Phospholipids in Salt Stress Response
Abstract
:1. Introduction
2. Salt Stress Signals in Plants
2.1. Salt Stress Perception
2.2. Ionic Signaling Pathway
2.3. Osmotic Signaling Pathway
2.4. ROS Signaling Pathways
2.5. Phytohormones in Salt Stress Response
2.6. Organelles in Salt Stress Response
3. PA in Salt Stress
3.1. PA Generation in Plants
3.2. PA Functions in Salt Stress
3.2.1. PA Level Increases under Salt Stress
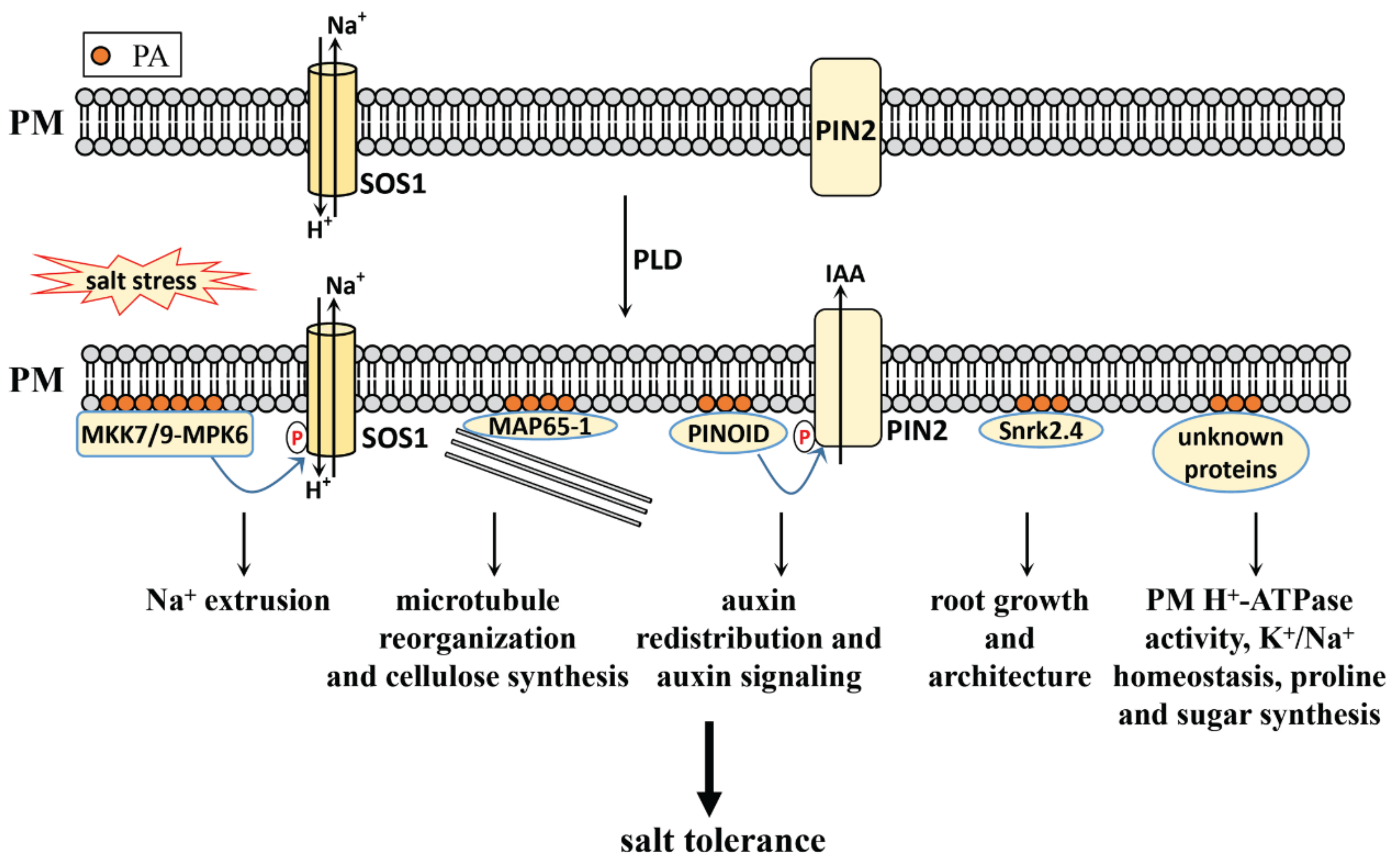
3.2.2. PLD-Derived PA in Salt Stress Response
3.2.3. PLC-DGK-Derived PA in Salt Stress Response
4. PI, PIP, and PIP2 in Salt Stress
4.1. PI, PIP, and PIP2 Generation in Plants
4.2. PI, PIP and PIP2 Function in Salt Stress
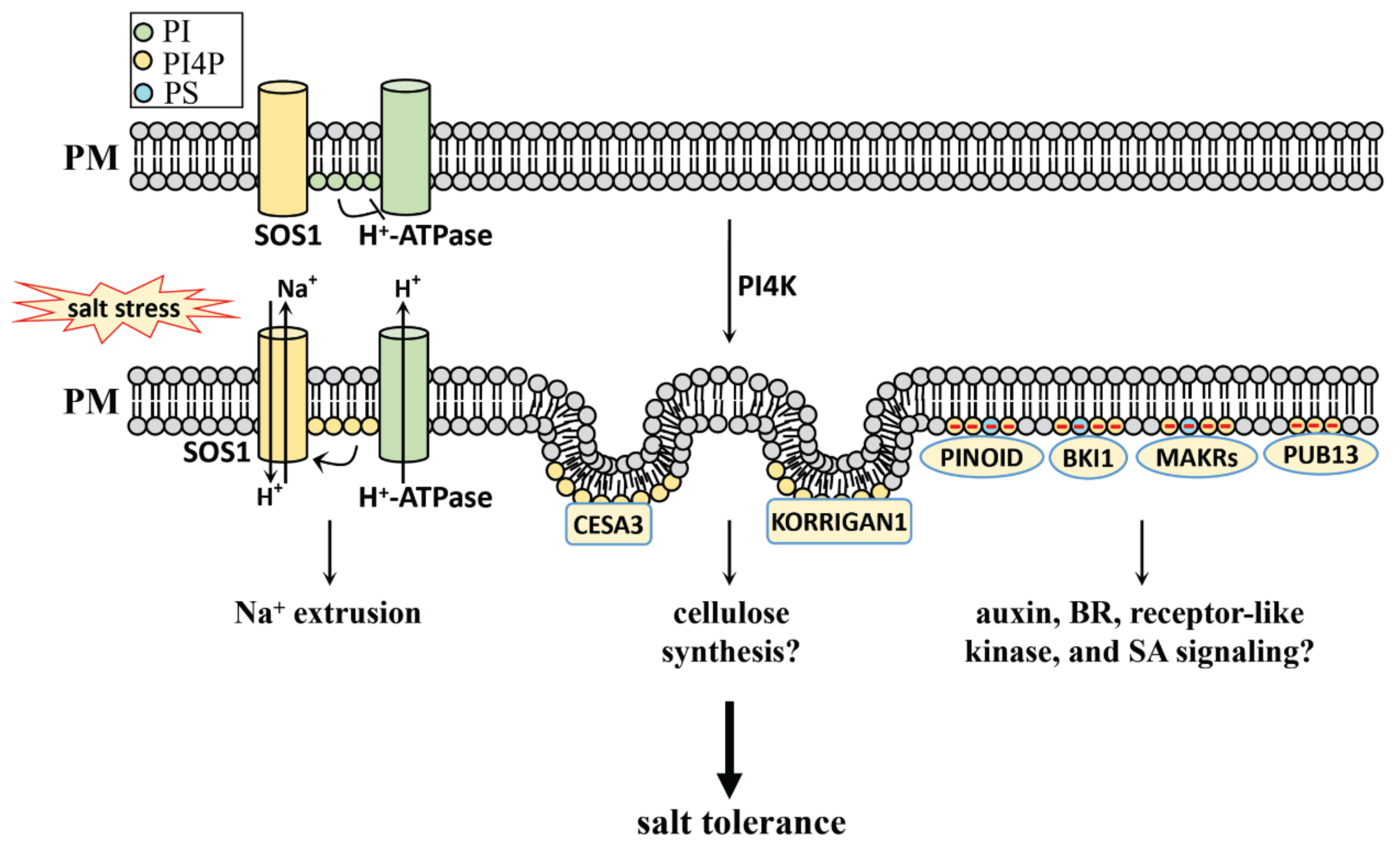
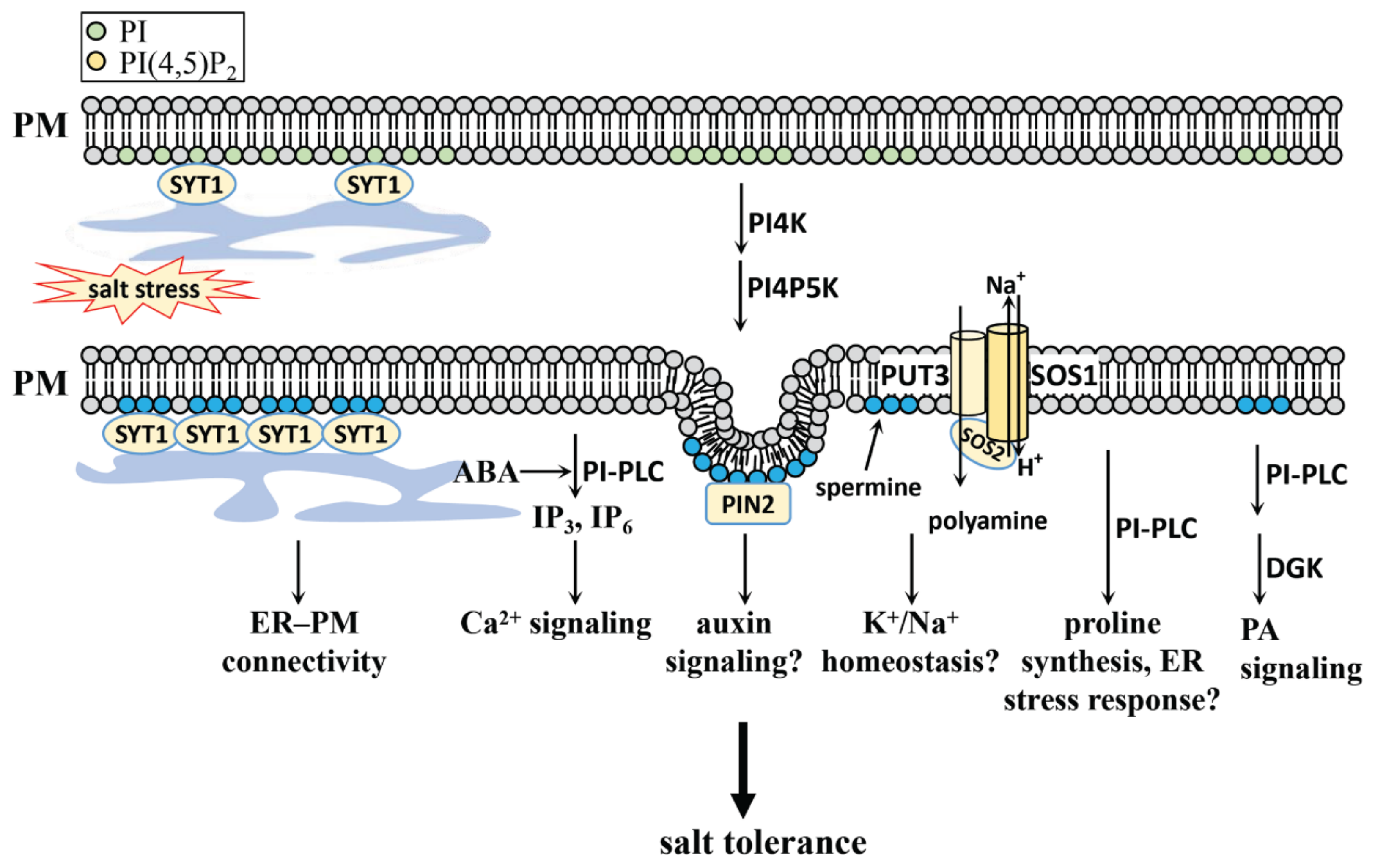
4.2.1. PI, PI4P, and PI(4,5)P2 in Salt Stress
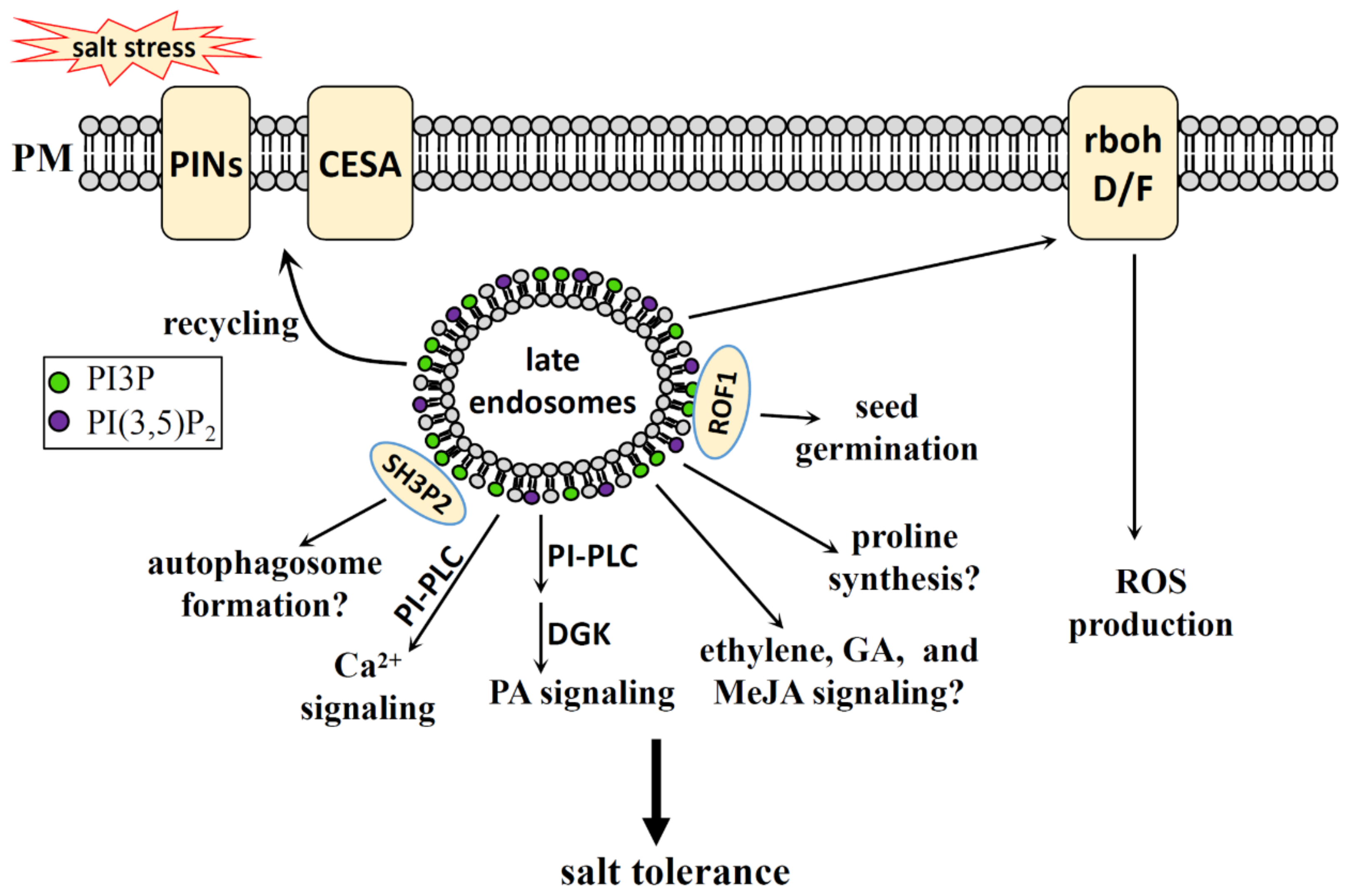
4.2.2. PI3P, PI(3,5)P2 in Salt Stress
4.2.3. PI-PLC in Salt Stress Response
5. PS in Salt Stress
5.1. PS Generation in Plants
5.2. PS Function in Salt Stress
6. PC, PE in Salt Stress
6.1. PC, PE Generation in Plants
6.2. PC, PE Function in Salt Stress
6.2.1. PC and PE in Salt Stress
6.2.2. NPC in Salt Stress
7. PG in Salt Stress
7.1. PG Generation in Plants
7.2. PG Function in Salt Stress
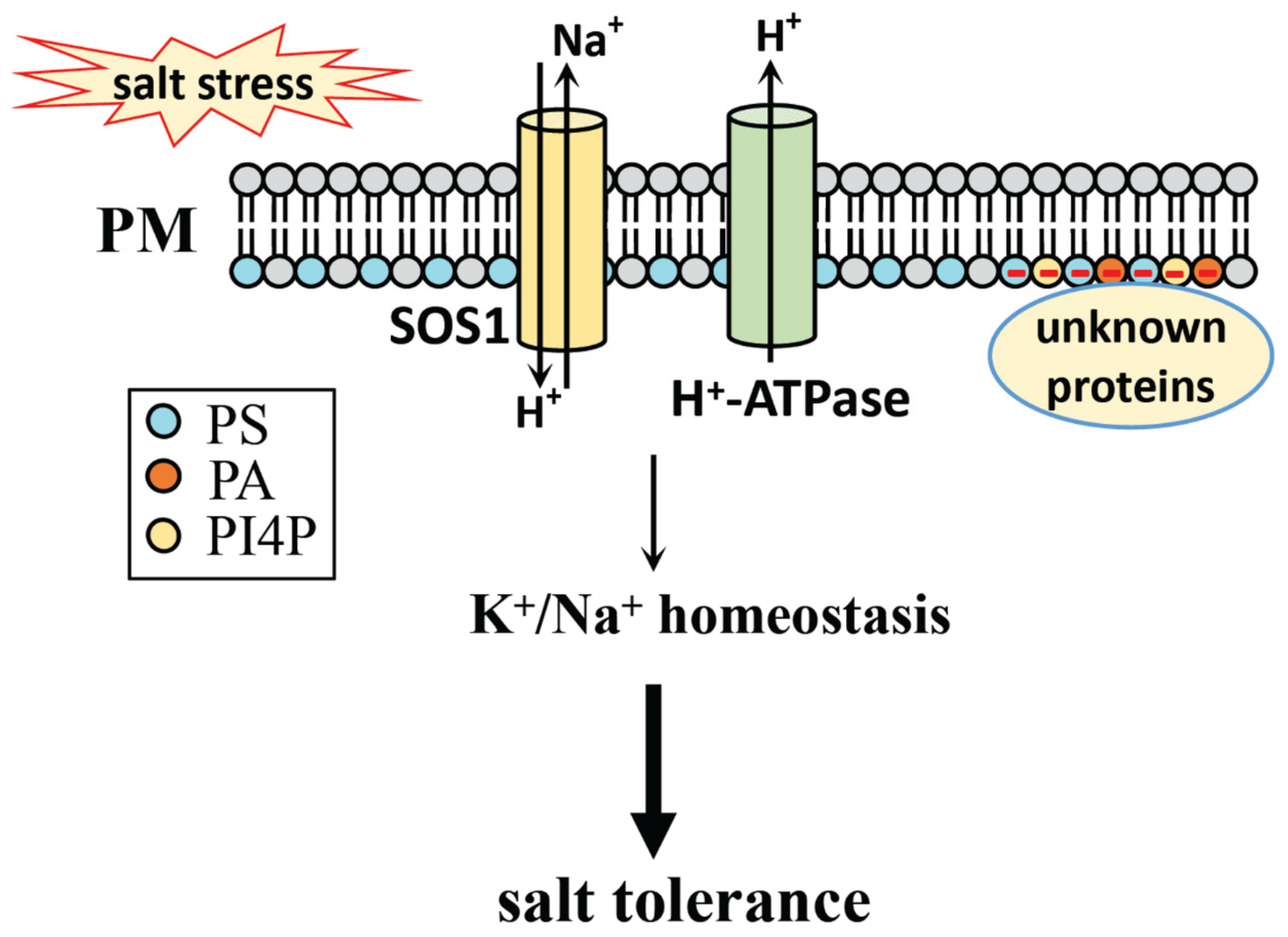
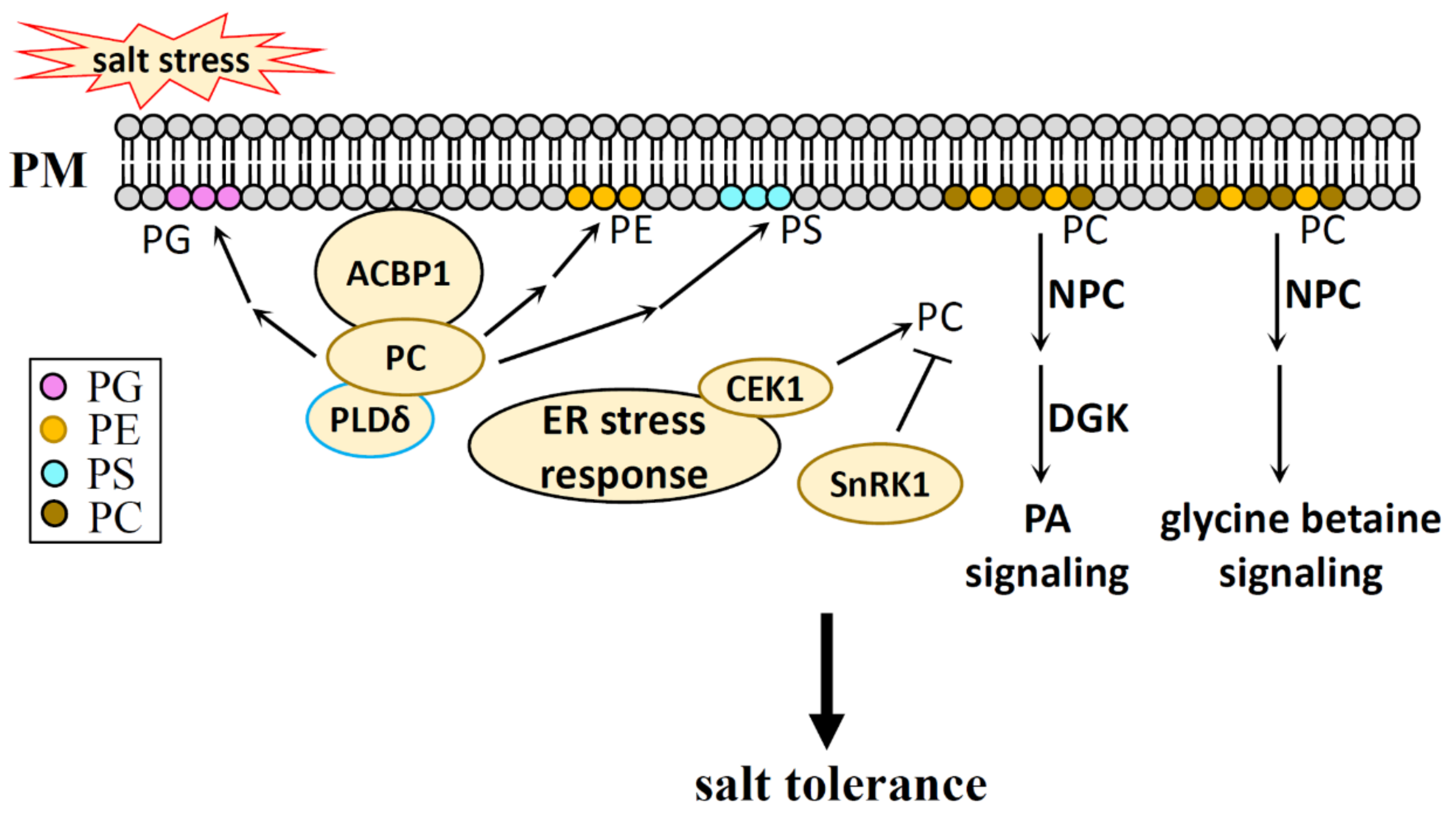
8. The Mechanism of Phospholipids in Salt Stress Response
8.1. Phospholipid Head Groups in Salt Stress
8.2. Phospholipid Fatty Acid Chains in Salt Stress
9. Conclusions and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant. Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant. Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Li, M.; Yao, S.; Cai, G.; Wang, X. Patatin-Related Phospholipase pPLAIIIγ Involved in Osmotic and Salt Tolerance in Arabidopsis. Plants 2020, 9, 650. [Google Scholar] [CrossRef]
- Peters, C.; Kim, S.-C.; Devaiah, S.; Li, M.; Wang, X. Non-specific phospholipase C5 and diacylglycerol promote lateral root development under mild salt stress in Arabidopsis. Plant Cell Environ. 2014, 37, 2002–2013. [Google Scholar] [CrossRef]
- Wang, P.; Shen, L.; Guo, J.; Jing, W.; Qu, Y.; Li, W.; Bi, R.; Xuan, W.; Zhang, Q.; Zhang, W. Phosphatidic Acid Directly Regulates PINOID-Dependent Phosphorylation and Activation of the PIN-FORMED2 Auxin Efflux Transporter in Response to Salt Stress. Plant Cell 2018, 31, 250–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, K.; Wang, B.; Zhang, J.; Li, Y.; Yang, H.; Ren, D. Arabidopsis phosphoinositide-specific phospholipase C 4 negatively regulates seedling salt tolerance. Plant Cell Environ. 2017, 40, 1317–1331. [Google Scholar] [CrossRef]
- Li, W.; Li, M.; Zhang, W.; Welti, R.; Wang, X. The plasma membrane-bound phospholipase Ddelta enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2004, 22, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Yao, H.; Jia, L.; Tan, J.; Xu, Z.; Zheng, W.; Xue, H. Phospholipase D-derived phosphatidic acid promotes root hair development under phosphorus deficiency by suppressing vacuolar degradation of PIN-FORMED2. New Phytol. 2019, 226, 142–155. [Google Scholar] [CrossRef]
- Pleskot, R.; Li, J.; Žárský, V.; Potocký, M.; Staiger, C.J. Regulation of cytoskeletal dynamics by phospholipase D and phosphatidic acid. Trends Plant. Sci. 2013, 18, 496–504. [Google Scholar] [CrossRef]
- Song, P.; Jia, Q.; Chen, L.; Jin, X.; Xiao, X.; Li, L.; Chen, H.; Qu, Y.; Su, Y.; Zhang, W.; et al. Involvement of Arabidopsis phospholipase D delta in regulation of ROS-mediated microtubule organization and stomatal movement upon heat shock. J. Exp. Bot. 2020, 71, 6555–6570. [Google Scholar] [CrossRef]
- Zhukovsky, M.A.; Filograna, A.; Luini, A.; Corda, D.; Valente, C. Phosphatidic acid in membrane rearrangements. FEBS Lett. 2019, 593, 2428–2451. [Google Scholar] [CrossRef] [Green Version]
- Steinhorst, L.; Kudla, J. How plants perceive salt. Nature 2019, 572, 318–320. [Google Scholar] [CrossRef] [Green Version]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, J.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 2019, 572, 341–346. [Google Scholar] [CrossRef]
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 Complex Generates and Fine-Tunes an AtANN4-Dependent Calcium Signature under Salt Stress. Dev. Cell 2019, 48, 697–709.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, D.; Guo, Y.; Schumaker, K.S.; Zhu, J.-K. The SOS3 Family of Calcium Sensors and SOS2 Family of Protein Kinases in Arabidopsis. Plant. Physiol. 2004, 134, 919–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Wang, C.; Xue, Y.; Liu, X.; Chen, S.; Song, C.; Yang, Y.; Guo, Y. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.-Y.; Ali, Z.; Park, H.J.; Park, S.J.; Cha, J.-Y.; Perez-Hormaeche, J.; Quintero, F.J.; Shin, G.; Kim, M.R.; Qiang, Z.; et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 2013, 4, 1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Lin, H.; Chen, S.; Becker, K.; Yang, Y.; Zhao, J.; Kudla, J.; Schumaker, K.S.; Guo, Y. Inhibition of the Arabidopsis Salt Overly Sensitive Pathway by 14-3-3 Proteins. Plant Cell 2014, 26, 1166–1182. [Google Scholar] [CrossRef] [Green Version]
- Fuglsang, A.T.; Guo, Y.; Cuin, T.A.; Qiu, Q.; Song, C.; Kristiansen, K.A.; Bych, K.; Schulz, A.; Shabala, S.; Schumaker, K.S.; et al. Arabidopsis Protein Kinase PKS5 Inhibits the Plasma Membrane H+-ATPase by Preventing Interaction with 14-3-3 Protein. Plant Cell 2007, 19, 1617–1634. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Du, W.; Yang, Y.; Schumaker, K.S.; Guo, Y. A Calcium-Independent Activation of the Arabidopsis SOS2-Like Protein Kinase24 by Its Interacting SOS3-Like Calcium Binding Protein1. Plant. Physiol. 2014, 164, 2197–2206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Wu, Y.; Ma, L.; Yang, Z.; Dong, Q.; Li, Q.; Ni, X.; Kudla, J.; Song, C.; Guo, Y. The Ca2+ Sensor SCaBP3/CBL7 Modulates Plasma Membrane H+-ATPase Activity and Promotes Alkali Tolerance in Arabidopsis. Plant Cell 2019, 31, 1367–1384. [Google Scholar] [CrossRef] [PubMed]
- Barajas-Lopez, J.D.D.; Moreno, J.R.; Gamez-Arjona, F.M.; Pardo, J.M.; Punkkinen, M.; Zhu, J.-K.; Quintero, F.J.; Fujii, H. Upstream kinases of plant SnRKs are involved in salt stress tolerance. Plant. J. 2017, 93, 107–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, T.; Cai, J.; Zhan, E.; Yang, Y.; Zhao, J.; Guo, Y.; Zhou, H. Stability and localization of 14-3-3 proteins are involved in salt tolerance in Arabidopsis. Plant. Mol. Biol. 2016, 92, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Nie, J.; Cao, C.; Jin, Y.; Yan, M.; Wang, F.; Liu, J.; Xiao, Y.; Liang, Y.; Zhang, W. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 2010, 188, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Yang, Y.; Wu, Y.; Liu, X.; Lei, X.; Guo, Y. A bioassay-guided fractionation system to identify endogenous small molecules that activate plasma membrane H+-ATPase activity in Arabidopsis. J. Exp. Bot. 2017, 68, 2951–2962. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Qin, Y.; Xie, C.; Zhao, F.; Zhao, J.; Liu, D.; Chen, S.; Fuglsang, A.T.; Palmgren, M.; Schumaker, K.S.; et al. The Arabidopsis Chaperone J3 Regulates the Plasma Membrane H+-ATPase through Interaction with the PKS5 Kinase. Plant Cell 2010, 22, 1313–1332. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhou, H.; Zhang, Y.; Li, Z.; Yang, Y.; Guo, Y. The GSK3-like Kinase BIN2 Is a Molecular Switch between the Salt Stress Response and Growth Recovery in Arabidopsis thaliana. Dev. Cell 2020, 55, 367–380.e6. [Google Scholar] [CrossRef]
- Brini, F.; Masmoudi, K. Ion Transporters and Abiotic Stress Tolerance in Plants. ISRN Mol. Biol. 2012, 2012, 1–13. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2017, 217, 523–539. [Google Scholar] [CrossRef] [Green Version]
- Adams, E.; Shin, R. Transport, signaling, and homeostasis of potassium and sodium in plants. J. Integr. Plant. Biol. 2014, 56, 231–249. [Google Scholar] [CrossRef]
- Chérel, I.; Gaillard, I. The Complex Fine-Tuning of K+ Fluxes in Plants in Relation to Osmotic and Ionic Abiotic Stresses. Int. J. Mol. Sci. 2019, 20, 715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.; Hu, J.; Zhou, X.-R.; Yuan, H.-J.; Kumar, T.; Luan, S.; Wang, S.-M. ZxAKT1 is essential for K+uptake and K+/Na+homeostasis in the succulent xerophyteZygophyllum xanthoxylum. Plant. J. 2017, 90, 48–60. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant. Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef] [PubMed]
- Fàbregas, N.; Yoshida, T.; Fernie, A.R. Role of Raf-like kinases in SnRK2 activation and osmotic stress response in plants. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Fujii, H.; Zhu, J.-K. Osmotic stress signaling via protein kinases. Cell. Mol. Life Sci. 2012, 69, 3165–3173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLoughlin, F.; Galvan-Ampudia, C.S.; Julkowska, M.M.; Caarls, L.; van der Does, D.; Laurière, C.; Munnik, T.; Haring, M.A.; Testerink, C. The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant. J. 2012, 72, 436–449. [Google Scholar] [CrossRef] [Green Version]
- Kawa, D.; Meyer, A.J.; Dekker, H.L.; Abd-El-Haliem, A.M.; Gevaert, K.; Van De Slijke, E.; Maszkowska, J.; Bucholc, M.; Dobrowolska, G.; De Jaeger, G.; et al. SnRK2 Protein Kinases and mRNA Decapping Machinery Control Root Development and Response to Salt. Plant. Physiol. 2019, 182, 361–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzywinska, E.; Kulik, A.; Bucholc, M.; Fernandez, M.A.; Rodriguez, P.L.; Dobrowolska, G. Protein phosphatase type 2C PP2CA together with ABI1 inhibits SnRK2.4 activity and regulates plant responses to salinity. Plant. Signal. Behav. 2016, 11, e1253647. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wu, S.; Liu, S.; Takano, T. The Arabidopsis sucrose non-fermenting-1-related protein kinase AtSnRK2.4 interacts with a transcription factor, AtMYB21, that is involved in salt tolerance. Plant Sci. 2021, 303, 110685. [Google Scholar] [CrossRef]
- De Col, V.; Fuchs, P.; Nietzel, T.; Elsässer, M.; Voon, C.P.; Candeo, A.; Seeliger, I.; Fricker, M.D.; Grefen, C.; Møller, I.M.; et al. ATP sensing in living plant cells reveals tissue gradients and stress dynamics of energy physiology. eLife 2017, 6, e26770. [Google Scholar] [CrossRef]
- Lang, T.; Deng, C.; Yao, J.; Zhang, H.; Wang, Y.; Deng, S. A salt-signaling network involving ethylene, extracellular ATP, hydrogen peroxide, and calcium mediates K+/Na+ homeostasis in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 8683. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Naguro, I.; Ichijo, H.; Watanabe, K. Mitogen-activated protein kinases as key players in osmotic stress signaling. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 2037–2052. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Wang, C.; Wang, H.; Li, L.; Wang, C. The Function of MAPK Cascades in Response to Various Stresses in Horticultural Plants. Front. Plant Sci. 2020, 11, 952. [Google Scholar] [CrossRef] [PubMed]
- Im, J.H.; Lee, H.; Kim, J.; Kim, H.B.; An, C.S. Soybean MAPK, GMK1 Is dually regulated by phosphatidic acid and hydrogen peroxide and translocated to nucleus during salt stress. Mol. Cells 2012, 34, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-M.; Woo, D.-H.; Kim, S.-H.; Lee, S.-Y.; Park, H.-Y.; Seok, H.-Y.; Chung, W.S.; Moon, Y.-H. Arabidopsis MKKK20 is involved in osmotic stress response via regulation of MPK6 activity. Plant Cell Rep. 2011, 31, 217–224. [Google Scholar] [CrossRef]
- Kim, S.-H.; Woo, D.-H.; Kim, J.-M.; Lee, S.-Y.; Chung, W.S.; Moon, Y.-H. Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem. Biophys. Res. Commun. 2011, 412, 150–154. [Google Scholar] [CrossRef]
- Shen, L.; Zhuang, B.; Wu, Q.; Zhang, H.; Nie, J.; Jing, W.; Yang, L.; Zhang, W. Phosphatidic acid promotes the activation and plasma membrane localization of MKK7 and MKK9 in response to salt stress. Plant. Sci. 2019, 287, 110190. [Google Scholar] [CrossRef]
- Zhang, J.; Zou, D.; Li, Y.; Sun, X.; Wang, N.-N.; Gong, S.-Y.; Zheng, Y.; Li, X.-B. GhMPK17, a Cotton Mitogen-Activated Protein Kinase, Is Involved in Plant Response to High Salinity and Osmotic Stresses and ABA Signaling. PLoS ONE 2014, 9, e95642. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Jing, W.; Zhang, W. The mitogen-activated protein kinase cascade MKK1–MPK4 mediates salt signaling in rice. Plant. Sci. 2014, 227, 181–189. [Google Scholar] [CrossRef]
- Katsuta, S.; Masuda, G.; Bak, H.; Shinozawa, A.; Kamiyama, Y.; Umezawa, T.; Takezawa, D.; Yotsui, I.; Taji, T.; Sakata, Y. Arabidopsis Raf-like kinases act as positive regulators of subclass III SnRK2 in osmostress signaling. Plant. J. 2020, 103, 634–644. [Google Scholar] [CrossRef]
- Jagodzik, P.; Tajdel-Zielinska, M.; Cieśla, A.; Marczak, M.; Ludwikow, A. Mitogen-Activated Protein Kinase Cascades in Plant Hormone Signaling. Front. Plant Sci. 2018, 9, 1387. [Google Scholar] [CrossRef]
- Ijaz, B.; Formentin, E.; Ronci, B.; Locato, V.; Barizza, E.; Hyder, M.Z.; Schiavo, F.L.; Yasmin, T. Salt tolerance in indica rice cell cultures depends on a fine tuning of ROS signalling and homeostasis. PLoS ONE 2019, 14, e0213986. [Google Scholar] [CrossRef]
- Wang, J.; Huang, R. Modulation of Ethylene and Ascorbic Acid on Reactive Oxygen Species Scavenging in Plant Salt Response. Front. Plant Sci. 2019, 10, 319. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Zhang, H.; Sun, L.; Jiao, Y.; Zhang, G.; Miao, C.; Hao, F. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 2011, 63, 305–317. [Google Scholar] [CrossRef]
- Sagi, M.; Fluhr, R. Production of Reactive Oxygen Species by Plant NADPH Oxidases. Plant. Physiol. 2006, 141, 336–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, H.; Guo, J.; Zhong, Y.; Hsu, C.; Zou, C.; Wang, P.; Zhu, J.; Shi, H. The plasma-membrane polyamine transporter PUT3 is regulated by the Na + /H + antiporter SOS1 and protein kinase SOS2. New Phytol. 2020, 226, 785–797. [Google Scholar] [CrossRef]
- Che-Othman, M.H.; Jacoby, R.P.; Millar, A.H.; Taylor, N.L. Wheat mitochondrial respiration shifts from the tricarboxylic acid cycle to the GABA shunt under salt stress. New Phytol. 2019, 225, 1166–1180. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Liu, L.; Wang, L.; Li, B.; Jin, C.; Lin, X. Melatonin: A master regulator of plant development and stress responses. J. Integr. Plant. Biol. 2020, 63, 126–145. [Google Scholar] [CrossRef]
- Wu, Y.; Jin, X.; Liao, W.; Hu, L.; Dawuda, M.M.; Zhao, X.; Tang, Z.; Gong, T.; Yu, J. 5-Aminolevulinic Acid (ALA) Alleviated Salinity Stress in Cucumber Seedlings by Enhancing Chlorophyll Synthesis Pathway. Front. Plant Sci. 2018, 9, 635. [Google Scholar] [CrossRef] [Green Version]
- Endler, A.; Kesten, C.; Schneider, R.; Zhang, Y.; Ivakov, A.; Froehlich, A.; Funke, N.; Persson, S. A Mechanism for Sustained Cellulose Synthesis during Salt Stress. Cell 2015, 162, 1353–1364. [Google Scholar] [CrossRef] [Green Version]
- Testerink, C.; Munnik, T. Phosphatidic acid: A multifunctional stress signaling lipid in plants. Trends Plant. Sci. 2005, 10, 368–375. [Google Scholar] [CrossRef]
- Hong, Y.; Pan, X.; Welti, R.; Wang, X. Phospholipase Dalpha3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 2008, 20, 803–816. [Google Scholar] [CrossRef] [Green Version]
- Arisz, S.; Testerink, C.; Munnik, T. Plant PA signaling via diacylglycerol kinase. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2009, 1791, 869–875. [Google Scholar] [CrossRef]
- Bargmann, B.O.R.; Laxalt, A.M.; ter Riet, B.; van Schooten, B.; Merquiol, E.; Testerink, C.; Haring, M.A.; Bartels, D.; Munnik, T. Multiple PLDs Required for High Salinity and Water Deficit Tolerance in Plants. Plant Cell Physiol. 2008, 50, 78–89. [Google Scholar] [CrossRef] [Green Version]
- Darwish, E.; Testerink, C.; Khalil, M.; El-Shihy, O.; Munnik, T. Phospholipid Signaling Responses in Salt-Stressed Rice Leaves. Plant Cell Physiol. 2009, 50, 986–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munnik, T.; Meijer, H.; Ter Riet, B.; Hirt, H.; Frank, W.; Bartels, D.; Musgrave, A. Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant. J. 2000, 22, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Zarza, X.; Shabala, L.; Fujita, M.; Shabala, S.; Haring, M.A.; Tiburcio, A.F.; Munnik, T. Extracellular Spermine Triggers a Rapid Intracellular Phosphatidic Acid Response in Arabidopsis, Involving PLDδ Activation and Stimulating Ion Flux. Front. Plant Sci. 2019, 10, 601. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Song, T.; Wallrad, L.; Kudla, J.; Wang, X.; Zhang, W. Tissue-specific accumulation of pH-sensing phosphatidic acid determines plant stress tolerance. Nat. Plants 2019, 5, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Bargmann, B.; Munnik, T. The role of phospholipase D in plant stress responses. Curr. Opin. Plant. Biol. 2006, 9, 515–522. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Lin, F.; Mao, T.; Nie, J.; Yan, M.; Yuan, M.; Zhang, W. Phosphatidic Acid Regulates Microtubule Organization by Interacting with MAP65-1 in Response to Salt Stress in Arabidopsis. Plant Cell 2012, 24, 4555–4576. [Google Scholar] [CrossRef] [Green Version]
- Galvan-Ampudia, C.S.; Julkowska, M.; Darwish, E.; Gandullo, J.; Korver, R.A.; Brunoud, G.; Haring, M.A.; Munnik, T.; Vernoux, T.; Testerink, C. Halotropism Is a Response of Plant Roots to Avoid a Saline Environment. Curr. Biol. 2013, 23, 2044–2050. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Qin, C.; Zhao, J.; Wang, X. Phospholipase D 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2004, 101, 9508–9513. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhu, H.; Zhang, Q.; Li, M.; Yan, M.; Wang, R.; Wang, L.; Welti, R.; Zhang, W.; Wang, X. Phospholipase dalpha1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 2009, 21, 2357–2377. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, S.R.; Pandey, S. Phosphatidic acid binding inhibits RGS 1 activity to affect specific signaling pathways in Arabidopsis. Plant. J. 2017, 90, 466–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Mishra, G.; Markham, J.E.; Li, M.; Tawfall, A.; Welti, R.; Wang, X. Connections between Sphingosine Kinase and Phospholipase D in the Abscisic Acid Signaling Pathway in Arabidopsis. J. Biol. Chem. 2012, 287, 8286–8296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, C.; Wang, P.; Song, H.; Jing, W.; Shen, L.; Zhang, Q.; Zhang, W. Phosphatidic acid binds to and regulates guanine nucleotide exchange factor 8 (GEF8) activity in Arabidopsis. Funct. Plant. Biol. 2017, 44, 1029. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Wang, R.; Jing, W.; Zhang, W. Rice Phospholipase Dα is Involved in Salt Tolerance by the Mediation of H+-ATPase Activity and Transcription. J. Integr. Plant. Biol. 2010, 53, 289–299. [Google Scholar] [CrossRef]
- Yu, H.Q.; Yong, T.M.; Li, H.J.; Liu, Y.P.; Zhou, S.F.; Fu, F.L.; Li, W.C. Overexpression of a phospholipase Dalpha gene from Ammopiptanthus nanus enhances salt tolerance of phospholipase Dalpha1-deficient Arabidopsis mutant. Planta 2015, 242, 1495–1509. [Google Scholar] [CrossRef] [PubMed]
- Ji, T.; Li, S.; Huang, M.; Di, Q.; Wang, X.; Wei, M.; Shi, Q.; Li, Y.; Gong, B.; Yang, F. Overexpression of cucumber phospholipase D alpha gene (CsPLDalpha) in tobacco enhanced salinity stress tolerance by regulating Na+-K+ balance and lipid peroxidation. Front. Plant Sci. 2017, 8, 499. [Google Scholar] [CrossRef] [PubMed]
- Sagar, S.; Singh, A. Emerging role of phospholipase C mediated lipid signaling in abiotic stress tolerance and development in plants. Plant Cell Rep. 2021, 1–11. [Google Scholar] [CrossRef]
- Takáč, T.; Novák, D.; Šamaj, J. Recent Advances in the Cellular and Developmental Biology of Phospholipases in Plants. Front. Plant Sci. 2019, 10, 362. [Google Scholar] [CrossRef]
- Ge, H.; Chen, C.; Jing, W.; Zhang, Q.; Wang, H.; Wang, R.; Zhang, W. The Rice Diacylglycerol Kinase Family: Functional Analysis Using Transient RNA Interference. Front. Plant Sci. 2012, 3, 60. [Google Scholar] [CrossRef] [Green Version]
- Heilmann, I. Phosphoinositide signaling in plant development. Development 2016, 143, 2044–2055. [Google Scholar] [CrossRef] [Green Version]
- Fatiha, A. Plant Lipid Metabolism, Advances in Lipid Metabolism; Baez, R.V., Ed.; IntechOpen: London, UK, 2018. [Google Scholar]
- Lee, Y.; Kim, E.-S.; Choi, Y.; Hwang, I.; Staiger, C.J.; Chung, Y.-Y.; Lee, Y. The Arabidopsis Phosphatidylinositol 3-Kinase Is Important for Pollen Development. Plant. Physiol. 2008, 147, 1886–1897. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Hu, W.; Li, F.; Marshall, R.S.; Zarza, X.; Munnik, T.; Vierstra, R.D. AUTOPHAGY-RELATED14 and Its Associated Phosphatidylinositol 3-Kinase Complex Promote Autophagy in Arabidopsis. Plant Cell 2020, 32, 3939–3960. [Google Scholar] [CrossRef]
- Heilmann, M.; Heilmann, I. Plant phosphoinositides—Complex networks controlling growth and adaptation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2015, 1851, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhao, C.Y.; Tan, S.T.; Xue, H.W. Arabidopsis type II phosphatidylinositol 4-Kinase PI4Kgamma5 regulates auxin biosynthesis and leaf margin development through interacting with membrane-bound transcription factor ANAC078. PLoS Gene 2016, 12, e1006252. [Google Scholar]
- Yang, Y.; Han, X.; Ma, L.; Wu, Y.; Liu, X.; Fu, H.; Liu, G.; Lei, X.; Guo, Y. Dynamic changes of phosphatidylinositol and phosphatidylinositol 4-phosphate levels modulate H+-ATPase and Na+/H+ antiporter activities to maintain ion homeostasis in Arabidopsis under salt stress. Mol. Plant. 2021. [Google Scholar] [CrossRef]
- Gonorazky, G.; Laxalt, A.M.; Dekker, H.L.; Rep, M.; Munnik, T.; Testerink, C.; de la Canal, L. Phosphatidylinositol 4-phosphate is associated to extracellular lipoproteic fractions and is detected in tomato apoplastic fluids. Plant. Biol. 2011, 14, 41–49. [Google Scholar] [CrossRef]
- Simon, M.L.A.; Platre, M.P.; Assil, S.; Van Wijk, R.; Chen, W.Y.; Chory, J.; Dreux, M.; Munnik, T.; Jaillais, Y. A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant. J. 2013, 77, 322–337. [Google Scholar] [CrossRef] [Green Version]
- Simon, M.L.A.; Platre, M.P.; Marquès-Bueno, M.M.; Armengot, L.; Stanislas, T.; Bayle, V.; Caillaud, M.-C.; Jaillais, Y. A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat. Plants 2016, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, N.; King, S.; Samuels, A.L.; McFarlane, H.E. Subcellular coordination of plant cell wall synthesis. Dev. Cell 2021, 56, 933–948. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, M.; Suda, Y.; Vernhettes, S.; Nakano, A.; Ueda, T. Phosphatidylinositol 3-Kinase and 4-Kinase Have Distinct Roles in Intracellular Trafficking of Cellulose Synthase Complexes in Arabidopsis thaliana. Plant Cell Physiol. 2014, 56, 287–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagashima, Y.; Ma, Z.; Liu, X.; Qian, X.; Zhang, X.; von Schaewen, A.; Koiwa, H. Multiple Quality Control Mechanisms in the ER and TGN Determine Subcellular Dynamics and Salt-Stress Tolerance Function of KORRIGAN1. Plant Cell 2019, 32, 470–485. [Google Scholar] [CrossRef] [PubMed]
- Rubilar-Hernandez, C.; Osorio-Navarro, C.; Cabello, F.; Norambuena, L. PI4KIIIbeta Activity Regulates Lateral Root Formation Driven by Endocytic Trafficking to the Vacuole. Plant Physiol. 2019, 181, 112–126. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Dabi, M.; Sapara, K.K.; Joshi, P.S.; Agarwal, P.K. Ectopic Expression of JcWRKY Transcription Factor Confers Salinity Tolerance via Salicylic Acid Signaling. Front. Plant Sci. 2016, 7, 1541. [Google Scholar] [CrossRef] [Green Version]
- Ahanger, M.A.; Aziz, U.; Alsahli, A.A.; Alyemeni, M.N.; Ahmad, P. Influence of Exogenous Salicylic Acid and Nitric Oxide on Growth, Photosynthesis, and Ascorbate-Glutathione Cycle in Salt Stressed Vigna angularis. Biomolecules 2019, 10, 42. [Google Scholar] [CrossRef] [Green Version]
- Jayakannan, M.; Bose, J.; Babourina, O.; Shabala, S.; Massart, A.; Poschenrieder, C.; Rengel, Z. The NPR1-dependent salicylic acid signalling pathway is pivotal for enhanced salt and oxidative stress tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 1865–1875. [Google Scholar] [CrossRef] [Green Version]
- Janda, M.; Sasek, V.; Ruelland, E. The Arabidopsis pi4kIIIbeta1beta2 double mutant is salicylic acid-overaccumulating: A new example of salicylic acid influence on plant stature. Plant Signal. Behav. 2014, 9, e977210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krinke, O.; Ruelland, E.; Valentová, O.; Vergnolle, C.; Renou, J.-P.; Taconnat, L.; Flemr, M.; Burketová, L.; Zachowski, A. Phosphatidylinositol 4-Kinase Activation Is an Early Response to Salicylic Acid in Arabidopsis Suspension Cells. Plant. Physiol. 2007, 144, 1347–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šašek, V.; Janda, M.; Delage, E.; Puyaubert, J.; Guivarc’h, A.; López Maseda, E.; Dobrev, P.I.; Caius, J.; Bóka, K.; Valentová, O.; et al. Constitutive salicylic acid accumulation in pi4kIIIβ1β2 Arabidopsis plants stunts rosette but not root growth. New Phytol. 2014, 203, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Antignani, V.; Klocko, A.L.; Bak, G.; Chandrasekaran, S.D.; Dunivin, T.; Nielsen, E. Recruitment of PLANT U-BOX13 and the PI4Kβ1/β2 phosphatidylinositol-4 kinases by the small GTPase RabA4B plays important roles during salicylic acid-mediated plant defense signaling in Arabidopsis. Plant Cell 2015, 27, 243–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeWald, D.B.; Torabinejad, J.; Jones, C.A.; Shope, J.C.; Cangelosi, A.R.; Thompson, J.E.; Prestwich, G.D.; Hama, H. Rapid Accumulation of Phosphatidylinositol 4,5-Bisphosphate and Inositol 1,4,5-Trisphosphate Correlates with Calcium Mobilization in Salt-Stressed Arabidopsis. Plant. Physiol. 2001, 126, 759–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribba, T.; Garrido-Vargas, F.; O’Brien, J.A. Auxin-mediated responses under salt stress: From developmental regulation to biotechnological applications. J. Exp. Bot. 2020, 71, 3843–3853. [Google Scholar] [CrossRef]
- Tejos, R.; Sauer, M.; Vanneste, S.; Palacios-Gomez, M.; Li, H.; Heilmann, M.; van Wijk, R.; Vermeer, J.; Heilmann, I.; Munnik, T.; et al. Bipolar Plasma Membrane Distribution of Phosphoinositides and Their Requirement for Auxin-Mediated Cell Polarity and Patterning in Arabidopsis. Plant Cell 2014, 26, 2114–2128. [Google Scholar] [CrossRef] [Green Version]
- Ischebeck, T.; Werner, S.; Krishnamoorthy, P.; Lerche, J.; Meijón, M.; Stenzel, I.; Löfke, C.; Wiessner, T.; Im, Y.J.; Perera, I.; et al. Phosphatidylinositol 4,5-Bisphosphate Influences PIN Polarization by Controlling Clathrin-Mediated Membrane Trafficking in Arabidopsis. Plant Cell 2013, 25, 4894–4911. [Google Scholar] [CrossRef] [Green Version]
- Mei, Y.; Jia, W.-J.; Chu, Y.-J.; Xue, H.-W. Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Res. 2011, 22, 581–597. [Google Scholar] [CrossRef] [Green Version]
- Heilmann, I. Towards understanding the function of stress-inducible PtdIns(4,5)P2 in plants. Commun. Integr. Biol. 2008, 1, 204–206. [Google Scholar] [CrossRef]
- König, S.; Ischebeck, T.; Lerche, J.; Stenzel, I.; Heilmann, I. Salt-stress-induced association of phosphatidylinositol 4,5-bisphosphate with clathrin-coated vesicles in plants. Biochem. J. 2008, 415, 387–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.; Vanneste, S.; Pérez-Sancho, J.; Benitez-Fuente, F.; Strelau, M.; Macho, A.P.; Botella, M.A.; Friml, J.; Rosado, A. Ionic stress enhances ER–PM connectivity via phosphoinositide-associated SYT1 contact site expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 1420–1429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, R.T.; Hasanuzzaman, M. Exogenous kinetin and putrescine synergistically mitigate salt stress in Luffa acutangula by modulating physiology and antioxidant defense. Physiol. Mol. Biol. Plants 2020, 26, 2125–2137. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jin, H.; Zhang, Q. The Effect of Exogenous Spermidine Concentration on Polyamine Metabolism and Salt Tolerance in Zoysiagrass (Zoysia japonica Steud) Subjected to Short-Term Salinity Stress. Front. Plant Sci. 2016, 7, 1221. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Yang, R.; Jian, N.; Wei, L.; Ye, L.; Wang, R.; Gao, H.; Zheng, Q. Putrescine metabolism modulates the biphasic effects of brassinosteroids on canola and Arabidopsis salt tolerance. Plant Cell Environ. 2020, 43, 1348–1359. [Google Scholar] [CrossRef]
- Liu, L.; Liu, D.; Wang, Z.; Zou, C.; Wang, B.; Zhang, H.; Gai, Z.; Zhang, P.; Wang, Y.; Li, C. Exogenous allantoin improves the salt tolerance of sugar beet by increasing putrescine metabolism and antioxidant activities. Plant. Physiol. Biochem. 2020, 154, 699–713. [Google Scholar] [CrossRef]
- Zarza, X.; Van Wijk, R.; Shabala, L.; Hunkeler, A.; Lefebvre, M.; Rodriguez-Villalón, A.; Shabala, S.; Tiburcio, A.F.; Heilmann, I.; Munnik, T. Lipid kinases PIP5K7 and PIP5K9 are required for polyamine-triggered K+efflux in Arabidopsis roots. Plant. J. 2020, 104, 416–432. [Google Scholar] [CrossRef]
- Leshem, Y.; Seri, L.; Levine, A. Induction of phosphatidylinositol 3-kinase-mediated endocytosis by salt stress leads to intracellular production of reactive oxygen species and salt tolerance. Plant. J. 2007, 51, 185–197. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, J.; Xing, D. Phosphatidylinositol 3-Kinase Plays a Vital Role in Regulation of Rice Seed Vigor via Altering NADPH Oxidase Activity. PLoS ONE 2012, 7, e33817. [Google Scholar] [CrossRef]
- Jung, J.-Y.; Kim, Y.-W.; Kwak, J.M.; Hwang, J.-U.; Young, J.; Schroeder, J.; Hwang, I.; Lee, Y. Phosphatidylinositol 3- and 4-Phosphate Are Required for Normal Stomatal Movements. Plant Cell 2002, 14, 2399–2412. [Google Scholar] [CrossRef] [Green Version]
- Park, K.-Y.; Jung, J.-Y.; Park, J.; Hwang, J.-U.; Kim, Y.-W.; Hwang, I.; Lee, Y. A Role for Phosphatidylinositol 3-Phosphate in Abscisic Acid-Induced Reactive Oxygen Species Generation in Guard Cells. Plant. Physiol. 2003, 132, 92–98. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Liu, X.; Zhang, X.; Qin, N.; Xu, K.; Yin, W.; Zheng, Y.; Song, Y.; Zeng, R.; Liu, J. Phosphoinositide 3-Kinase Promotes Oxidative Burst, Stomatal Closure and Plant Immunity in Bacterial Invasion. Front. Plant Sci. 2020, 10, 1740. [Google Scholar] [CrossRef]
- Avin-Wittenberg, T. Autophagy and its role in plant abiotic stress management. Plant Cell Environ. 2019, 42, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Xia, F.-N.; Xiao, S. Autophagy in plants: Physiological roles and post-translational regulation. J. Integr. Plant. Biol. 2021, 63, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xiong, Y.; Bassham, D.C. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009, 5, 954–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, L.; Zhang, P.; Zhu, R.; Fu, J.; Su, J.; Zheng, J.; Wang, Z.; Wang, D.; Gong, Q. Autophagy Is Rapidly Induced by Salt Stress and Is Required for Salt Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1459. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, X.; Wang, H.; Lam, S.K.; Gao, C.; Wang, X.; Cai, Y.; Jiang, L. A BAR-Domain Protein SH3P2, Which Binds to Phosphatidylinositol 3-Phosphate and ATG8, Regulates Autophagosome Formation in Arabidopsis. Plant Cell 2013, 25, 4596–4615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dek, M.S.P.; Padmanabhan, P.; Sherif, S.; Subramanian, J.; Paliyath, A.G. Upregulation of Phosphatidylinositol 3-Kinase (PI3K) Enhances Ethylene Biosynthesis and Accelerates Flower Senescence in Transgenic Nicotiana tabacum L. Int. J. Mol. Sci. 2017, 18, 1533. [Google Scholar] [CrossRef]
- Villasuso, A.L.; Racagni, G.E.; Machado, E.E. Phosphatidylinositol kinases as regulators of GA-stimulated α-amylase secretion in barley (Hordeum vulgare). Physiol. Plant. 2008, 133, 157–166. [Google Scholar] [CrossRef]
- Liu, J.; Ji, Y.; Zhou, J.; Xing, D. Phosphatidylinositol 3-Kinase Promotes V-ATPase Activation and Vacuolar Acidification and Delays Methyl Jasmonate-Induced Leaf Senescence. Plant. Physiol. 2016, 170, 1714–1731. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zhou, J.; Xing, D. A pivotal role of phosphatidylinositol 3-kinase in delaying of methyl jasmonate-induced leaf senescence. Plant. Signal. Behav. 2016, 11, e1147642. [Google Scholar] [CrossRef] [Green Version]
- Delgado, C.; Mora-Poblete, F.; Ahmar, S.; Chen, J.-T.; Figueroa, C. Jasmonates and Plant Salt Stress: Molecular Players, Physiological Effects, and Improving Tolerance by Using Genome-Associated Tools. Int. J. Mol. Sci. 2021, 22, 3082. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yin, C.; Ma, B.; Chen, S.; Zhang, J. Ethylene signaling in rice and Arabidopsis: New regulators and mechanisms. J. Integr. Plant. Biol. 2020, 63, 102–125. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-G.; Park, C.-M. Gibberellic acid-mediated salt signaling in seed germination. Plant. Signal. Behav. 2008, 3, 877–879. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Zhang, P.; Li, C.; Xia, G. The moss jasmonate ZIM-domain protein PnJAZ1 confers salinity tolerance via crosstalk with the abscisic acid signalling pathway. Plant. Sci. 2018, 280, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Leprince, A.-S.; Magalhaes, N.; de Vos, D.; Bordenave, M.; Crilat, E.; Clement, G.; Meyer, C.; Munnik, T.; Savoure, A.; Clément, G.; et al. Involvement of Phosphatidylinositol 3-kinase in the regulation of proline catabolism in Arabidopsis thaliana. Front. Plant Sci. 2015, 5, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirano, T.; Munnik, T.; Sato, M.H. Phosphatidylinositol 3-phosphate 5-kinase, FAB1/PIKfyve kinase mediates endosome maturation to establish endosome-cortical microtubule interaction in Arabidopsis. Plant Physiol. 2015, 169, 1961–1974. [Google Scholar] [CrossRef] [Green Version]
- Hirano, T.; Sato, M.H. Arabidopsis FAB1A/B is possibly involved in the recycling of auxin transporters. Plant Signal. Behav. 2011, 6, 583–585. [Google Scholar] [CrossRef] [Green Version]
- Karali, D.; Oxley, D.; Runions, J.; Ktistakis, N.; Farmaki, T. The Arabidopsis thaliana Immunophilin ROF1 Directly Interacts with PI(3)P and PI(3,5)P2 and Affects Germination under Osmotic Stress. PLoS ONE 2012, 7, e48241. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Yuan, S.; Cao, H.; Lam, S.M.; Shui, G.; Hong, Y.; Wang, X. Phosphatidylinositol-hydrolyzing phospholipase C4 modulates rice response to salt and drought. Plant Cell Environ. 2018, 42, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kanwar, P.; Pandey, A.; Tyagi, A.K.; Sopory, S.K.; Kapoor, S.; Pandey, G.K. Comprehensive Genomic Analysis and Expression Profiling of Phospholipase C Gene Family during Abiotic Stresses and Development in Rice. PLoS ONE 2013, 8, e62494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Wang, F.; Yan, P.; Jing, W.; Zhang, C.; Kudla, J.; Zhang, W. A phosphoinositide-specific phospholipase C pathway elicits stress-induced Ca 2+ signals and confers salt tolerance to rice. New Phytol. 2017, 214, 1172–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodas-Junco, B.; Racagni-Di-Palma, G.; Canul-Chan, M.; Usorach, J.; Hernández-Sotomayor, S. Link between Lipid Second Messengers and Osmotic Stress in Plants. Int. J. Mol. Sci. 2021, 22, 2658. [Google Scholar] [CrossRef]
- Sanchez, J.P.; Chua, N.H. Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell 2001, 13, 1143–1154. [Google Scholar] [CrossRef] [Green Version]
- Hirayama, T.; Ohto, C.; Mizoguchi, T.; Shinozaki, K. A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1995, 92, 3903–3907. [Google Scholar] [CrossRef] [Green Version]
- Hunt, L.; Mills, L.N.; Pical, C.; Leckie, C.P.; Aitken, F.L.; Kopka, J.; Mueller-Roeber, B.; McAinsh, M.R.; Hetherington, A.; Gray, J.E. Phospholipase C is required for the control of stomatal aperture by ABA. Plant. J. 2003, 34, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Mills, L.N.; Hunt, L.; Leckie, C.P.; Aitken, F.L.; Wentworth, M.; McAinsh, M.R.; Gray, J.E.; Hetherington, A.M. The effects of manipulating phospholipase C on guard cell ABA-signalling. J. Exp. Bot. 2003, 55, 199–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staxén, I.; Pical, C.; Montgomery, L.T.; Gray, J.E.; Hetherington, A.; McAinsh, M.R. Abscisic acid induces oscillations in guard-cell cytosolic free calcium that involve phosphoinositide-specific phospholipase C. Proc. Natl. Acad. Sci. USA 1999, 96, 1779–1784. [Google Scholar] [CrossRef] [Green Version]
- Van Wijk, R.; Zhang, Q.; Zarza, X.; Lamers, M.; Marquez, F.R.; Guardia, A.; Scuffi, D.; García-Mata, C.; Ligterink, W.; Haring, M.A.; et al. Role for Arabidopsis PLC7 in Stomatal Movement, Seed Mucilage Attachment, and Leaf Serration. Front. Plant Sci. 2018, 9, 1721. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; van Wijk, R.; Shahbaz, M.; Roels, W.; Van Schooten, B.; Vermeer, J.; Zarza, X.; Guardia, A.; Scuffi, D.; García-Mata, C.; et al. Arabidopsis Phospholipase C3 is Involved in Lateral Root Initiation and ABA Responses in Seed Germination and Stomatal Closure. Plant Cell Physiol. 2017, 59, 469–486. [Google Scholar] [CrossRef]
- Georges, F.; Das, S.; Ray, H.; Bock, C.; Nokhrina, K.; Kolla, V.A.; Keller, W. Over-expression of Brassica napus phosphatidylinositol-phospholipase C2 in canola induces significant changes in gene expression and phytohormone distribution patterns, enhances drought tolerance and promotes early flowering and maturation. Plant Cell Environ. 2009, 32, 1664–1681. [Google Scholar] [CrossRef]
- Chen, X.; Li, L.; Xu, B.; Zhao, S.; Lu, P.; He, Y.; Ye, T.; Feng, Y.-Q.; Wu, Y. Phosphatidylinositol-specific phospholipase C2 functions in auxin-modulated root development. Plant Cell Environ. 2018, 42, 1441–1457. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; van Wijk, R.; Zarza, X.; Shahbaz, M.; Van Hooren, M.; Guardia, A.; Scuffi, D.; García-Mata, C.; Ende, W.V.D.; Hoffmann-Benning, S.; et al. Knock-Down of Arabidopsis PLC5 Reduces Primary Root Growth and Secondary Root Formation While Overexpression Improves Drought Tolerance and Causes Stunted Root Hair Growth. Plant Cell Physiol. 2018, 59, 2004–2019. [Google Scholar] [CrossRef] [PubMed]
- Kanehara, K.; Yu, C.-Y.; Cho, Y.; Cheong, W.-F.; Torta, F.; Shui, G.; Wenk, M.R.; Nakamura, Y. Arabidopsis AtPLC2 Is a Primary Phosphoinositide-Specific Phospholipase C in Phosphoinositide Metabolism and the Endoplasmic Reticulum Stress Response. PLoS Genet. 2015, 11, e1005511. [Google Scholar] [CrossRef] [Green Version]
- Parre, E.; Ghars, M.A.; Leprince, A.-S.; Thiery, L.; Lefebvre, D.; Bordenave, M.; Richard, L.; Mazars, C.; Abdelly, C.; Savouré, A. Calcium Signaling via Phospholipase C Is Essential for Proline Accumulation upon Ionic but Not Nonionic Hyperosmotic Stresses in Arabidopsis. Plant. Physiol. 2007, 144, 503–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djafi, N.; Vergnolle, C.; Cantrel, C.; Wietrzyñski, W.; Delage, E.; Cochet, F.; Puyaubert, J.; Soubigou-Taconnat, L.; Gey, D.; Collin, S.; et al. The Arabidopsis DREB2 genetic pathway is constitutively repressed by basal phosphoinositide-dependent phospholipase C coupled to diacylglycerol kinase. Front. Plant Sci. 2013, 4, 307. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Zhao, J.; Guo, L.; Kim, S.-C.; Deng, X.; Wang, G.; Zhang, G.; Li, M.; Wang, X. Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 2016, 62, 55–74. [Google Scholar] [CrossRef] [Green Version]
- Yamaoka, Y.; Yu, Y.; Mizoi, J.; Fujiki, Y.; Saito, K.; Nishijima, M.; Lee, Y.; Nishida, I. Phosphatidylserine synthase1 is required for microspore development in Arabidopsis thaliana. Plant. J. 2011, 67, 648–661. [Google Scholar] [CrossRef]
- Lv, S.; Tai, F.; Guo, J.; Jiang, P.; Lin, K.; Wang, D.; Zhang, X.; Li, Y. Phosphatidylserine Synthase from Salicornia europaea Is Involved in Plant Salt Tolerance by Regulating Plasma Membrane Stability. Plant Cell Physiol. 2020, 62, 66–79. [Google Scholar] [CrossRef]
- Yu, Y.; Kou, M.; Gao, Z.; Liu, Y.; Xuan, Y.; Liu, Y.; Tang, Z.; Cao, Q.; Li, Z.; Sun, J. Involvement of Phosphatidylserine and Triacylglycerol in the Response of Sweet Potato Leaves to Salt Stress. Front. Plant Sci. 2019, 10, 1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Xuan, Y.; Bian, X.; Zhang, L.; Pan, Z.; Kou, M.; Cao, Q.; Tang, Z.; Li, Q.; Ma, D.; et al. Overexpression of phosphatidylserine synthase IbPSS1 affords cellular Na+ homeostasis and salt tolerance by activating plasma membrane Na+/H+ antiport activity in sweet potato roots. Hortic. Res. 2020, 7, 131. [Google Scholar] [CrossRef] [PubMed]
- Platre, M.P.; Noack, L.; Doumane, M.; Bayle, V.; Simon, M.L.A.; Maneta-Peyret, L.; Fouillen, L.; Stanislas, T.; Armengot, L.; Pejchar, P.; et al. A Combinatorial Lipid Code Shapes the Electrostatic Landscape of Plant Endomembranes. Dev. Cell 2018, 45, 465–480.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Platre, M.P.; Bayle, V.; Armengot, L.; Bareille, J.; Marquès-Bueno, M.D.M.; Creff, A.; Maneta-Peyret, L.; Fiche, J.-B.; Nollmann, M.; Miège, C.; et al. Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science 2019, 364, 57–62. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Lu, H.; Li, W.; Yuan, M.; Fu, Y. A ROP2-RIC1 pathway fine-tunes microtubule reorganization for salt tolerance in Arabidopsis. Plant Cell Environ. 2017, 40, 1127–1142. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Taylor, M.C.; Barrow, R.A.; Croyal, M.; Masle, J. Loss of Phosphoethanolamine N-Methyltransferases Abolishes Phosphatidylcholine Synthesis and Is Lethal. Plant. Physiol. 2018, 179, 124–142. [Google Scholar] [CrossRef] [Green Version]
- Omoto, E.; Iwasaki, Y.; Miyake, H.; Taniguchi, M. Salinity induces membrane structure and lipid changes in maize mesophyll and bundle sheath chloroplasts. Physiol. Plant. 2015, 157, 13–23. [Google Scholar] [CrossRef]
- Pical, C.; Westergren, T.; Dove, S.K.; Larsson, C.; Sommarin, M. Salinity and Hyperosmotic Stress Induce Rapid Increases in Phosphatidylinositol 4,5-Bisphosphate, Diacylglycerol Pyrophosphate, and Phosphatidylcholine in Arabidopsis thaliana Cells. J. Biol. Chem. 1999, 274, 38232–38240. [Google Scholar] [CrossRef] [Green Version]
- Tasseva, G.; Richard, L.; Zachowski, A. Regulation of phosphatidylcholine biosynthesis under salt stress involves choline kinases in Arabidopsis thaliana. FEBS Lett. 2004, 566, 115–120. [Google Scholar] [CrossRef]
- Qiao, K.; Wang, M.; Takano, T.; Liu, S. Overexpression of acyl-CoA-binding protein 1 (ChACBP1) from saline-alkali-tolerant Chlorella sp. enhances stress tolerance in Arabidopsis. Front. Plant Sci. 2018, 9, 1772. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, J.; Yan, B.; Wei, Y.; Ge, S.; Li, J.; Han, Y.; Li, Z.; Zhao, C.; Xu, J. The Adjustment of Membrane Lipid Metabolism Pathways in Maize Roots Under Saline–Alkaline Stress. Front. Plant Sci. 2021, 12, 635327. [Google Scholar] [CrossRef]
- Caldo, K.M.P.; Xu, Y.; Falarz, L.; Jayawardhane, K.; Acedo, J.; Chen, G. Arabidopsis CTP:phosphocholine cytidylyltransferase 1 is phosphorylated and inhibited by sucrose nonfermenting 1–related protein kinase 1 (SnRK1). J. Biol. Chem. 2019, 294, 15862–15874. [Google Scholar] [CrossRef]
- Lin, Y.; Kanehara, K.; Nakamura, Y. Arabidopsis CHOLINE/ETHANOLAMINE KINASE 1 (CEK1) is a primary choline kinase localized at the endoplasmic reticulum (ER) and involved in ER stress tolerance. New Phytol. 2019, 223, 1904–1917. [Google Scholar] [CrossRef]
- Yu, C.-Y.; Nguyen, V.C.; Chuang, L.; Kanehara, K. Membrane glycerolipid equilibrium under endoplasmic reticulum stress in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2018, 500, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Kocourková, D.; Krčková, Z.; Pejchar, P.; Veselková, S.; Valentová, O.; Wimalasekera, R.; Scherer, G.F.E.; Martinec, J. The phosphatidylcholine-hydrolysing phospholipase C NPC4 plays a role in response of Arabidopsis roots to salt stress. J. Exp. Bot. 2011, 62, 3753–3763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, C.; Li, M.; Narasimhan, R.; Roth, M.; Welti, R.; Wang, X. Nonspecific Phospholipase C NPC4 Promotes Responses to Abscisic Acid and Tolerance to Hyperosmotic Stress in Arabidopsis. Plant Cell 2010, 22, 2642–2659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Li, M.; Zhang, X.; Yang, Q.; Huang, B. Up-regulation of lipid metabolism and glycine betaine synthesis are associated with choline-induced salt tolerance in halophytic seashore paspalum. Plant Cell Environ. 2019, 43, 159–173. [Google Scholar] [CrossRef]
- Summers, P.S.; Weretilnyk, E.A. Choline Synthesis in Spinach in Relation to Salt Stress. Plant. Physiol. 1993, 103, 1269–1276. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Lyu, W.; Gao, Y.; Zhang, X.; Sun, Y.; Huang, B. Choline-Mediated Lipid Reprogramming as a Dominant Salt Tolerance Mechanism in Grass Species Lacking Glycine Betaine. Plant Cell Physiol. 2020, 61, 2018–2030. [Google Scholar] [CrossRef]
- Xu, C.; Härtel, H.; Wada, H.; Hagio, M.; Yu, B.; Eakin, C.; Benning, C. The pgp1 Mutant Locus of Arabidopsis Encodes a Phosphatidylglycerolphosphate Synthase with Impaired Activity. Plant. Physiol. 2002, 129, 594–604. [Google Scholar] [CrossRef] [Green Version]
- Frentzen, M. Phosphatidylglycerol and sulfoquinovosyldiacylglycerol: Anionic membrane lipids and phosphate regulation. Curr. Opin. Plant. Biol. 2004, 7, 270–276. [Google Scholar] [CrossRef]
- Han, X.; Shi, Y.; Liu, G.; Guo, Y.; Yang, Y. Activation of ROP6 GTPase by Phosphatidylglycerol in Arabidopsis. Front. Plant Sci. 2018, 9, 347. [Google Scholar] [CrossRef]
- Kobayashi, K.; Endo, K.; Wada, H. Multiple Impacts of Loss of Plastidic Phosphatidylglycerol Biosynthesis on Photosynthesis during Seedling Growth of Arabidopsis. Front. Plant Sci. 2016, 7, 336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.-J.; Wang, N.; Bao, J.-J.; Zhu, H.-X.; Wang, L.-J.; Chen, X.-Y. Lipidomic Analysis Reveals the Importance of GIPCs in Arabidopsis Leaf Extracellular Vesicles. Mol. Plant. 2020, 13, 1523–1532. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Yang, Y.; Zhao, F.; Zhang, T.; Yu, X. An improved protein lipid overlay assay for studying lipid-protein interactions. Plant Methods 2020, 16, 1–11. [Google Scholar] [CrossRef]
- Hirano, T.; Stecker, K.; Munnik, T.; Xu, H.; Sato, M.H. Visualization of Phosphatidylinositol 3,5-Bisphosphate Dynamics by a Tandem ML1N-Based Fluorescent Protein Probe in Arabidopsis. Plant Cell Physiol. 2017, 58, 1185–1195. [Google Scholar] [CrossRef] [PubMed]
- Vermeer, J.; Thole, J.M.; Goedhart, J.; Nielsen, E.; Munnik, T.; Gadella, T.W., Jr. Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. Plant. J. 2009, 57, 356–372. [Google Scholar] [CrossRef] [Green Version]
- Vermeer, J.E.; van Leeuwen, W.; Tobena-Santamaria, R.; Laxalt, A.M.; Jones, D.R.; Divecha, N.; Gadella, T.W., Jr.; Munnik, T. Visualization of PtdIns3P dynamics in living plant cells. Plant J. 2006, 47, 687–700. [Google Scholar] [CrossRef]
- Vermeer, J.E.M.; Munnik, T. Imaging lipids in living plants. In Lipid Signaling in Plants; Munnik, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 185–199. [Google Scholar]
- Vermeer, J.; van Wijk, R.; Goedhart, J.; Geldner, N.; Chory, J.; Gadella, T.; Munnik, T. In Vivo Imaging of Diacylglycerol at the Cytoplasmic Leaflet of Plant Membranes. Plant Cell Physiol. 2017, 58, 1196–1207. [Google Scholar] [CrossRef] [Green Version]
- De Jong, F.; Munnik, T. Attracted to membranes: Lipid-binding domains in plants. Plant Physiol. 2021, 185, 707–723. [Google Scholar] [CrossRef]
- Poyton, M.F.; Pullanchery, S.; Sun, S.; Yang, T.; Cremer, P.S. Zn2+ Binds to Phosphatidylserine and Induces Membrane Blebbing. J. Am. Chem. Soc. 2020, 142, 18679–18686. [Google Scholar] [CrossRef]
- Xu, J.; Chen, D.; Yan, X.; Chen, J.; Zhou, C. Global characterization of the photosynthetic glycerolipids from a marine diatom Stephanodiscus sp. by ultra performance liquid chromatography coupled with electrospray ionization-quadrupole-time of flight mass spectrometry. Anal. Chim. Acta 2010, 663, 60–68. [Google Scholar] [CrossRef]
- Diehl, B.W.; Herling, H.; Riedl, I.; Heinz, E. 13C-NMR analysis of the positional distribution of fatty acids in plant glycolipids. Chem. Phys. Lipids 1995, 77, 147–153. [Google Scholar] [CrossRef]
- Murakami, Y.; Tsuyama, M.; Kobayashi, Y.; Kodama, H.; Iba, K. Trienoic Fatty Acids and Plant Tolerance of High Temperature. Science 2000, 287, 476–479. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Itoh, K.; Yamada, T.; Cerveny, K.L.; Suzuki, T.L.; Macdonald, P.; Frohman, M.A.; Ramachandran, R.; Iijima, M.; Sesaki, H. Coincident Phosphatidic Acid Interaction Restrains Drp1 in Mitochondrial Division. Mol. Cell 2016, 63, 1034–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- König, S.; Mosblech, A.; Heilmann, I. Stress-inducible and constitutive phosphoinositide pools have distinctive fatty acid patterns in Arabidopsis thaliana. FASEB J. 2007, 21, 1958–1967. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, X.; Yang, Y. Phospholipids in Salt Stress Response. Plants 2021, 10, 2204. https://doi.org/10.3390/plants10102204
Han X, Yang Y. Phospholipids in Salt Stress Response. Plants. 2021; 10(10):2204. https://doi.org/10.3390/plants10102204
Chicago/Turabian StyleHan, Xiuli, and Yongqing Yang. 2021. "Phospholipids in Salt Stress Response" Plants 10, no. 10: 2204. https://doi.org/10.3390/plants10102204
APA StyleHan, X., & Yang, Y. (2021). Phospholipids in Salt Stress Response. Plants, 10(10), 2204. https://doi.org/10.3390/plants10102204




