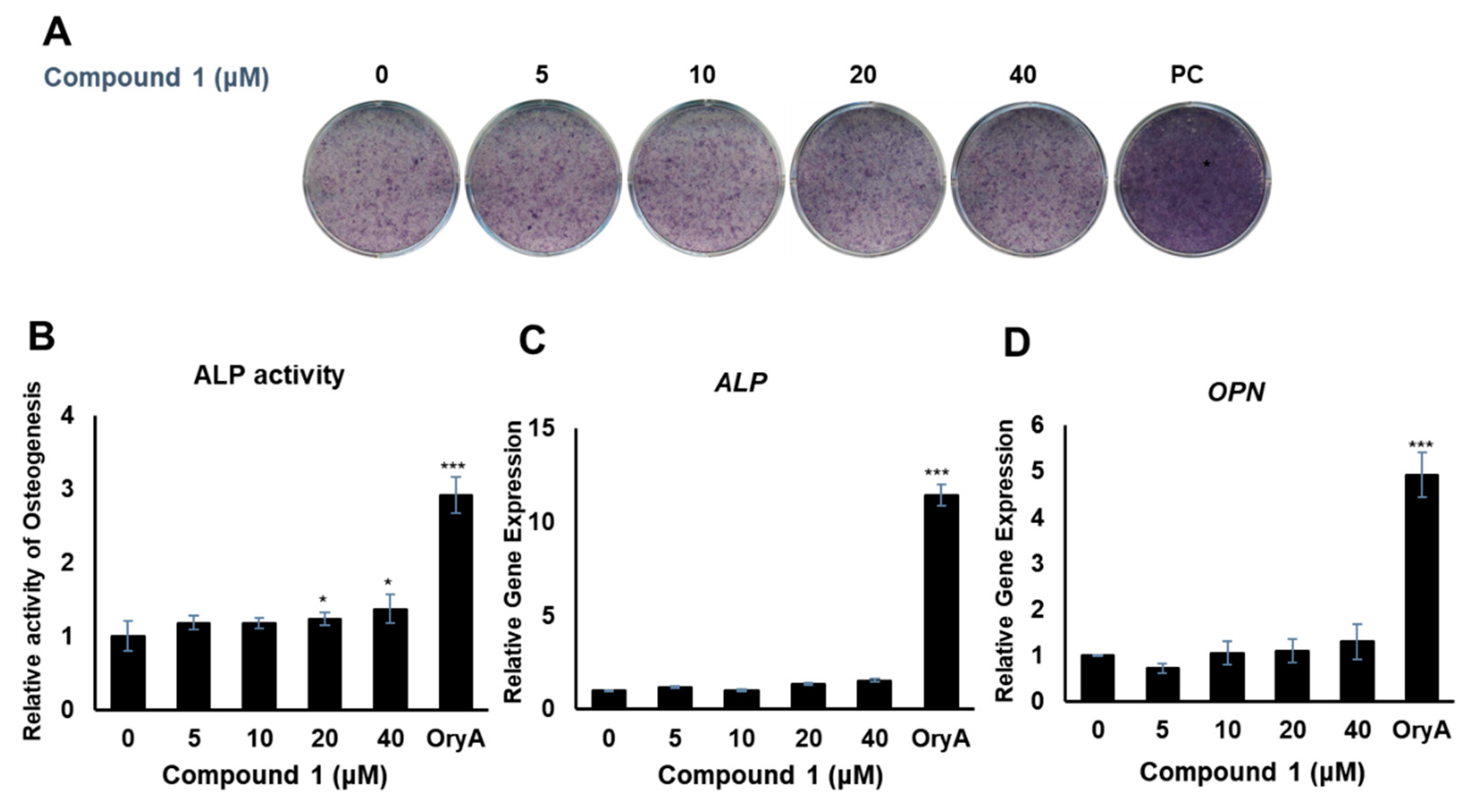
2.3. Evaluation of the Biological Activities of Compounds 1–4
3. Materials and Methods
3.1. Plant Material
3.2. Extraction and Isolation
3.3. Cell Culture and Differentiation
3.4. Oil Red O Staining
3.5. Alkaline Phosphatase Staining
3.6. mRNA Isolation and Real-Time PCR
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Meng, J.; Bai, Z.; Huang, W.; Liu, Y.; Wang, P.; Nie, S.; Huang, X. Polysaccharide from white kidney bean can improve hyperglycemia and hyperlipidemia in diabetic rats. Bioact. Carbohydr. Diet. Fibre 2020, 24, 100222. [Google Scholar] [CrossRef]
- Mojica, L.; de Mejía, E.G. Characterization and comparison of protein and peptide profiles and their biological activities of improved common bean cultivars (Phaseolus vulgaris L.) from Mexico and Brazil. Plant Foods Hum. Nutr. 2015, 70, 105–112. [Google Scholar] [CrossRef]
- Neil, E.S.; McGinley, J.N.; Fitzgerald, V.K.; Lauck, C.A.; Tabke, J.A.; Streeter-McDonald, M.R.; Yao, L.; Broeckling, C.D.; Weir, T.L.; Foster, M.T. White kidney bean (Phaseolus Vulgaris L.) consumption reduces fat accumulation in a polygenic mouse model of obesity. Nutrients 2019, 11, 2780. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Liu, Q.; Cui, Y.; Li, D.; Wang, H.; Ng, T.B. Isolation and characterization of a Phaseolus vulgaris trypsin inhibitor with antiproliferative activity on leukemia and lymphoma cells. Molecules 2017, 22, 187. [Google Scholar] [CrossRef] [PubMed]
- E Lacerda, R.R.; do Nascimento, E.S.; de Lacerda, J.T.J.G.; da Silva Pinto, L.; Rizzi, C.; Bezerra, M.M.; Pinto, I.R.; Pereira Filho, S.M.; Pinto, V.d.P.T.; Cristino Filho, G. Lectin from seeds of a Brazilian lima bean variety (Phaseolus lunatus L. var. cascavel) presents antioxidant, antitumour and gastroprotective activities. Int. J. Biol. Macromol. 2017, 95, 1072–1081. [Google Scholar] [PubMed]
- Jabir, A.S.; Iraby, A.G. Studying the effect of anti-amylase inhibitor extracted from white kidney bean (Phaseolus vulgaris) in treat diabetes and obesity in an affected mice. Int. J. Curr. Microbiol. App. Sci 2014, 3, 97–106. [Google Scholar]
- Barrett, M.L.; Udani, J.K. A proprietary alpha-amylase inhibitor from white bean (Phaseolus vulgaris): A review of clinical studies on weight loss and glycemic control. Nutr. J. 2011, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Park, Y.H.; Lee, J.S.; Jeong, H.I.; Lee, K.W.; Kang, T.H. Anti-obesity effect of DKB-117 through the inhibition of pancreatic lipase and α-amylase activity. Nutrients 2020, 12, 3053. [Google Scholar] [CrossRef]
- MacMillan, J.; Pryce, R. Further investigations of gibberellins in Phaseolus multiflorus by combined gas chromatography-mass spectrometry-the occurence of gibberellin A20 (pharbitis gibberellin) and the structure of compound b. Tetrahedron Lett. 1968, 9, 1537–1542. [Google Scholar] [CrossRef]
- MacMillan, J.; Seaton, J.; Suter, P. Plant hormones—I: Isolation of gibberellin A1 and gibberellin A5 from Phaseolus multiflorus. Tetrahedron 1960, 11, 60–66. [Google Scholar] [CrossRef]
- MacMillan, J.; Pryce, R. Plant hormones—IX: Phaseic acid, a relative of abscisic acid from seed of Phaseolus multiflorus. Possible structures. Tetrahedron 1969, 25, 5893–5901. [Google Scholar] [CrossRef]
- MacMillan, J.; Suter, P. The structure of a C12-acid from the seed of Phaseolus multiflorus. Tetrahedron 1967, 23, 2417–2419. [Google Scholar] [CrossRef]
- Lee, S.R.; Kang, H.; Yoo, M.J.; Yu, J.S.; Lee, S.; Yi, S.A.; Beemelmanns, C.; Lee, J.; Kim, K.H. Anti-adipogenic pregnane steroid from a Hydractinia-associated fungus, Cladosporium sphaerospermum SW67. Nat. Prod. Sci. 2020, 26, 230–235. [Google Scholar]
- Yu, J.S.; Park, M.; Pang, C.; Rashan, L.; Jung, W.H.; Kim, K.H. Antifungal phenols from Woodfordia uniflora collected in Oman. J. Nat. Prod. 2020, 83, 2261–2268. [Google Scholar] [CrossRef]
- Lee, S.; Lee, D.; Ryoo, R.; Kim, J.-C.; Park, H.B.; Kang, K.S.; Kim, K.H. Calvatianone, a sterol possessing a 6/5/6/5-fused ring system with a contracted tetrahydrofuran B-ring, from the fruiting bodies of Calvatia nipponica. J. Nat. Prod. 2020, 83, 2737–2742. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.A.; Park, E.-J.; Lee, D.; Song, J.H.; Lee, H.L.; Kim, K.H.; Kim, Y.; Jung, K.; Kang, K.S.; Yoo, J.-E. Estrogenic activity of sanguiin H-6 through activation of estrogen receptor α coactivator-binding site. Nat. Prod. Sci. 2019, 25, 28–33. [Google Scholar] [CrossRef]
- Yu, J.S.; Li, C.; Kwon, M.; Oh, T.; Lee, T.H.; Kim, D.H.; Ahn, J.S.; Ko, S.-K.; Kim, C.S.; Cao, S. Herqueilenone A, a unique rearranged benzoquinone-chromanone from the hawaiian volcanic soil-associated fungal strain Penicillium herquei FT729. Bioorg. Chem. 2020, 105, 104397. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.W.; Kim, J.; Kim, H.; Jang, W.; Kim, K.H. Mushrooms: An important source of natural bioactive compounds. Nat. Prod. Sci. 2020, 26, 118–131. [Google Scholar]
- Lee, S.; Ryoo, R.; Choi, J.H.; Kim, J.-H.; Kim, S.-H.; Kim, K.H. Trichothecene and tremulane sesquiterpenes from a hallucinogenic mushroom Gymnopilus junonius and their cytotoxicity. Arch. Pharm. Res. 2020, 43, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Bolling, B.W.; Parkin, K.L. Phenolic derivatives from soy flour ethanol extract are potent in vitro quinone reductase (QR) inducing agents. J. Agric. Food Chem. 2008, 56, 10473–10480. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Makboul, M.; Attia, A.; Ali, D. Chromones and flavans from Pancratium maritimum. Phytochemistry 1990, 29, 625–627. [Google Scholar] [CrossRef]
- Kasai, T.; Sakamura, S. NMR spectra of glutamic acid-containing dipeptides in relation to sequence determination. Agric. Biol. Chem. 1973, 37, 2155–2157. [Google Scholar] [CrossRef]
- Venditti, A. What is and what should never be: Artifacts, improbable phytochemicals, contaminants and natural products. Nat. Prod. Res. 2020, 34, 1014–1031. [Google Scholar] [CrossRef] [PubMed]
- What is Bitrex? Available online: https://www.bitrex.com/about-bitrex/what-is-bitrex (accessed on 3 October 2021).
- Kwiatkowski, A.; Czerwicka, M.; Smulko, J.; Stepnowski, P. Detection of denatonium benzoate (Bitrex) remnants in noncommercial alcoholic beverages by raman spectroscopy. J. Forensic Sci. 2014, 59, 1358–1363. [Google Scholar] [CrossRef]
- Sibert, J.R.; Frude, N. Bittering agents in the prevention of accidental poisoning: children’s reactions to denatonium benzoate (Bitrex). Arch. Emerg. Med. 1991, 8, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.B.; Benkusky, N.A.; Sen, B.; Rubin, J.; Pike, J.W. Epigenetic plasticity drives adipogenic and osteogenic differentiation of marrow-derived mesenchymal stem cells. J. Biol. Chem. 2016, 291, 17829–17847. [Google Scholar] [CrossRef] [Green Version]
- Yi, S.A.; Lee, J.; Park, S.K.; Kim, J.Y.; Park, J.W.; Lee, M.G.; Nam, K.H.; Park, J.H.; Oh, H.; Kim, S. Fermented ginseng extract, BST204, disturbs adipogenesis of mesenchymal stem cells through inhibition of S6 kinase 1 signaling. J. Ginseng Res. 2020, 44, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Ciuffreda, M.C.; Malpasso, G.; Musarò, P.; Turco, V.; Gnecchi, M. Protocols for in vitro differentiation of human mesenchymal stem cells into osteogenic, chondrogenic and adipogenic lineages. In Mesenchymal Stem Cells; Springer: New York, NY, USA, 2016; pp. 149–158. [Google Scholar]
- Kang, M.-H.; Lee, S.-J.; Lee, M.-H. Bone remodeling effects of Korean Red Ginseng extracts for dental implant applications. J. Ginseng Res. 2020, 44, 823–832. [Google Scholar] [CrossRef] [PubMed]
 ) and HMBC (
) and HMBC (  ) correlations of compound 1. A and B represent two partial structures.
) correlations of compound 1. A and B represent two partial structures.
 ) and HMBC (
) and HMBC (  ) correlations of compound 1. A and B represent two partial structures.
) correlations of compound 1. A and B represent two partial structures.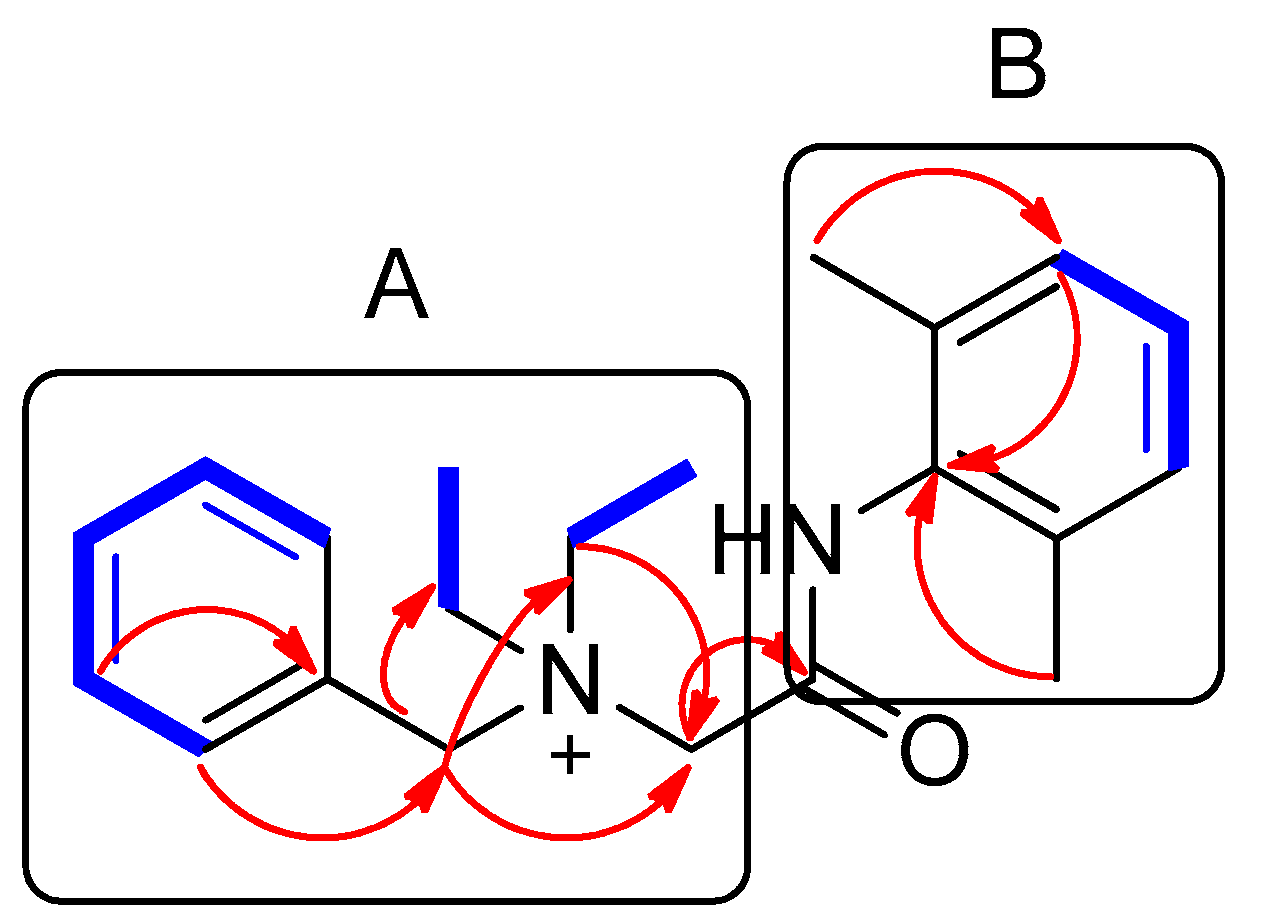
| Position | Denatonium (1) | |
|---|---|---|
| δH (J in Hz) | δC | |
| 1 | 128.7 C | |
| 2 | 7.64, d (7.5) | 134.1 CH |
| 3 | 7.58, t (7.5) | 130.7 CH |
| 4 | 7.62, t (7.5) | 132.3 CH |
| 5 | 7.58, t (7.5) | 130.7 CH |
| 6 | 7.64, d (7.5) | 134.1 CH |
| 7 | 4.94, s | 63.4 CH2 |
| 8 | 4.16, s | 55.7 CH2 |
| 9 | 164.1 C | |
| 10 | 134.2 C | |
| 11 | 136.7 C | |
| 12 | 7.17, d (7.0) | 129.5 CH |
| 13 | 7.18, t (7.0) | 129.1 CH |
| 14 | 7.17, d (7.0) | 129.5 CH |
| 15 | 136.7 C | |
| 16 | 2.30, s | 18.7 CH3 |
| 17 | 2.30, s | 18.7 CH3 |
| 1′ | 3.67, m | 54.9 CH2 |
| 2′ | 1.56, t (7.5) | 8.4 CH3 |
| 1″ | 3.67, m | 54.9 CH2 |
| 2″ | 1.56, t (7.5) | 8.4 CH3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.H.; Hong, J.-H.; Park, K.H.; Kim, S.-H.; Kim, J.-C.; Kim, D.H.; Park, Y.H.; Lee, K.W.; Kim, J.K.; Kim, K.H. Phytochemical Investigation of Bioactive Compounds from White Kidney Beans (Fruits of Phaseolus multiflorus var. Albus): Identification of Denatonium with Osteogenesis-Inducing Effect. Plants 2021, 10, 2205. https://doi.org/10.3390/plants10102205
Lee YH, Hong J-H, Park KH, Kim S-H, Kim J-C, Kim DH, Park YH, Lee KW, Kim JK, Kim KH. Phytochemical Investigation of Bioactive Compounds from White Kidney Beans (Fruits of Phaseolus multiflorus var. Albus): Identification of Denatonium with Osteogenesis-Inducing Effect. Plants. 2021; 10(10):2205. https://doi.org/10.3390/plants10102205
Chicago/Turabian StyleLee, Yong Hoon, Joo-Hyun Hong, Kun Hee Park, Seon-Hee Kim, Jin-Chul Kim, Do Hoon Kim, Yu Hwa Park, Kye Wan Lee, Jung Kyu Kim, and Ki Hyun Kim. 2021. "Phytochemical Investigation of Bioactive Compounds from White Kidney Beans (Fruits of Phaseolus multiflorus var. Albus): Identification of Denatonium with Osteogenesis-Inducing Effect" Plants 10, no. 10: 2205. https://doi.org/10.3390/plants10102205
APA StyleLee, Y. H., Hong, J.-H., Park, K. H., Kim, S.-H., Kim, J.-C., Kim, D. H., Park, Y. H., Lee, K. W., Kim, J. K., & Kim, K. H. (2021). Phytochemical Investigation of Bioactive Compounds from White Kidney Beans (Fruits of Phaseolus multiflorus var. Albus): Identification of Denatonium with Osteogenesis-Inducing Effect. Plants, 10(10), 2205. https://doi.org/10.3390/plants10102205