Activity of Serpins in Context to Hydrophobic Interaction
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.2. Description of the Model Used
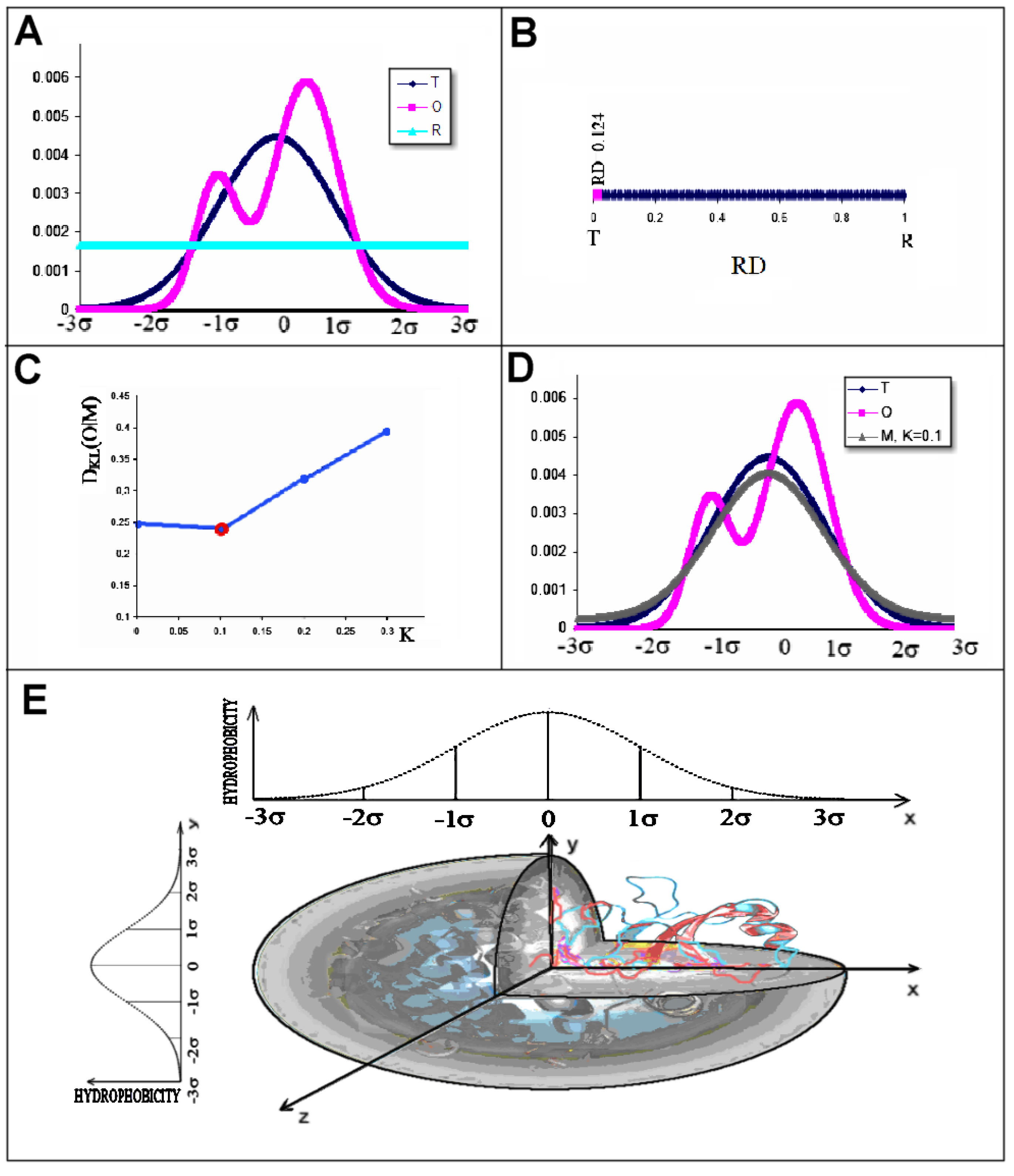
- RD indicates the degree of restoration of the micelle-like arrangement in the hydrophobicity distribution. The hydrophobicity distribution in the micelle structure (RD < 0.5) was treated as idealised and consistent with a centric hydrophobic nucleus and a polar surface. The higher the RD value in the range [0–1], the more the hydrophobicity distribution in the protein body approaches an aligned distribution with a uniform layout of comparable hydrophobicity levels. This means deprivation of the hydrophobic nucleus.
- K indicates the degree to which the environment is different from the polar aqueous environment. The higher the K parameter value, the higher the contribution of non-aqueous factors to the formation of the protein structure from the perspective of hydrophobicity distribution.
3. Results
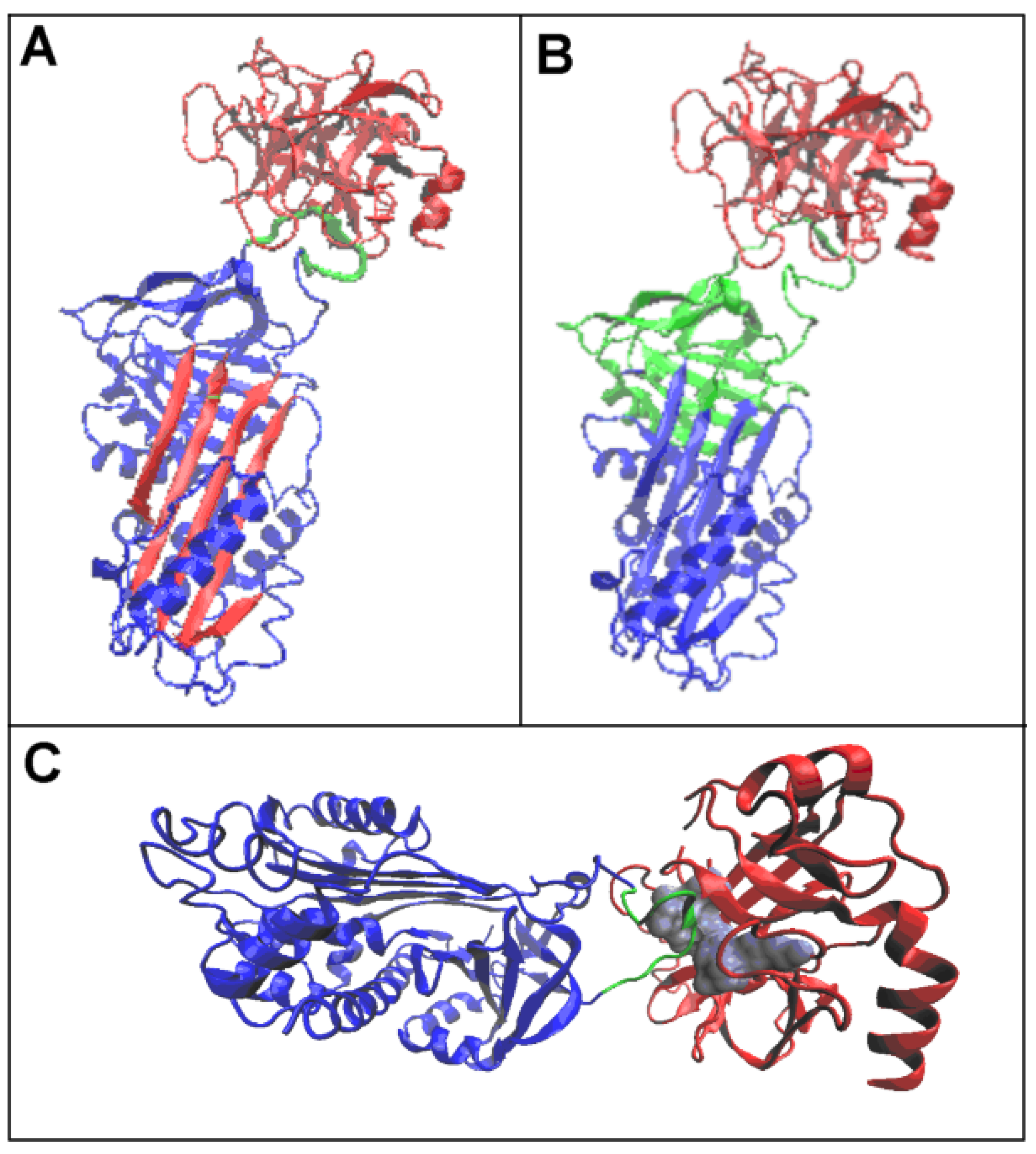
3.1. Status of Complexes and Chains Present in Them
3.2. Structure of the Serpin Chain
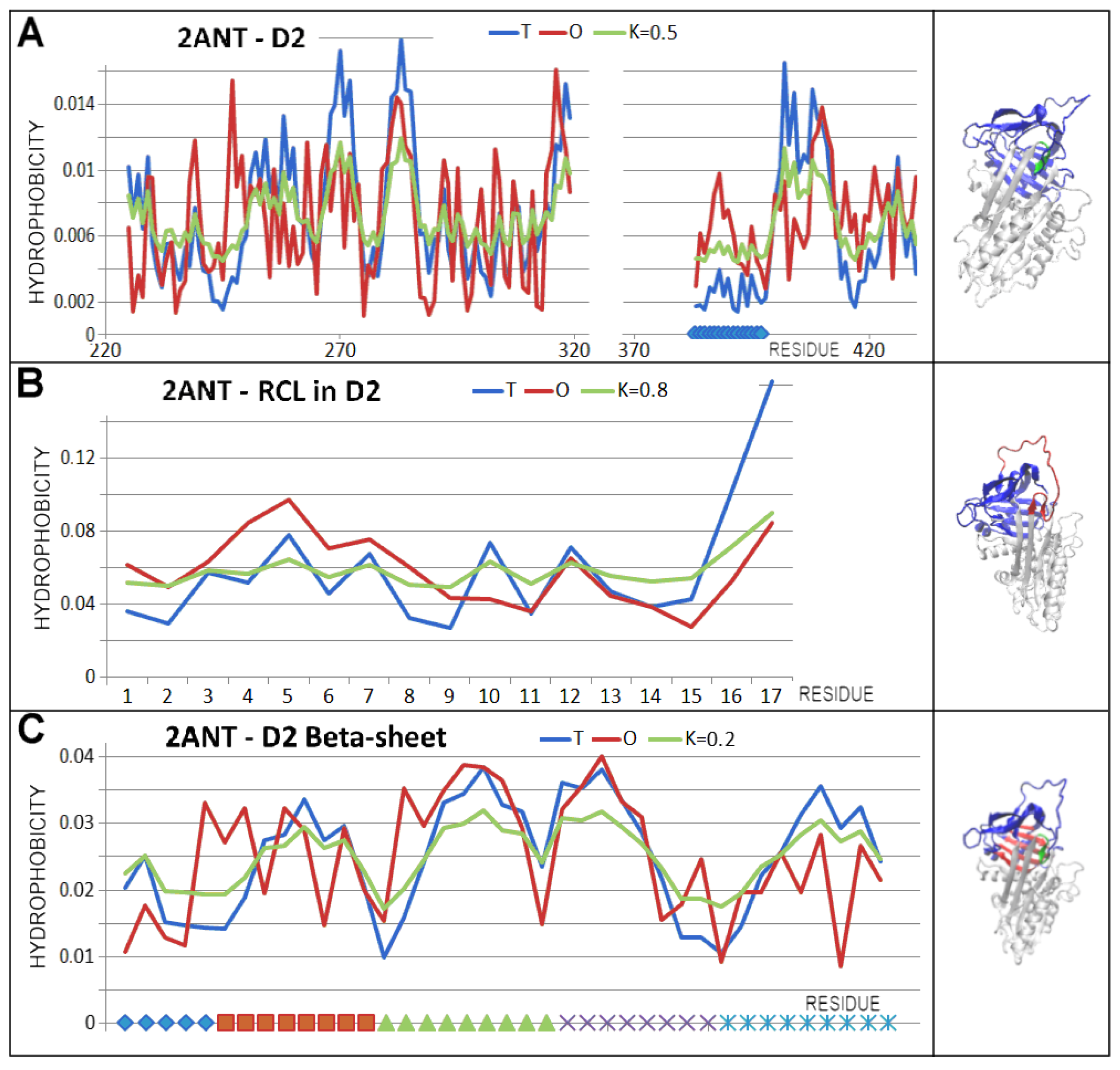
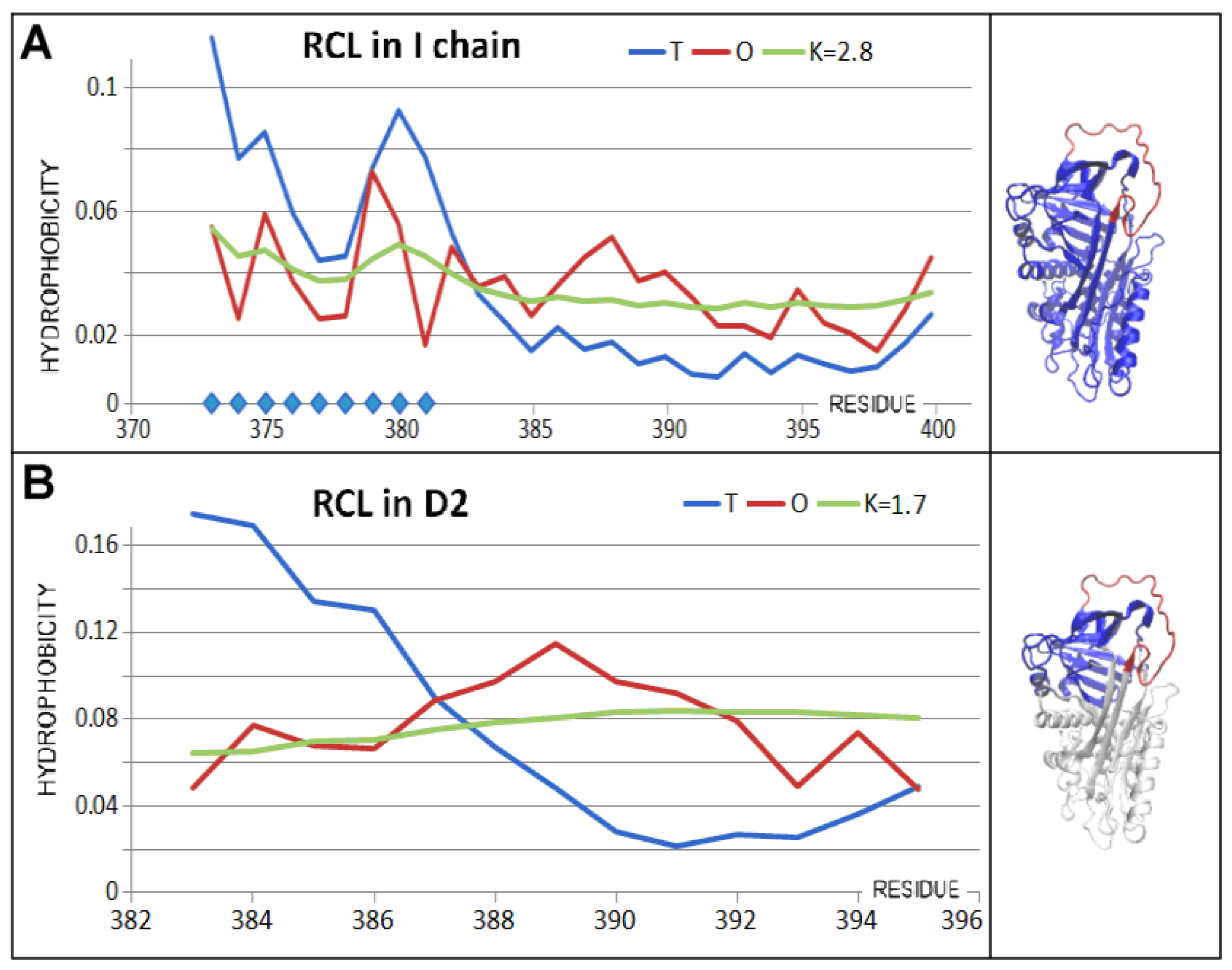
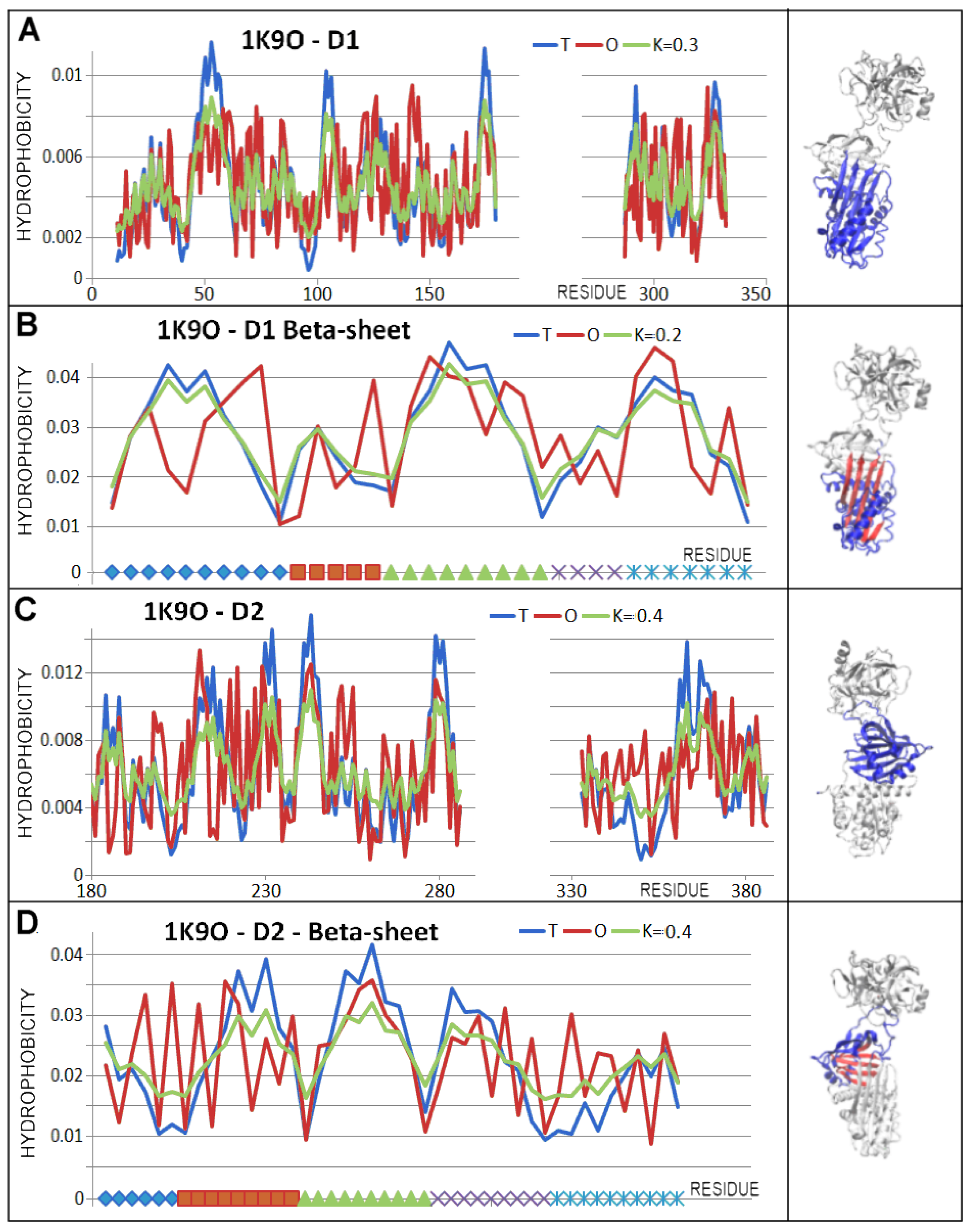
3.3. Status of the Domains
3.4. Status of Beta-Sheets
3.5. Comparable Analysis of Discussed Structural Forms
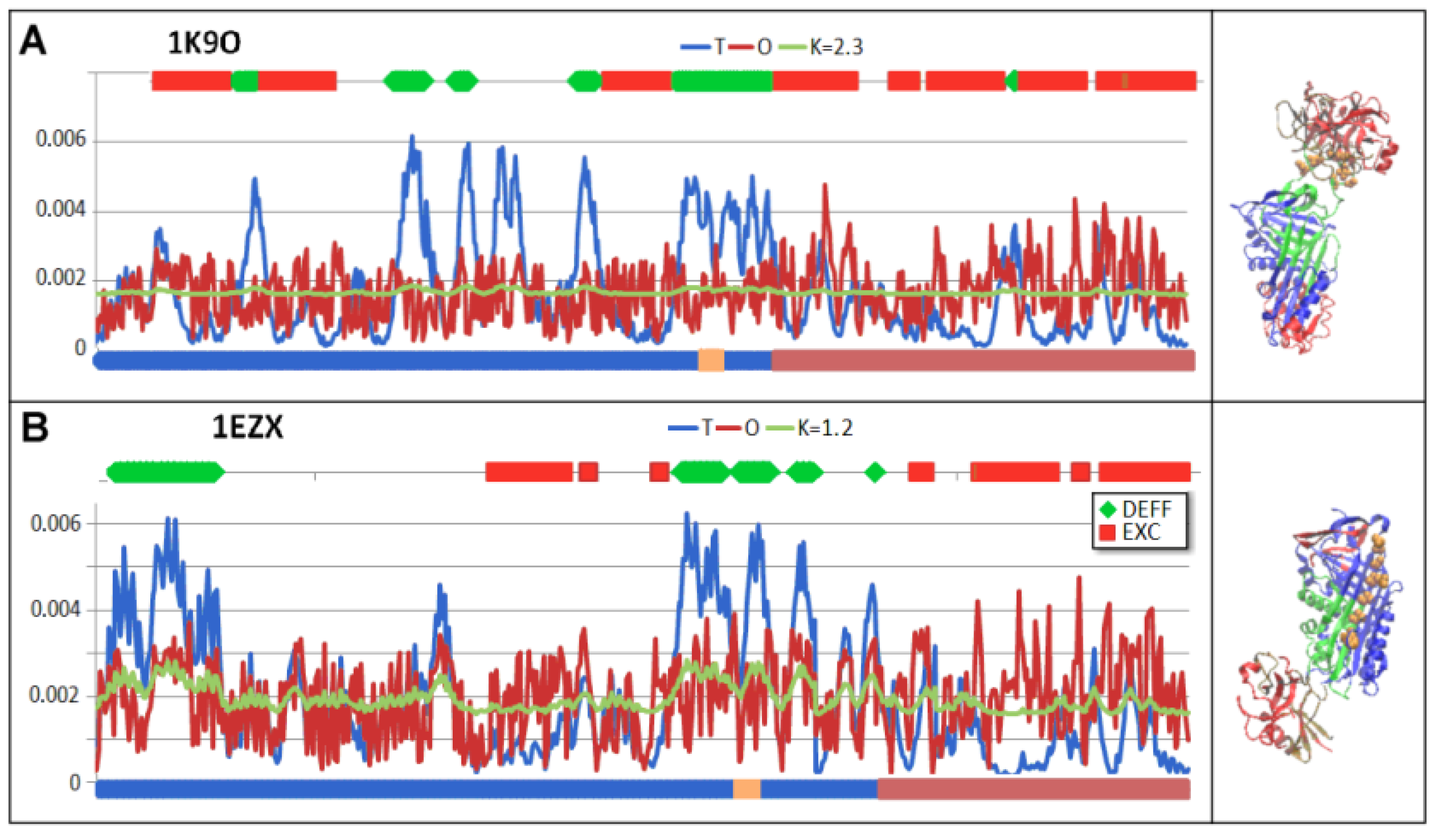
3.6. Comparison of T, O and M Distribution with Mobility of Residues in Discussed Structural Forms
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FOD-M | Fuzzy oil drop model—modified |
| RD | Relative Distance |
References
- Cano-Martínez, A.; Rubio-Ruiz, M.E.; Guarner-Lans, V. Homeostasis and evolution in relation to regeneration and repair. J. Physiol. 2024, 602, 2627–2648. [Google Scholar] [CrossRef]
- Bechtel, W.; Bich, L. Situating homeostasis in organisms: Maintaining organization through time. J. Physiol. 2024, 602, 6003–6020. [Google Scholar] [CrossRef]
- Castagna, D.; Gourdet, B.; Hjerpe, R.; MacFaul, P.; Novak, A.; Revol, G.; Rochette, E.; Jordan, A. To homeostasis and beyond! Recent advances in the medicinal chemistry of heterobifunctional derivatives. Prog. Med. Chem. 2024, 63, 61–160. [Google Scholar] [CrossRef]
- Giordano, G.; Cuba Samaniego, C.; Franco, E.; Blanchini, F. Computing the structural influence matrix for biological systems. J. Math. Biol. 2016, 72, 1927–1958. [Google Scholar] [CrossRef]
- Antoneli, F.; Golubitsky, M.; Jin, J.; Stewart, I. Homeostasis in input-output networks: Structure, Classification and Applications. Math. Biosci. 2025, 384, 109435. [Google Scholar] [CrossRef]
- Lucas, A.; Yaron, J.R.; Zhang, L.; Macaulay, C.; McFadden, G. Serpins: Development for Therapeutic Applications. Methods Mol. Biol. 2018, 1826, 255–265. [Google Scholar] [CrossRef]
- Recordati, G.; Bellini, T.G. A definition of internal constancy and homeostasis in the context of non-equilibrium thermodynamics. Exp. Physiol. 2004, 89, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.J. Serpins in cartilage and osteoarthritis: What do we know? Biochem. Soc. Trans. 2021, 49, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Cazzolli, G.; Wang, F.; a Beccara, S.; Gershenson, A.; Faccioli, P.; Wintrode, P.L. Serpin latency transition at atomic resolution. Proc. Natl. Acad. Sci. USA 2014, 111, 15414–15419. [Google Scholar] [CrossRef] [PubMed]
- Fa, M.; Bergström, F.; Hägglöf, P.; Wilczynska, M.; Johansson, L.B.; Ny, T. The structure of a serpin-protease complex revealed by intramolecular distance measurements using donor-donor energy migration and mapping of interaction sites. Structure 2000, 8, 397–405. [Google Scholar] [CrossRef]
- Harrop, S.J.; Jankova, L.; Coles, M.; Jardine, D.; Whittaker, J.S.; Gould, A.R.; Meister, A.; King, G.C.; Mabbutt, B.C.; Curmi, P.M. The crystal structure of plasminogen activator inhibitor 2 at 2.0 A resolution: Implications for serpin function. Structure 1999, 7, 43–54. [Google Scholar] [CrossRef]
- Mahon, B.P.; McKenna, R. Methods for Determining and Understanding Serpin Structure and Function: X-Ray Crystallography. Methods Mol. Biol. 2018, 1826, 9–39. [Google Scholar] [CrossRef]
- Patston, P.A.; Church, F.C.; Olson, S.T. Serpin-ligand interactions. Methods 2004, 32, 93–109. [Google Scholar] [CrossRef]
- Wei, A.; Rubin, H.; Cooperman, B.S.; Christianson, D.W. Crystal structure of an uncleaved serpin reveals the conformation of an inhibitory reactive loop. Nat. Struct. Biol. 1994, 1, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Law, R.H.; Zhang, Q.; McGowan, S.; Buckle, A.M.; Silverman, G.A.; Wong, W.; Rosado, C.J.; Langendorf, C.G.; Pike, R.N.; Bird, P.I.; et al. An overview of the serpin superfamily. Genome Biol. 2006, 7, 216. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Antalis, T.M.; Lawrence, D.A. Serpin mutagenesis. Methods 2004, 32, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Ellisdon, A.M.; Zhang, Q.; Henstridge, M.A.; Johnson, T.K.; Warr, C.G.; Law, R.H.; Whisstock, J.C. High resolution structure of cleaved Serpin 42 Da from Drosophila melanogaster. BMC Struct. Biol. 2014, 14, 14. [Google Scholar] [CrossRef]
- Loebermann, H.; Tokuoka, R.; Deisenhofer, J.; Huber, R. Human alpha 1-proteinase inhibitor. Crystal structure analysis of two crystal modifications, molecular model and preliminary analysis of the implications for function. J. Mol. Biol. 1984, 177, 531–557. [Google Scholar] [CrossRef]
- Elliott, P.R.; Pei, X.Y.; Dafforn, T.R.; Lomas, D.A. Topography of a 2.0 A structure of alpha1-antitrypsin reveals targets for rational drug design to prevent conformational disease. Protein Sci. 2000, 9, 1274–1281. [Google Scholar] [CrossRef]
- Geiger, M.; Wahlmüller, F.; Furtmüller, M. The Serpin Family. Proteins with Multiple Functions in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar] [CrossRef]
- Whisstock, J.C.; Pike, R.N.; Jin, L.; Skinner, R.; Pei, X.Y.; Carrell, R.W.; Lesk, A.M. Conformational changes in serpins: II. The mechanism of activation of antithrombin by heparin. J. Mol. Biol. 2000, 301, 1287–1305. [Google Scholar] [CrossRef]
- Whisstock, J.C.; Skinner, R.; Carrell, R.W.; Lesk, A.M. Conformational changes in serpins: I. The native and cleaved conformations of alpha(1)-antitrypsin. J. Mol. Biol. 2000, 296, 685–699. [Google Scholar] [CrossRef]
- Gettins, P.G. Serpin structure, mechanism, and function. Chem. Rev. 2002, 102, 4751–4804. [Google Scholar] [CrossRef]
- Rein, C.M.; Desai, U.R.; Church, F.C. Serpin-glycosaminoglycan interactions. Methods Enzymol. 2011, 501, 105–137. [Google Scholar] [CrossRef]
- Ren, Z.; Bao, J.; Zhao, S.; Pozzi, N.; Wedner, H.J.; Atkinson, J.P. N-glycosylation in the SERPIN domain of the C1-esterase inhibitor in hereditary angioedema. J. Clin. Investig. 2025, 10, e185548. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.J.; Huber, R.; Bode, W.; Engh, R.A. Structural aspects of serpin inhibition. FEBS Lett. 1994, 344, 117–124. [Google Scholar] [CrossRef]
- Simonovic, M.; Gettins, P.G.W.; Volz, K. Crystal structure of viral serpin crmA provides insights into its mechanism of cysteine proteinase inhibition. Protein Sci. 2000, 9, 1423–1427. [Google Scholar] [CrossRef] [PubMed]
- Pippel, J.; Kuettner, E.B.; Ulbricht, D.; Daberger, J.; Schultz, S.; Heiker, J.T.; Sträter, N. Crystal structure of cleaved vaspin (serpinA12). Biol. Chem. 2016, 397, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Stephens, R.; Mirza, G.; Kanai, H.; Ragoussis, J.; Bird, P.I. A serpin gene cluster on human chromosome 6p25 contains PI6, PI9 and ELANH2 which have a common structure almost identical to the 18q21 ovalbumin serpin genes. Cytogenet. Cell Genet. 1998, 82, 273–277. [Google Scholar] [CrossRef]
- van Gent, D.; Sharp, P.; Morgan, K.; Kalsheker, N. Serpins: Structure, function and molecular evolution. Int. J. Biochem. Cell Biol. 2003, 35, 1536–1547. [Google Scholar] [CrossRef]
- Ludeman, J.P.; Whisstock, J.C.; Hopkins, P.C.; Le Bonniec, B.F.; Bottomley, S.P. Structure of a serpin-enzyme complex probed by cysteine substitutions and fluorescence spectroscopy. Biophys. J. 2001, 80, 491–497. [Google Scholar] [CrossRef]
- Perkins, S.J. Three-dimensional structure and molecular modelling of C1-inhibitor. Behring Inst. Mitt. 1993, 93, 63–80. [Google Scholar]
- Bird, P.I.; Whisstock, J.C. Serpin structure and evolution. Preface. Methods Enzymol. 2011, 501, xvii–xviii. [Google Scholar] [CrossRef]
- Irving, J.A.; Steenbakkers, P.J.; Lesk, A.M.; Op den Camp, H.J.; Pike, R.N.; Whisstock, J.C. Serpins in prokaryotes. Mol. Biol. Evol. 2002, 19, 1881–1890. [Google Scholar] [CrossRef]
- Janciauskiene, S.; Lechowicz, U.; Pelc, M.; Olejnicka, B.; Chorostowska-Wynimko, J. Diagnostic and therapeutic value of human serpin family proteins. Biomed. Pharmacother. 2024, 175, 116618. [Google Scholar] [CrossRef]
- Fluhr, R.; Lampl, N.; Roberts, T.H. Serpin protease inhibitors in plant biology. Physiol. Plant. 2012, 145, 95–102. [Google Scholar] [CrossRef]
- Cohen, M.; Davydov, O.; Fluhr, R. Plant serpin protease inhibitors: Specificity and duality of function. J. Exp. Bot. 2019, 70, 2077–2085. [Google Scholar] [CrossRef]
- Li, M.; Christen, J.M.; Dittmer, N.T.; Cao, X.; Zhang, X.; Jiang, H.; Kanost, M.R. The Manduca sexta serpinome: Analysis of serpin genes and proteins in the tobacco hornworm. Insect Biochem. Mol. Biol. 2018, 102, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Han, Y.; Chen, X.; Zhang, W. Structure-function relationship of SW-AT-1, a serpin-type protease inhibitor in silkworm. PLoS ONE 2014, 9, e99013. [Google Scholar] [CrossRef] [PubMed]
- Meekins, D.A.; Kanost, M.R.; Michel, K. Serpins in arthropod biology. Semin. Cell Dev. Biol. 2017, 62, 105–119. [Google Scholar] [CrossRef]
- An, C.; Lovell, S.; Kanost, M.R.; Battaile, K.P.; Michel, K. Crystal structure of native Anopheles gambiae serpin-2, a negative regulator of melanization in mosquitoes. Proteins 2011, 79, 1999–2003. [Google Scholar] [CrossRef]
- Beinrohr, L.; Harmat, V.; Dobó, J.; Lörincz, Z.; Gál, P.; Závodszky, P. C1 inhibitor serpin domain structure reveals the likely mechanism of heparin potentiation and conformational disease. J. Biol. Chem. 2007, 282, 21100–21109. [Google Scholar] [CrossRef]
- Goulas, T.; Ksiazek, M.; Garcia-Ferrer, I.; Sochaj-Gregorczyk, A.M.; Waligorska, I.; Wasylewski, M.; Potempa, J.; Gomis-Rüth, F.X. A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome. J. Biol. Chem. 2017, 292, 10883–10898. [Google Scholar] [CrossRef]
- Kostin, N.N.; Bobik, T.V.; Shurdova, E.M.; Ziganshin, R.H.; Surina, E.A.; Shagin, D.A.; Shagina, I.A.; Knorre, V.D.; Isaev, V.A.; Rudenskaya, G.N.; et al. Cloning and characterization of serpin from red king crab Paralithodes camtschaticus. Fish Shellfish. Immunol. 2018, 81, 99–107. [Google Scholar] [CrossRef]
- Kummer, J.A.; Strik, M.C.; Bladergroen, B.A.; Hack, C.E. Production, characterization, and use of serpin antibodies. Methods 2004, 32, 141–149. [Google Scholar] [CrossRef]
- Lane, D.A.; Caso, R. Antithrombin: Structure, genomic organization, function and inherited deficiency. Baillieres Clin. Haematol. 1989, 2, 961–998. [Google Scholar] [CrossRef] [PubMed]
- Marszal, E.; Shrake, A. Serpin crystal structure and serpin polymer structure. Arch. Biochem. Biophys. 2006, 453, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, S.V. Serpin receptor 1: A hepatic receptor that mediates the clearance of antithrombin III-proteinase complexes. Am. J. Med. 1989, 87, 10S–14S. [Google Scholar] [CrossRef] [PubMed]
- Mahon, B.P.; Ambadapadi, S.; Yaron, J.R.; Lomelino, C.L.; Pinard, M.A.; Keinan, S.; Kurnikov, I.; Macaulay, C.; Zhang, L.; Reeves, W.; et al. Crystal Structure of Cleaved Serp-1, a Myxomavirus-Derived Immune Modulating Serpin: Structural Design of Serpin Reactive Center Loop Peptides with Improved Therapeutic Function. Biochemistry 2018, 57, 1096–1107. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Yamagishi, S.I.; Sata, M. Structure-function relationships of PEDF. Curr. Mol. Med. 2010, 10, 302–311. [Google Scholar] [CrossRef]
- Wang, W.J.; Wang, J.; Ouyang, C.; Chen, C.; Xu, X.F.; Ye, X.Q. Overview of serpin B9 and its roles in cancer (Review). Oncol. Rep. 2021, 46, 190. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.H.; Kibler, K.; Kraberger, S.; Varsani, A.; Turk, J.; Elmadbouly, N.; Aliskevich, E.; Spaccarelli, L.; Estifanos, B.; et al. Viral anti-inflammatory serpin reduces immuno-coagulopathic pathology in SARS-CoV-2 mouse models of infection. EMBO Mol. Med. 2023, 15, e17376. [Google Scholar] [CrossRef]
- Sillitoe, I.; Bordin, N.; Dawson, N.; Waman, V.P.; Ashford, P.; Scholes, H.M.; Pang, C.S.M.; Woodridge, L.; Rauer, C.; Sen, N.; et al. CATH: Increased structural coverage of functional space. Nucleic Acids Res. 2021, 49, D266–D273. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Cech, A.L.; Belmares, R.; Bergstrom, R.C.; Tong, Y.; Corey, D.R.; Kanost, M.R.; Goldsmith, E.J. The structure of a Michaelis serpin-protease complex. Nat. Struct. Biol. 2001, 8, 979–983. [Google Scholar] [CrossRef] [PubMed]
- Skinner, R.; Abrahams, J.P.; Whisstock, J.C.; Lesk, A.M.; Carrell, R.W.; Wardell, M.R. The 2.6 A structure of antithrombin indicates a conformational change at the heparin binding site. J. Mol. Biol. 1997, 266, 601–609. [Google Scholar] [CrossRef]
- Huntington, J.A.; Read, R.J.; Carrell, R.W. Structure of a serpin-protease complex shows inhibition by deformation. Nature 2000, 407, 923–926. [Google Scholar] [CrossRef] [PubMed]
- Engh, R.; Löbermann, H.; Schneider, M.; Wiegand, G.; Huber, R.; Laurell, C.B. The S variant of human alpha 1-antitrypsin, structure and implications for function and metabolism. Protein Eng. 1989, 2, 407–415. [Google Scholar] [CrossRef]
- Roterman, I.; Konieczny, L. Protein Is an Intelligent Micelle. Entropy 2023, 25, 850. [Google Scholar] [CrossRef]
- Levitt, M.A. A simplified representation of protein conformations for rapid simulation of protein folding. J. Mol. Biol. 1976, 104, 59–107. [Google Scholar] [CrossRef]
- Kalinowska, B.; Banach, M.; Konieczny, L.; Roterman, I. Application of Divergence Entropy to Characterize the Structure of the Hydrophobic Core in DNA Interacting Proteins. Entropy 2015, 17, 1477–1507. [Google Scholar] [CrossRef]
- Kullback, S.; Leibler, R.A. On information and sufficiency. Ann. Math. Stat. 1951, 22, 79–86. [Google Scholar] [CrossRef]
- Banach, M.; Stapor, K.; Konieczny, L.; Fabian, P.; Roterman, I. Downhill, Ultrafast and Fast Folding Proteins Revised. Int. J. Mol. Sci. 2020, 21, 7632. [Google Scholar] [CrossRef] [PubMed]
- Roterman, I.; Stapor, K.; Dułak, D.; Szoniec, G.; Konieczny, L. Aquaporins as Membrane Proteins: The Current Status. Front. Biosci. 2025, 17, 27967. [Google Scholar] [CrossRef]
- Roterman, I.; Stapor, K.; Konieczny, L. Transmembrane proteins-Different anchoring systems. Proteins 2024, 92, 593–609. [Google Scholar] [CrossRef]
- Roterman, I.; Stapor, K.; Dułak, D.; Konieczny, L. Heat Shock Protein and Disaggregase Influencing the Casein Structuralisation. Int. J. Mol. Sci. 2025, 26, 6360. [Google Scholar] [CrossRef]
- Roterman, I.; Stapor, K.; Dułak, D.; Konieczny, L. External Force Field for Protein Folding in Chaperonins-Potential Application in In Silico Protein Folding. ACS Omega 2024, 9, 18412–18428. [Google Scholar] [CrossRef]
- Roterman, I.; Stapor, K.; Konieczny, L. Ab initio protein structure prediction: The necessary presence of external force field as it is delivered by Hsp40 chaperone. BMC Bioinform. 2023, 24, 418. [Google Scholar] [CrossRef]
- Lomas, D.A.; Mahadeva, R. α1-Antitrypsin polymerization and the serpinopathies: Pathobiology and prospects for therapy. J. Clin. Invest. 2002, 110, 1585–1590. [Google Scholar] [CrossRef] [PubMed]
- Engh, R.A.; Schulze, A.J.; Huber, R.; Bode, W. Serpin structures. Behring Inst. Mitt. 1993, 41–62. [Google Scholar] [PubMed]
- Granzin, J.; Huang, Y.; Topbas, C.; Huang, W.; Wu, Z.; Misra, S.; Hazen, S.L.; Blanton, R.E.; Lee, X.; Weiergräber, O.H. Three-dimensional structure of a schistosome serpin revealing an unusual configuration of the helical subdomain. Acta Crystallogr. D Biol. Crystallogr. 2012, 68 Pt 6, 686–694. [Google Scholar] [CrossRef]
- Heit, C.; Jackson, B.C.; McAndrews, M.; Wright, M.W.; Thompson, D.C.; Silverman, G.A.; Nebert, D.W.; Vasiliou, V. Update of the human and mouse SERPIN gene superfamily. Hum. Genomics. 2013, 7, 22. [Google Scholar] [CrossRef]
- Huntington, J.A.; Stein, P.E. Structure and properties of ovalbumin. J. Chromatogr. B Biomed. Sci. Appl. 2001, 756, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Irving, J.A.; Cabrita, L.D.; Rossjohn, J.; Pike, R.N.; Bottomley, S.P.; Whisstock, J.C. The 1.5 A crystal structure of a prokaryote serpin: Controlling conformational change in a heated environment. Structure 2003, 11, 387–397. [Google Scholar] [CrossRef]
- Kascakova, B.; Kotal, J.; Havlickova, P.; Vopatkova, V.; Prudnikova, T.; Grinkevich, P.; Kuty, M.; Chmelar, J.; Kuta Smatanova, I. Conformational transition of the Ixodes ricinus salivary serpin Iripin-4. Acta Crystallogr. D Struct. Biol. 2023, 79 Pt 5, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Korpula-Mastalerz, R.; Dubin, A. The intracellular serpin family. Acta Biochim. Pol. 1996, 43, 419–429. [Google Scholar] [CrossRef]
- Li, J.; Wang, Z.; Canagarajah, B.; Jiang, H.; Kanost, M.; Goldsmith, E.J. The structure of active serpin 1K from Manduca sexta. Structure 1999, 7, 103–109. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pearce, M.C.; Cabrita, L.D.; Rubin, H.; Gore, M.G.; Bottomley, S.P. Identification of residual structure within denatured antichymotrypsin: Implications for serpin folding and misfolding. Biochem. Biophys. Res. Commun. 2004, 324, 729–735. [Google Scholar] [CrossRef]
- Potempa, J.; Korzus, E.; Travis, J. The serpin superfamily of proteinase inhibitors: Structure, function, and regulation. J. Biol. Chem. 1994, 269, 15957–15960. [Google Scholar] [CrossRef]
- Read, R.J.; Zhou, A.; Stein, P.E. Solving serpin crystal structures. Methods Enzymol. 2011, 501, 49–61. [Google Scholar] [CrossRef]
- Singh, P.; Jairajpuri, M.A. Structure function analysis of serpin super-family: “A computational approach”. Protein Pept. Lett. 2014, 21, 714–721. [Google Scholar] [CrossRef]
- Song, J.; Matthews, A.Y.; Reboul, C.F.; Kaiserman, D.; Pike, R.N.; Bird, P.I.; Whisstock, J.C. Predicting serpin/protease interactions. Methods Enzymol. 2011, 501, 237–273. [Google Scholar] [CrossRef]
- Stein, P.; Chothia, C. Serpin tertiary structure transformation. J. Mol. Biol. 1991, 221, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Whisstock, J.C.; Bottomley, S.P. Molecular gymnastics: Serpin structure, folding and misfolding. Curr. Opin. Struct. Biol. 2006, 16, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Devlin, G.L.; Bottomley, S.P. A protein family under ‘stress’—Serpin stability, folding and misfolding. Front. Biosci. 2005, 10, 288–299. [Google Scholar] [CrossRef]
- Yamasaki, M.; Takahashi, N.; Hirose, M. Crystal structure of S-ovalbumin as a non-loop-inserted thermostabilized serpin form. J. Biol. Chem. 2003, 278, 35524–35530. [Google Scholar] [CrossRef]
- Olson, S.T.; Swanson, R.; Day, D.; Verhamme, I.; Kvassman, J.; Shore, J.D. Resolution of Michaelis complex, acylation, and conformational change steps in the reactions of the serpin, plasminogen activator inhibitor-1, with tissue plasminogen activator and trypsin. Biochemistry 2001, 40, 11742–11756. [Google Scholar] [CrossRef]
- Huntington, J.A.; Yamasaki, M. Serpin polymerization in vitro. Methods Enzymol. 2011, 501, 379–420. [Google Scholar] [CrossRef]
- Yamasaki, M.; Sendall, T.J.; Harris, L.E.; Lewis, G.M.; Huntington, J.A. Loop-sheet mechanism of serpin polymerization tested by reactive center loop mutations. J. Biol. Chem. 2010, 285, 30752–30758. [Google Scholar] [CrossRef]
- Johnson, D.J.; Langdown, J.; Li, W.; Luis, S.A.; Baglin, T.P.; Huntington, J.A. Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation. J. Biol. Chem. 2006, 281, 35478–35486. [Google Scholar] [CrossRef]
- Olivieri, C.; Li, G.C.; Wang, Y.; Manu, V.S.; Walker, C.; Kim, J.; Camilloni, C.; De Simone, A.; Vendruscolo, M.; Bernlohr, D.A.; et al. ATP-competitive inhibitors modulate the substrate binding cooperativity of a kinase by altering its conformational entropy. Sci. Adv. 2022, 8, eabo0696. [Google Scholar] [CrossRef]
- Weisberg, E.; Manley, P.W.; Breitenstein, W.; Brüggen, J.; Cowan-Jacob, S.W.; Ray, A.; Huntly, B.; Fabbro, D.; Fendrich, G.; Hall-Meyers, E.; et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell 2005, 7, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, M.; Li, W.; Johnson, D.J.D.; Huntington, J.A. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature 2008, 455, 1255–1258. [Google Scholar] [CrossRef]
- Marijanovic, E.M.; Fodor, J.; Riley, B.T.; Porebski, B.T.; Costa, M.G.S.; Kass, I.; Hoke, D.E.; McGowan, S.; Buckle, A.M. Reactive centre loop dynamics and serpin specificity. Sci. Rep. 2019, 9, 3870. [Google Scholar] [CrossRef]
- Roterman, I.; Stapor, K.; Dułak, D.; Konieczny, L. Domain swapping: A mathematical model for quantitative assessment of structural effects. FEBS Open Bio. 2024, 14, 2006–2025. [Google Scholar] [CrossRef]
- Elliott, P.R.; Lomas, D.A.; Carrell, R.W.; Abrahams, J.P. Inhibitory conformation of the reactive loop of alpha 1-antitrypsin. Nat. Struct. Biol. 1996, 3, 676–681. [Google Scholar] [CrossRef]
- Huntington, J.A. Serpin structure, function and dysfunction. J. Thromb. Haemost. 2011, 9 (Suppl. S1), 26–34. [Google Scholar] [CrossRef] [PubMed]
- Engh, R.A.; Huber, R.; Bode, W.; Schulze, A.J. Divining the serpin inhibition mechanism: A suicide substrate ‘springe’? Trends Biotechnol. 1995, 13, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Dygut, J.; Kalinowska, B.; Banach, M.; Piwowar, M.; Konieczny, L.; Roterman, I. Structural Interface Forms and Their Involvement in Stabilization of Multidomain Proteins or Protein Complexes. Int. J. Mol. Sci. 2016, 17, 1741. [Google Scholar] [CrossRef] [PubMed]
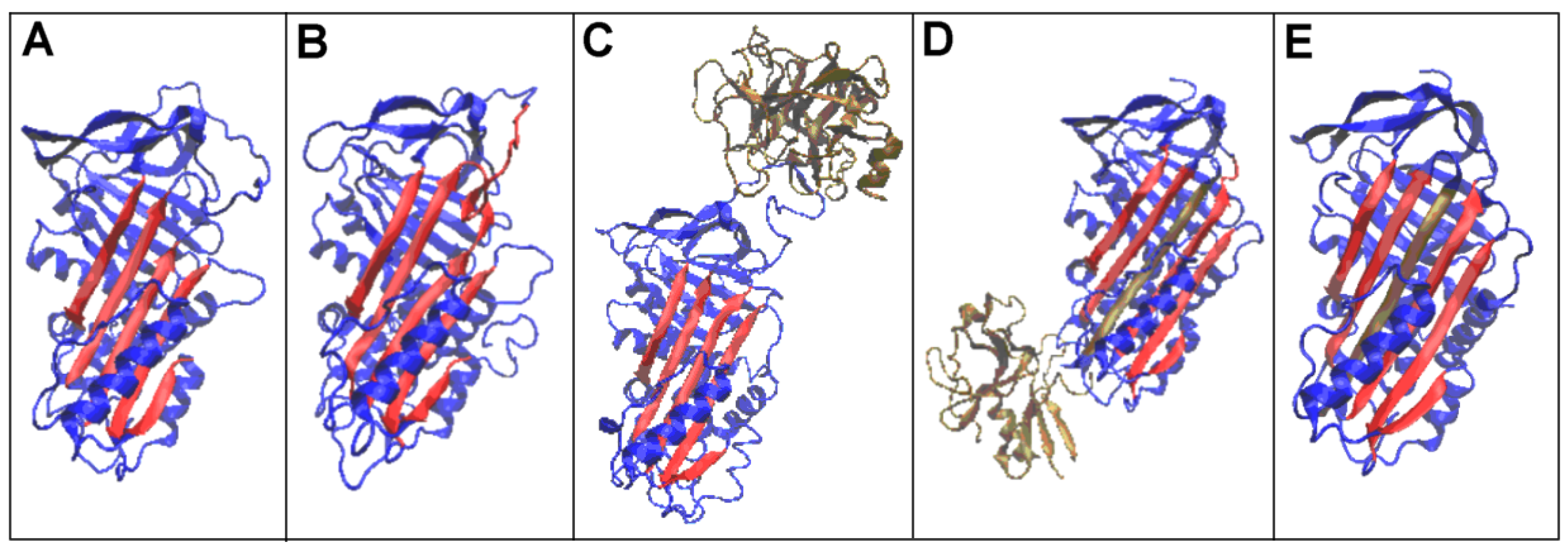
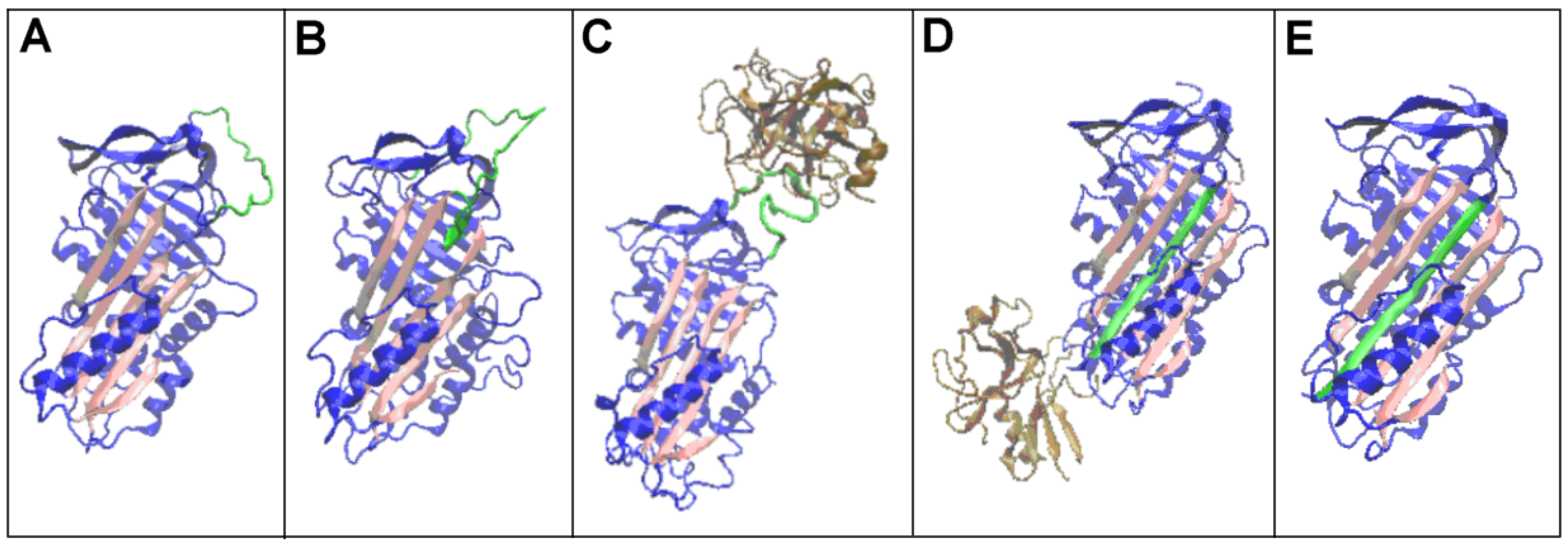
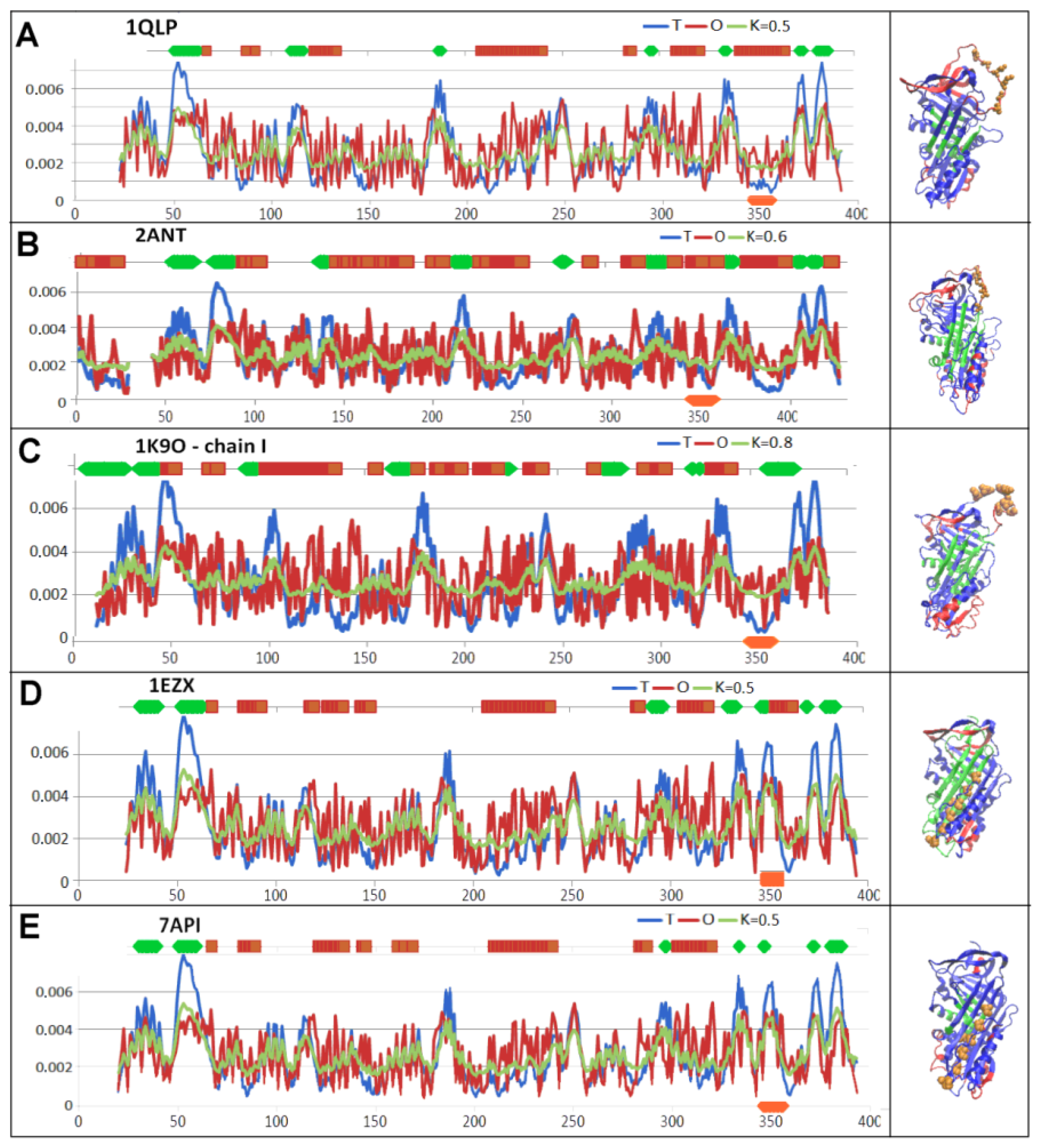
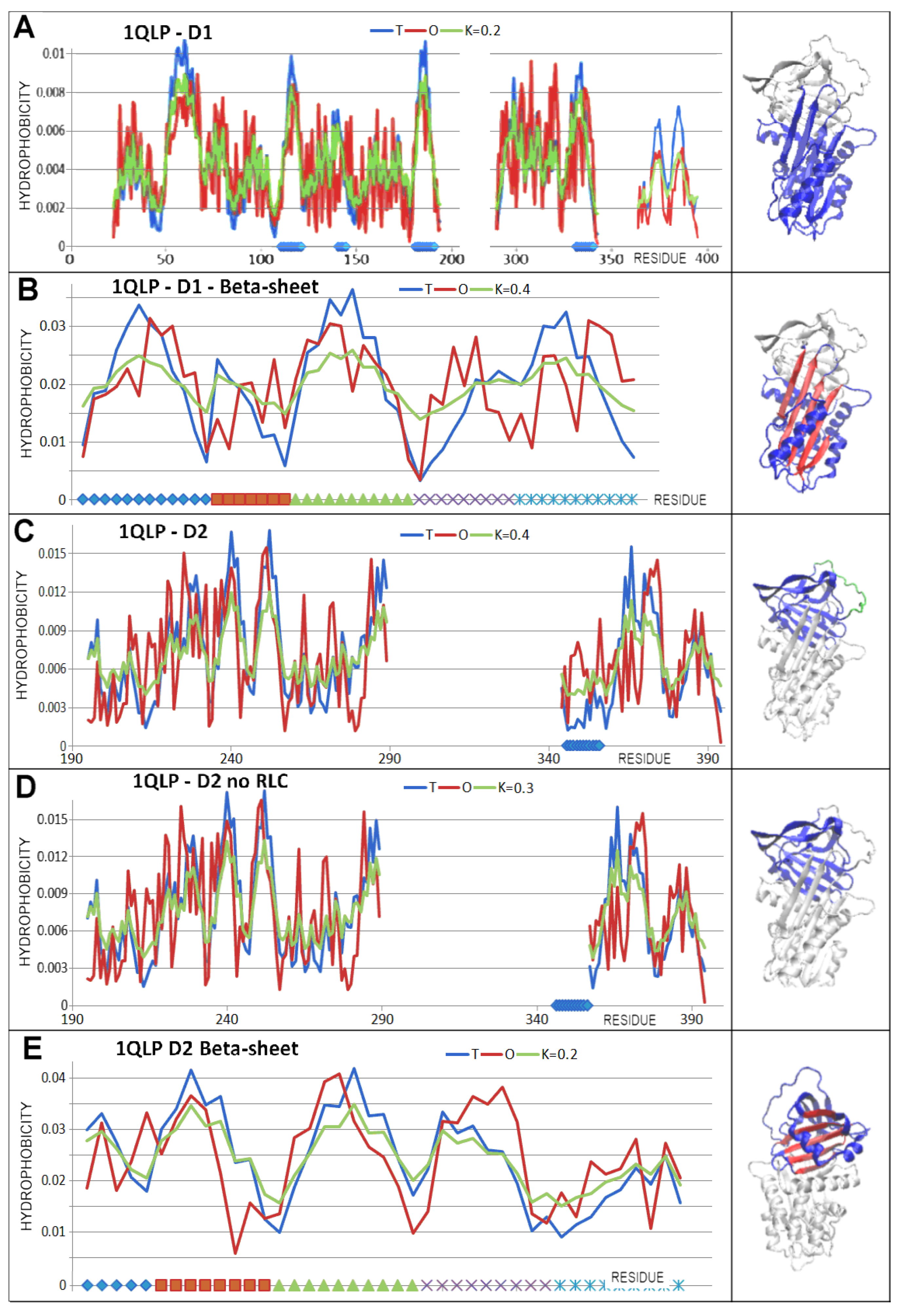
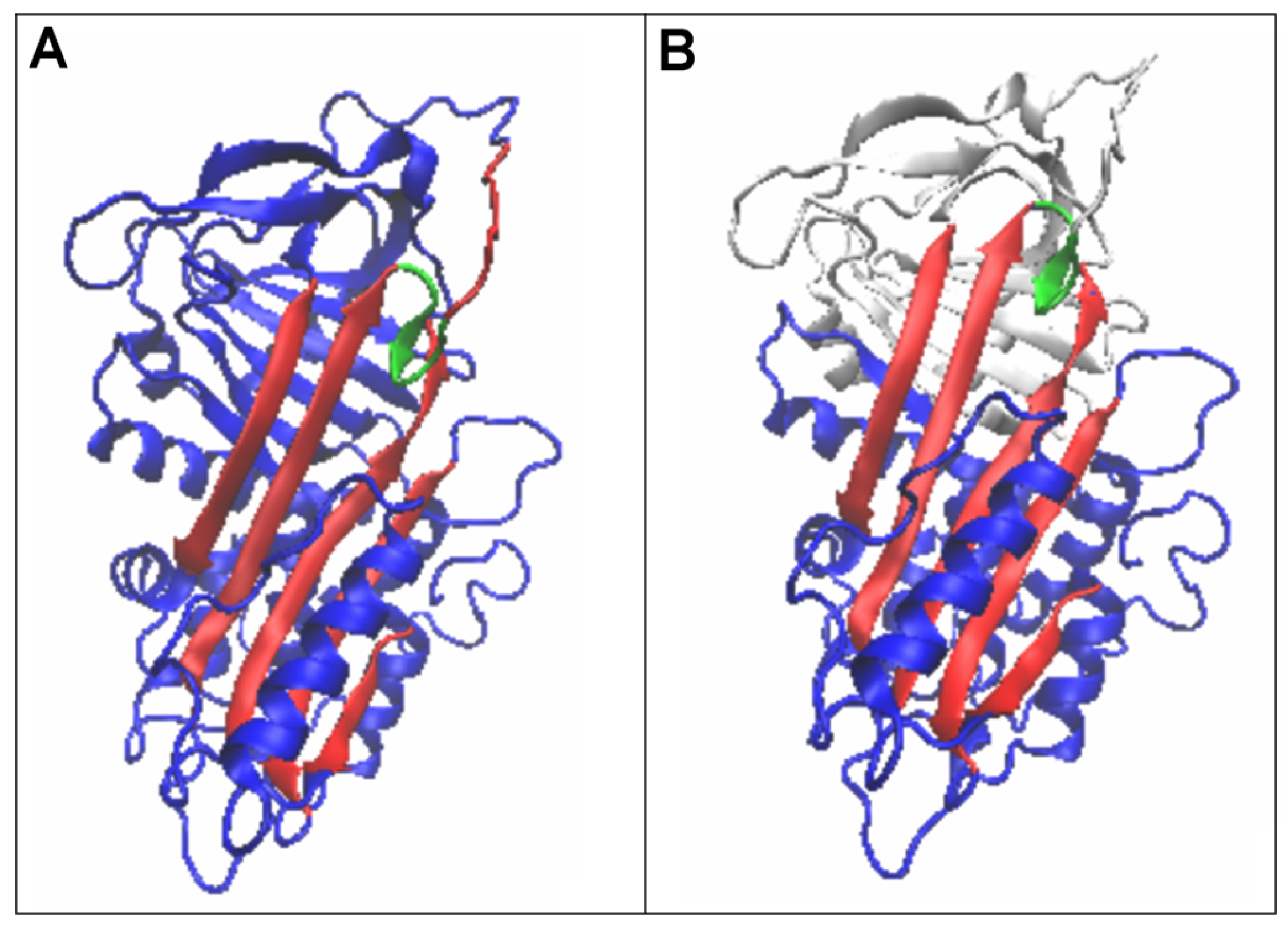
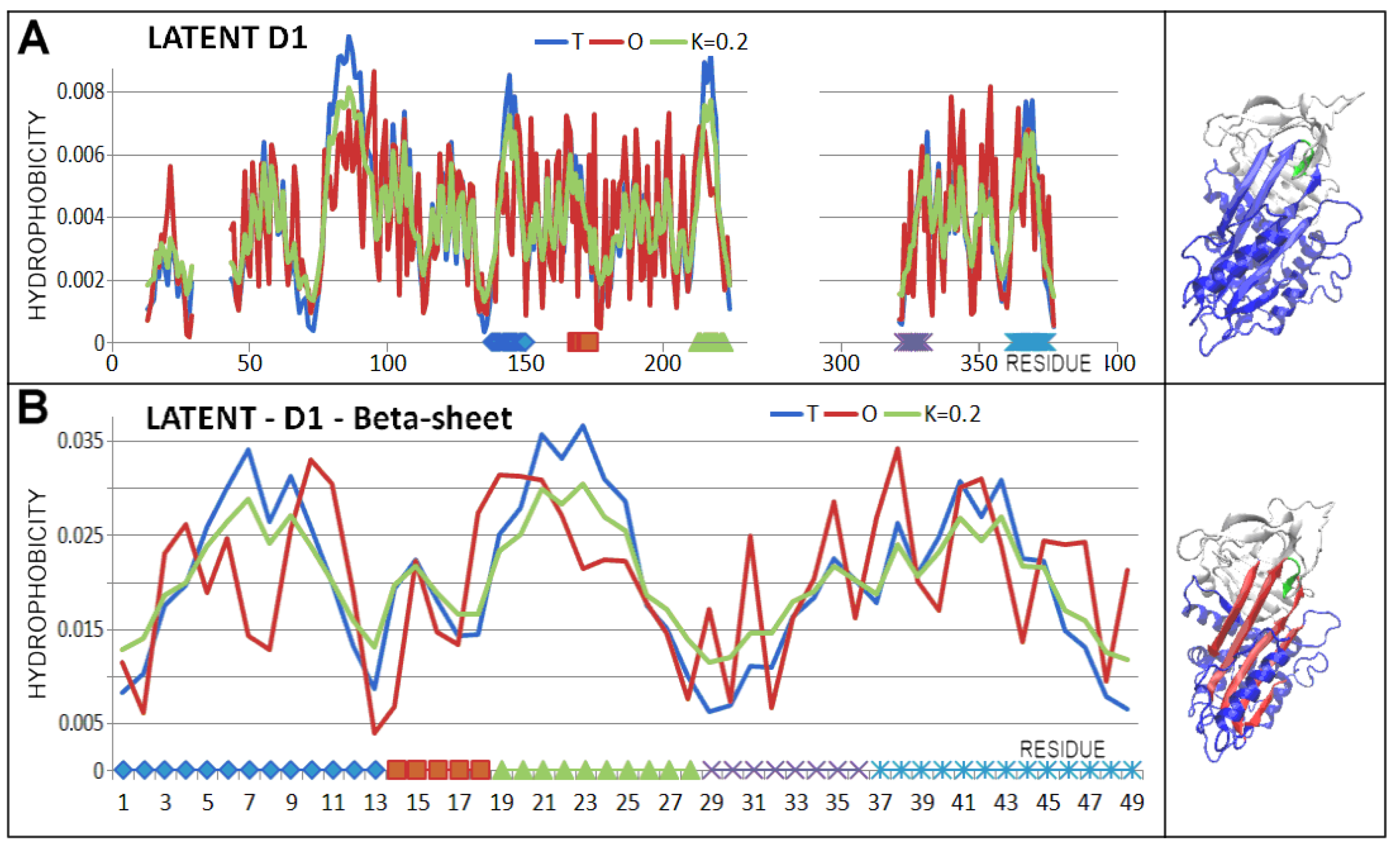
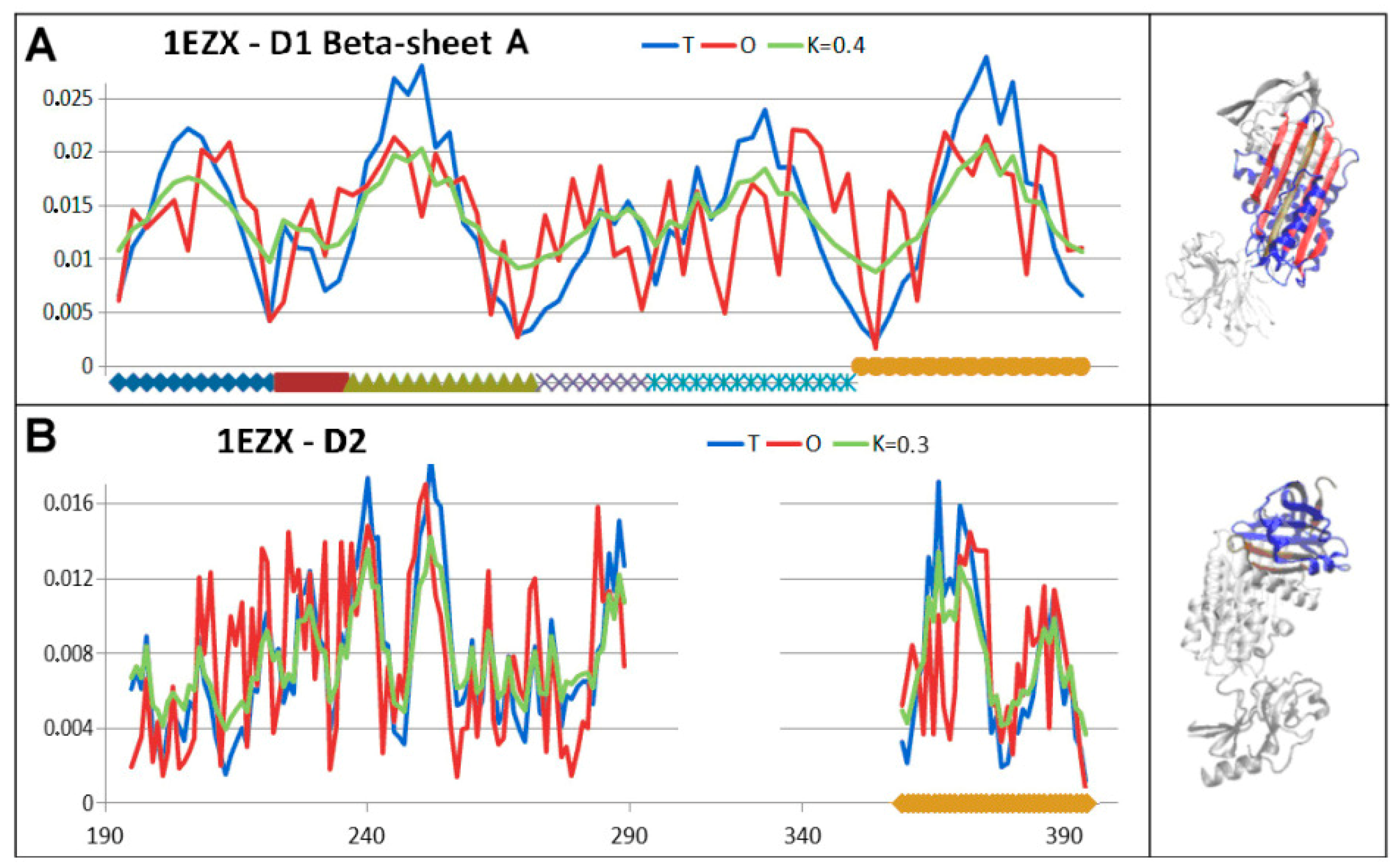
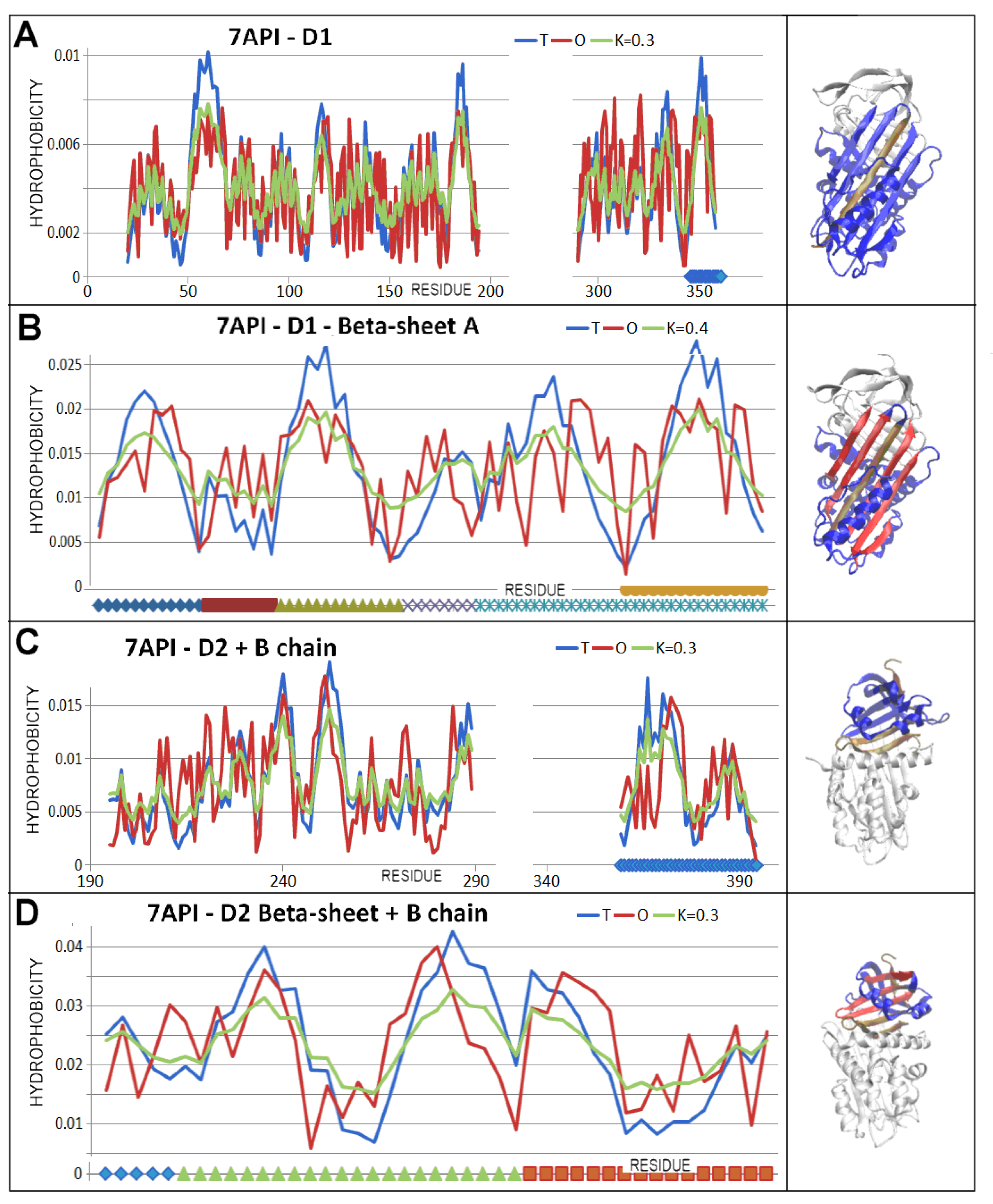
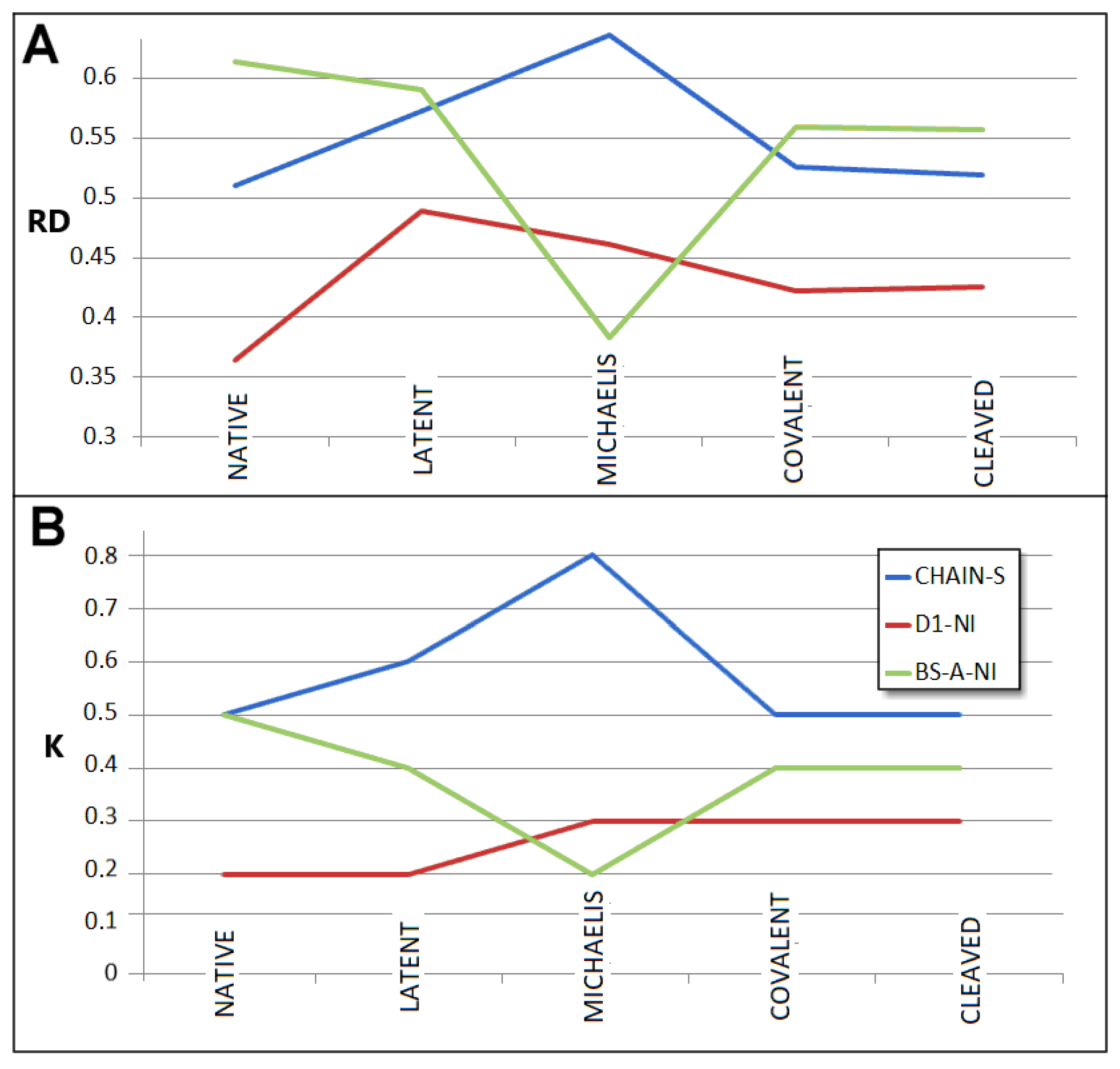
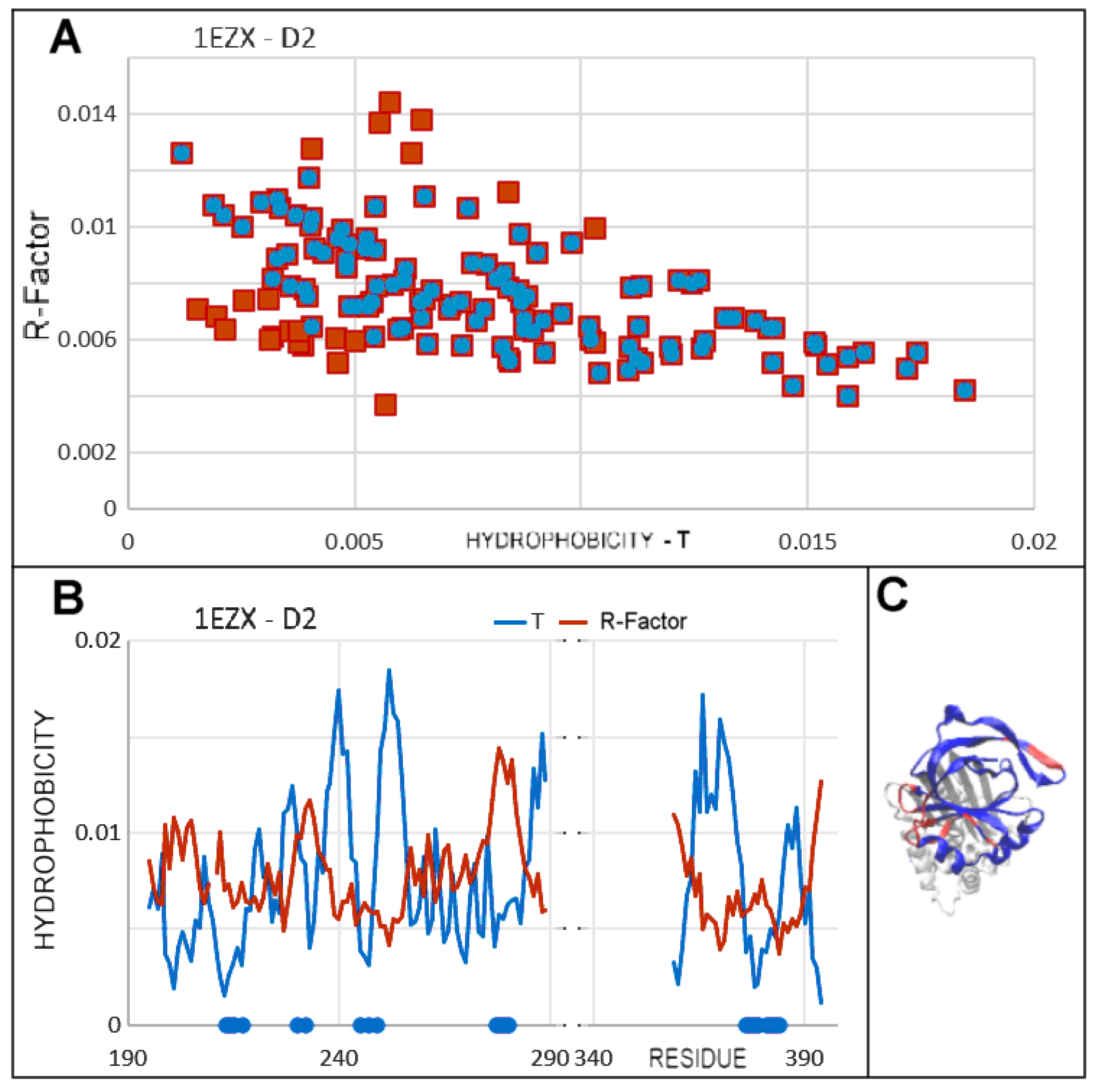
| PDB—ID | Structural Form | Ref. |
|---|---|---|
| 1QLP | Human native state α1-antitrypsin | [19] |
| 2ANT | Latent AT III (M*) | [55] |
| 1K9O | Michaelis non-covalent complex with anionic trypsine-2 | [54] |
| 1EZX | Acyl-enzyme complex with serine protease | [56] |
| 7API | Cleaved | [57] |
| PDB ID | Domain 1—Beta-Sheet A | Domain 2—Beta-Sheet B |
|---|---|---|
| 1QLP | 112–121 | 2218–232 |
| 141–145 | 236–244 | |
| 182–190 | 247–255 | |
| 291–298 | 370–376 | |
| 330–340 | 382–388 | |
| 2ANT | 140–149 | 256–262 |
| 167–173 | 268–273 | |
| 213–221 | 278–285 | |
| 323–329 | 408–414 | |
| 364–375 | 419–426 | |
| 1K9O | 101–109 | 217–222 |
| 129–133 | 227–233 | |
| 172–179 | 239–246 | |
| 287–291 | 367–372 | |
| 326–332 | 377–383 | |
| 1EZX | 111–121 | 227–232 |
| 182–193 | 237–243 | |
| 291–298 | 248–255 | |
| 327–340 | 370–376 * | |
| 344–357 * | 382–388 * | |
| 7API | 110–121 | 228–232 |
| 141–145 | 238–244 | |
| 182–193 | 248–255 | |
| 291–298 | 370–375 * | |
| 327–340 | 381–388 * | |
| 344–357 * |
| FORM | PDB ID | Chain in Complex | Chain—Individual | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Serpin | Protease | Serpin | Protease | ||||||||
| RD | K | RD | K | RD | K | RD | K | RD | K | ||
| NATIVE | 1QLP | 0.510 | 0.5 | ||||||||
| LATENT | 2ANT | 0.572 | 0.6 | ||||||||
| MICHAELIS | 1K9O | 0.636 | 0.8 | ||||||||
| ACYL-ENZYME | 1EZX | 0.787 | 2.3 | 0.778 | 1.6 | 0.711 | 1.3 | 0.526 | 0.5 | 0.447 | 0.4 |
| CLEAVED | 7API | 0.726 | 1.2 | 0.590 | 0.6 | 0.779 | 1.7 | 0.519 | 0.5 | 0.497 | 0.4 |
| Form | PDB—ID | Domain 1 | Domain 2 | Beta-Sheet A in D1 | Beta-Sheet B in D2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No Incorp. | Incorporated | No Incorp. | Incorporated | ||||||||||
| RD | K | RD | K | RD | K | RD | K | RD | K | RD | K | ||
| NATIVE | 1QLP | 0.364 | 0.2 | 0.488 | 0.4 | 0.613 | 0.5 | 0.429 | 0.2 | ||||
| LATENT | 2ANT | 0.489 | 0.2 | 0.396 | 0.2 | 0.578 | 0.4 | 0.590 | 0.4 | 0.584 | 0.4 | 0.483 | 0.2 |
| MICHAELIS | 1K9O | 0.461 | 0.3 | 0.514 | 0.2 | 0.483 | 0.2 | 0.598 | 0.4 | ||||
| COV.S-P.C | 1EZX | 0.422 | 0.3 | 0.426 | 0.3 | 0.444 | 0.3 | 0.559 | 0.4 | 0.548 | 0.4 | 0.447 | 0.2 |
| CLEAVED | 7API | 0.425 | 0.3 | 0.416 | 0.3 | 0.446 | 0.3 | 0.557 | 0.4 | 0.539 | 0.4 | 0.481 | 0.3 |
| FORM | PDB ID | Distribution | |||
|---|---|---|---|---|---|
| Domain | T | O | M | ||
| NATIVE | 1QLP | D1 | −0.494 | −0.549 | −0.494 |
| D2 | −0.664 | −0.491 | −0.664 | ||
| LATENT | 2ANT | D1 | −0.503 | −0.492 | −0.503 |
| D2 | −0.469 | −0.375 | −0.469 | ||
| MICHAELIS | 1K9O | D1 | −0.375 | −0.204 | −0.375 |
| D2 | −0.403 | −0.150 | −0.403 | ||
| COV.S-P.C | 1EZX | D1 | −0.585 | −0.578 | −0.585 |
| D2 | −0.483 | −0.527 | −0.483 | ||
| CLEAVED | 7API | D1 | −0.509 | −0.523 | −0.509 |
| D2 | −0.495 | −0.576 | −0.495 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roterman, I.; Stapor, K.; Zemanek, G.; Dulak, D.; Konieczny, L. Activity of Serpins in Context to Hydrophobic Interaction. Biomolecules 2025, 15, 1615. https://doi.org/10.3390/biom15111615
Roterman I, Stapor K, Zemanek G, Dulak D, Konieczny L. Activity of Serpins in Context to Hydrophobic Interaction. Biomolecules. 2025; 15(11):1615. https://doi.org/10.3390/biom15111615
Chicago/Turabian StyleRoterman, Irena, Katarzyna Stapor, Grzegorz Zemanek, Dawid Dulak, and Leszek Konieczny. 2025. "Activity of Serpins in Context to Hydrophobic Interaction" Biomolecules 15, no. 11: 1615. https://doi.org/10.3390/biom15111615
APA StyleRoterman, I., Stapor, K., Zemanek, G., Dulak, D., & Konieczny, L. (2025). Activity of Serpins in Context to Hydrophobic Interaction. Biomolecules, 15(11), 1615. https://doi.org/10.3390/biom15111615







