Amylase Binding to Oral Streptococci: A Key Interaction for Human Oral Microbial Ecology, Adaptation and Fitness
Abstract
1. Introduction
2. The Genetic Basis and Diversity of ABPs
2.1. abpA-srtB Operon: System for Processing and Display in ABS
2.2. Multiple ABPs: Convergent Evolution and Horizontal Gene Transfer
3. Functional Characterization of ABPs
3.1. Canonical Functions: Adhesion and Nutrition
3.2. Potential Roles for AbpA Beyond Adhesion and Nutrition
3.3. AbpA-CTM Axis: Pathway for Oxidative Stress Resistance
4. Ecological and Evolutionary Considerations
4.1. Co-Evolution: Interactions of Diet, Genes and Microbiome
4.2. Amylase-Binding Bacteria in Other Mammals
5. Future Directions
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
Abbreviations
| ABS | Amylase-Binding Streptococci |
| HSAmy | Human Alpha-amylase |
| AbpA | Amylase-binding Protein A |
| AbpB | Amylase-binding Protein B |
References
- Scannapieco, F.A.; Ruhl, S. The Oral Environment. In Oral Microbiology and Immunology, 3rd ed.; Richard, J., Lamont, G.N.H., Koo, H., Jenkinson, H.F., Eds.; ASM Press: Washington, DC, USA, 2019. [Google Scholar]
- Scannapieco, F.A. Saliva-bacterium interactions in oral microbial ecology. Crit. Rev. Oral Biol. Med. 1994, 5, 203–248. [Google Scholar] [CrossRef] [PubMed]
- Nikitkova, A.E.; Haase, E.M.; Scannapieco, F.A. Taking the starch out of oral biofilm formation: Molecular basis and functional significance of salivary alpha-amylase binding to oral streptococci. Appl. Environ. Microbiol. 2013, 79, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Douglas, C.W. The binding of human salivary alpha-amylase by oral strains of streptococcal bacteria. Arch. Oral Biol. 1983, 28, 567–573. [Google Scholar] [CrossRef]
- Scannapieco, F.A.; Bergey, E.J.; Reddy, M.S.; Levine, M.J. Characterization of salivary alpha-amylase binding to Streptococcus sanguis. Infect. Immun. 1989, 57, 2853–2863. [Google Scholar] [CrossRef]
- Rogers, J.D.; Palmer, R.J., Jr.; Kolenbrander, P.E.; Scannapieco, F.A. Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation. Infect. Immun. 2001, 69, 7046–7056. [Google Scholar] [CrossRef] [PubMed]
- Ragunath, C.; Manuel, S.G.; Venkataraman, V.; Sait, H.B.; Kasinathan, C.; Ramasubbu, N. Probing the role of aromatic residues at the secondary saccharide-binding sites of human salivary alpha-amylase in substrate hydrolysis and bacterial binding. J. Mol. Biol. 2008, 384, 1232–1248. [Google Scholar] [CrossRef]
- Scannapieco, F.A.; Bhandary, K.; Ramasubbu, N.; Levine, M.J. Structural relationship between the enzymatic and streptococcal binding sites of human salivary alpha-amylase. Biochem. Biophys. Res. Commun. 1990, 173, 1109–1115. [Google Scholar] [CrossRef]
- Tanzer, J.M.; Baranowski, L.K.; Rogers, J.D.; Haase, E.M.; Scannapieco, F.A. Oral colonization and cariogenicity of Streptococcus gordonii in specific pathogen-free TAN:SPFOM(OM)BR rats consuming starch or sucrose diets. Arch. Oral Biol. 2001, 46, 323–333. [Google Scholar] [CrossRef]
- Tanzer, J.M.; Grant, L.; Thompson, A.; Li, L.; Rogers, J.D.; Haase, E.M.; Scannapieco, F.A. Amylase-binding proteins A (AbpA) and B (AbpB) differentially affect colonization of rats’ teeth by Streptococcus gordonii. Microbiology 2003, 149 Pt 9, 2653–2660. [Google Scholar] [CrossRef]
- Tanzer, J.M.; Thompson, A.M.; Grant, L.P.; Vickerman, M.M.; Scannapieco, F.A. Streptococcus gordonii’s sequenced strain CH1 glucosyltransferase determines persistent but not initial colonization of teeth of rats. Arch. Oral Biol. 2008, 53, 133–140. [Google Scholar] [CrossRef][Green Version]
- Tanzer, J.M.; Thompson, A.; Sharma, K.; Vickerman, M.M.; Haase, E.M.; Scannapieco, F.A. Streptococcus mutans out-competes Streptococcus gordonii in vivo. J. Dent. Res. 2012, 91, 513–519. [Google Scholar] [CrossRef]
- Fellows Yates, J.A.; Velsko, I.M.; Aron, F.; Posth, C.; Hofman, C.A.; Austin, R.M.; Parker, C.E.; Mann, A.E.; Nagele, K.; Arthur, K.W.; et al. The evolution and changing ecology of the African hominid oral microbiome. Proc. Natl. Acad. Sci. USA 2021, 118, e2021655118. [Google Scholar] [CrossRef] [PubMed]
- Velsko, I.M.; Warinner, C. Streptococcus abundance and oral site tropism in humans and non-human primates reflects host and lifestyle differences. NPJ Biofilms Microbiomes 2025, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Haase, E.M.; Feng, X.; Pan, J.; Miecznikowski, J.C.; Scannapieco, F.A. Dynamics of the Streptococcus gordonii Transcriptome in Response to Medium, Salivary alpha-Amylase, and Starch. Appl. Environ. Microbiol. 2015, 81, 5363–5374. [Google Scholar] [CrossRef]
- Nikitkova, A.E.; Haase, E.M.; Vickerman, M.M.; Gill, S.R.; Scannapieco, F.A. Response of fatty acid synthesis genes to the binding of human salivary amylase by Streptococcus gordonii. Appl. Environ. Microbiol. 2012, 78, 1865–1875. [Google Scholar] [CrossRef]
- Kreth, J.; Merritt, J.; Qi, F. Bacterial and host interactions of oral streptococci. DNA Cell Biol. 2009, 28, 397–403. [Google Scholar] [CrossRef]
- Rogers, J.D.; Haase, E.M.; Brown, A.E.; Douglas, C.W.; Gwynn, J.P.; Scannapieco, F.A. Identification and analysis of a gene (abpA) encoding a major amylase-binding protein in Streptococcus gordonii. Microbiology 1998, 144 Pt 5, 1223–1233. [Google Scholar] [CrossRef][Green Version]
- Liang, X.; Liu, B.; Zhu, F.; Scannapieco, F.A.; Haase, E.M.; Matthews, S.; Wu, H. A distinct sortase SrtB anchors and processes a streptococcal adhesin AbpA with a novel structural property. Sci. Rep. 2016, 6, 30966. [Google Scholar] [CrossRef]
- Ton-That, H.; Marraffini, L.A.; Schneewind, O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim. Biophys. Acta 2004, 1694, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Haase, E.M.; Kou, Y.; Sabharwal, A.; Liao, Y.C.; Lan, T.; Lindqvist, C.; Scannapieco, F.A. Comparative genomics and evolution of the amylase-binding proteins of oral streptococci. BMC Microbiol. 2017, 17, 94. [Google Scholar]
- Douglas, C.W.; Pease, A.A.; Whiley, R.A. Amylase-binding as a discriminator among oral streptococci. FEMS Microbiol. Lett. 1990, 54, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tanzer, J.M.; Scannapieco, F.A. Identification and analysis of the amylase-binding protein B (AbpB) and gene (abpB) from Streptococcus gordonii. FEMS Microbiol. Lett. 2002, 212, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Nikitkova, A.E.; Haase, E.M.; Scannapieco, F.A. Effect of starch and amylase on the expression of amylase-binding protein A in Streptococcus gordonii. Mol. Oral Microbiol. 2012, 27, 284–294. [Google Scholar] [CrossRef]
- Briggs, N.S.; Bruce, K.E.; Naskar, S.; Winkler, M.E.; Roper, D.I. The pneumococcal divisome: Dynamic control of Streptococcus pneumoniae cell division. Front. Microbiol. 2021, 12, 737396. [Google Scholar] [CrossRef] [PubMed]
- Fleurie, A.; Lesterlin, C.; Manuse, S.; Zhao, C.; Cluzel, C.; Lavergne, J.P.; Franz-Wachtel, M.; Macek, B.; Combet, C.; Kuru, E.; et al. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 2014, 516, 259–262. [Google Scholar] [CrossRef]
- Berg, K.H.; Stamsas, G.A.; Straume, D.; Havarstein, L.S. Effects of low PBP2b levels on cell morphology and peptidoglycan composition in Streptococcus pneumoniae R6. J. Bacteriol. 2013, 195, 4342–4354. [Google Scholar] [CrossRef]
- Scannapieco, F.A.; Haraszthy, G.G.; Cho, M.I.; Levine, M.J. Characterization of an amylase-binding component of Streptococcus gordonii G9B. Infect. Immun. 1992, 60, 4726–4733. [Google Scholar] [CrossRef]
- Andisi, V.F.; Hinojosa, C.A.; de Jong, A.; Kuipers, O.P.; Orihuela, C.J.; Bijlsma, J.J. Pneumococcal gene complex involved in resistance to extracellular oxidative stress. Infect. Immun. 2012, 80, 1037–1049. [Google Scholar] [CrossRef]
- Saleh, M.; Bartual, S.G.; Abdullah, M.R.; Jensch, I.; Asmat, T.M.; Petruschka, L.; Pribyl, T.; Gellert, M.; Lillig, C.H.; Antelmann, H.; et al. Molecular architecture of Streptococcus pneumoniae surface thioredoxin-fold lipoproteins crucial for extracellular oxidative stress resistance and maintenance of virulence. EMBO Mol. Med. 2013, 5, 1852–1870. [Google Scholar] [CrossRef]
- Jalal, N.; Lee, S.F. The MsrAB reducing pathway of Streptococcus gordonii is needed for oxidative stress tolerance, biofilm formation, and oral colonization in mice. PLoS ONE 2020, 15, e0229375. [Google Scholar] [CrossRef]
- Kim, H.Y. The methionine sulfoxide reduction system: Selenium utilization and methionine sulfoxide reductase enzymes and their functions. Antioxid. Redox Signal 2013, 19, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M. Effector Molecules and Regulatory Proteins: Applications. Trends Biotechnol. 2016, 34, 777–780. [Google Scholar] [CrossRef]
- Liu, Y.; Burne, R.A. Multiple two-component systems modulate alkali generation in Streptococcus gordonii in response to environmental stresses. J. Bacteriol. 2009, 191, 7353–7362. [Google Scholar] [CrossRef]
- Zhu, L.; Kreth, J. The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid. Med. Cell Longev. 2012, 2012, 717843. [Google Scholar] [CrossRef]
- Bolognini, D.; Halgren, A.; Lou, R.N.; Raveane, A.; Rocha, J.L.; Guarracino, A.; Soranzo, N.; Chin, C.S.; Garrison, E.; Sudmant, P.H. Recurrent evolution and selection shape structural diversity at the amylase locus. Nature 2024, 634, 617–625. [Google Scholar] [CrossRef]
- Perry, G.H.; Dominy, N.J.; Claw, K.G.; Lee, A.S.; Fiegler, H.; Redon, R.; Werner, J.; Villanea, F.A.; Mountain, J.L.; Misra, R.; et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007, 39, 1256–1260. [Google Scholar] [CrossRef]
- Pajic, P.; Pavlidis, P.; Dean, K.; Neznanova, L.; Romano, R.A.; Garneau, D.; Daugherity, E.; Globig, A.; Ruhl, S.; Gokcumen, O. Independent amylase gene copy number bursts correlate with dietary preferences in mammals. eLife 2019, 8, e44628. [Google Scholar] [CrossRef]
- Adler, C.J.; Dobney, K.; Weyrich, L.S.; Kaidonis, J.; Walker, A.W.; Haak, W.; Bradshaw, C.J.; Townsend, G.; Soltysiak, A.; Alt, K.W.; et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat. Genet. 2013, 45, 450–455.e1. [Google Scholar] [CrossRef]
- Scannapieco, F.A.; Solomon, L.; Wadenya, R.O. Emergence in human dental plaque and host distribution of amylase-binding streptococci. J. Dent. Res. 1994, 73, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Votsch, D.; Willenborg, M.; Weldearegay, Y.B.; Valentin-Weigand, P. Streptococcus suis—The “Two Faces” of a Pathobiont in the Porcine Respiratory Tract. Front. Microbiol. 2018, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Kouki, A.; Haataja, S.; Loimaranta, V.; Pulliainen, A.T.; Nilsson, U.J.; Finne, J. Identification of a novel streptococcal adhesin P (SadP) protein recognizing galactosyl-alpha1-4-galactose-containing glycoconjugates: Convergent evolution of bacterial pathogens to binding of the same host receptor. J. Biol. Chem. 2011, 286, 38854–38864. [Google Scholar] [CrossRef]
- Ferrando, M.L.; Fuentes, S.; de Greeff, A.; Smith, H.; Wells, J.M. ApuA, a multifunctional alpha-glucan-degrading enzyme of Streptococcus suis, mediates adhesion to porcine epithelium and mucus. Microbiology 2010, 156 Pt 9, 2818–2828. [Google Scholar] [CrossRef]
- Wang, C.; Li, M.; Feng, Y.; Zheng, F.; Dong, Y.; Pan, X.; Cheng, G.; Dong, R.; Hu, D.; Feng, X.; et al. The involvement of sortase A in high virulence of STSS-causing Streptococcus suis serotype 2. Arch. Microbiol. 2009, 191, 23–33. [Google Scholar] [CrossRef]
- Jenkinson, H.F. Cell-surface proteins of Streptococcus sanguis associated with cell hydrophobicity and coaggregation properties. J. Gen. Microbiol. 1986, 132, 1575–1589. [Google Scholar] [CrossRef]
- Loo, C.Y.; Corliss, D.A.; Ganeshkumar, N. Streptococcus gordonii biofilm formation: Identification of genes that code for biofilm phenotypes. J. Bacteriol. 2000, 182, 1374–1382. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.D.; Scannapieco, F.A. Catabolite repression and regulation of the amylase-binding protein gene of Streptococcus gordonii. J. Bacteriol. 2001, 183, 3521–3525. [Google Scholar] [CrossRef]
- Sulavik, M.C.; Tardif, G.; Clewell, D.B. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J. Bacteriol. 1992, 174, 3577–3586. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jakubovics, N.S.; Smith, A.W.; Jenkinson, H.F. Oxidative stress tolerance is manganese (Mn(2+)) regulated in Streptococcus gordonii. Microbiology 2002, 148 Pt 10, 3255–3263. [Google Scholar] [CrossRef]
- Flannagan, R.S.; Jaumouille, V.; Grinstein, S. The cell biology of phagocytosis. Annu. Rev. Pathol. 2012, 7, 61–98. [Google Scholar] [CrossRef]
- Vaudaux, P.; Waldvogel, F.A. Gentamicin antibacterial activity in the presence of human polymorphonuclear leukocytes. Antimicrob. Agents Chemother. 1979, 16, 743–749. [Google Scholar] [CrossRef] [PubMed]
| ABP families | |
| ABP family | Features |
| AbpA | ~20 kDa; amylase-binding adhesin; SrtB anchored; novel sorting motif |
| AbpB | ~87 kDa, dipeptidase; binds amylase but not essential for surface binding |
| Other | ~20 to 87 kDa; homologous to peptidoglycan-binding proteins, glutamine ABC transporters and choline-binding proteins |
| AbpA-SrtB operon and processing | |
| Component | |
| abpA | Co-transcription with srtB |
| srtB | Dedicated sortase that anchors AbpA via novel motif; distinct from SrtA |
| Functional roles of ABPs | |
| Function | |
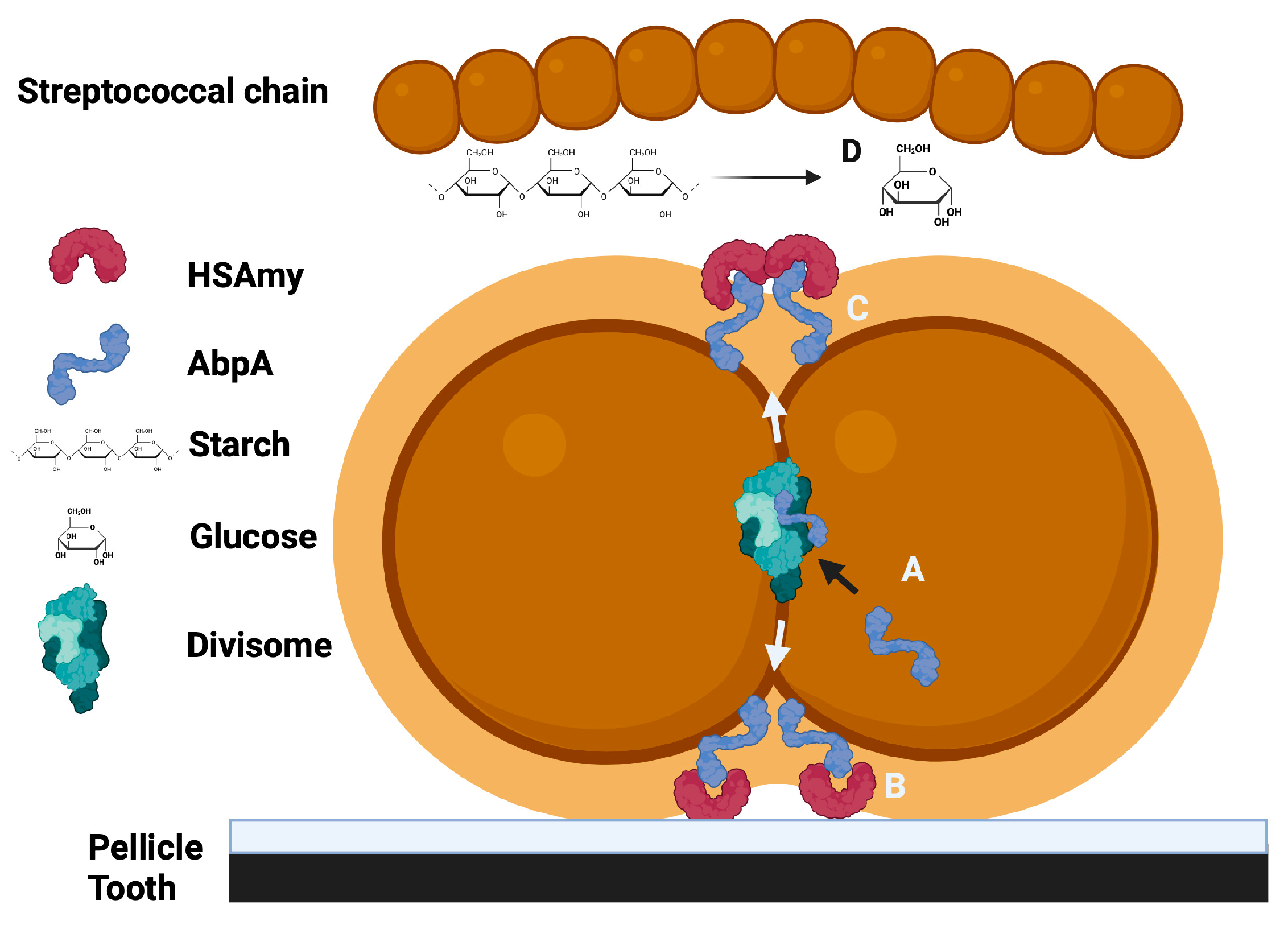
| Adhesion | AbpA enhances adhesion to HSAmy-coated surfaces |
| Nutrition | Starch is converted to maltose and maltotriose and AbpA- mutants cannot grow on starch only |
| Regulation | Amylase triggers gene expression changes via AbpA; increase in fatty acid synthesis and stress tolerance |
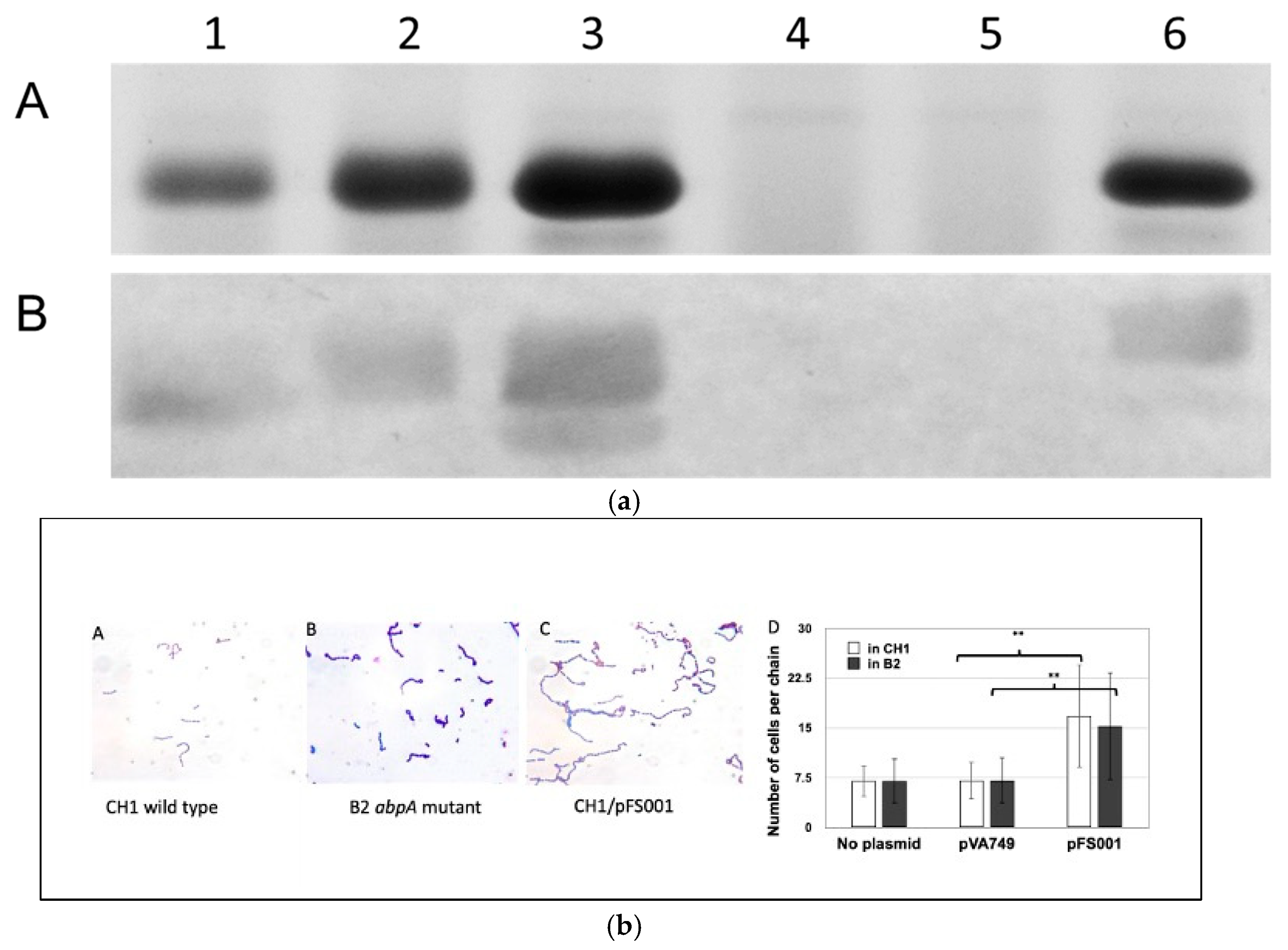
| Cell division | Putative divisome links (FtsZ, PBP2b); septal localization; microscopic observation of AbpA at septum and chain length change in AbpA- mutants |
| AbpA and oxidative stress | |
| Gene | |
| ccdA1 | Redox homeostasis; downregulated in AbpA- mutants |
| tlpA (etrx1) | Thioredoxin-like; downregulated in AbpA- mutants |
| msrB (msrAB) | Repairs methionine sulfoxide; AbpA- mutants sensitive to external H2O2 |
| Histidine kinase/response regulator | Sense and regulate; operon-like; gradient downregulation |
| Ecological and evolutionary aspects | |
| Evidence | Observation and implication |
| AMY1 copy number | High-starch groups have increased AMY1 and salivary amylase; selects for ABS niche |
| Ancient human calculus | ABS present in Neanderthals/Late Pleistocene humans and predates agriculture |
| Non-human mammals | ABS abundant in several amylase-secretors; absent otherwise; physiologic adaptation of microbiome |
| S. suis analogs | SadP/ApuA (SrtA) with alpha-amylase activity; convergence without AbpA/SrtB |
| Sequence | Protein BLAST (2.7.1) | Max Score | Matched Sequence | SGO# | Recurrence |
|---|---|---|---|---|---|
| TSNNNLL | cell division protein (FtsZ) | 21.8 | SNNNLL | SGO_0675 | (1/64 phages) |
| IVTQIPM | beta-ketoacyl-ACP reductase (FabG) | 18 | QIPM | SGO_1693 | (1/64 phages) |
| TGSTRPW | beta-ketoacyl-ACP reductase (FabG) | 17.6 | TGSTR | SGO_1693 | (4/64 phages) |
| EKKNMMN | sensor histidine kinase | 16.8 | +MMN | SGO_1174 | (3/64 phages) |
| SSHSVQR | penicillin-binding protein 2B (PBP2b) | 16.3 | SHSVQ+R | SGO_1449 | (20/64 phages) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabharwal, A.; Haase, E.M.; Scannapieco, F.A. Amylase Binding to Oral Streptococci: A Key Interaction for Human Oral Microbial Ecology, Adaptation and Fitness. Biomolecules 2025, 15, 1616. https://doi.org/10.3390/biom15111616
Sabharwal A, Haase EM, Scannapieco FA. Amylase Binding to Oral Streptococci: A Key Interaction for Human Oral Microbial Ecology, Adaptation and Fitness. Biomolecules. 2025; 15(11):1616. https://doi.org/10.3390/biom15111616
Chicago/Turabian StyleSabharwal, Amarpreet, Elaine M. Haase, and Frank A. Scannapieco. 2025. "Amylase Binding to Oral Streptococci: A Key Interaction for Human Oral Microbial Ecology, Adaptation and Fitness" Biomolecules 15, no. 11: 1616. https://doi.org/10.3390/biom15111616
APA StyleSabharwal, A., Haase, E. M., & Scannapieco, F. A. (2025). Amylase Binding to Oral Streptococci: A Key Interaction for Human Oral Microbial Ecology, Adaptation and Fitness. Biomolecules, 15(11), 1616. https://doi.org/10.3390/biom15111616






