Comparative Analysis of Panax ginseng Berries from Seven Cultivars Using UPLC-QTOF/MS and NMR-Based Metabolic Profiling
Abstract
1. Introduction
2. Materials and Methods
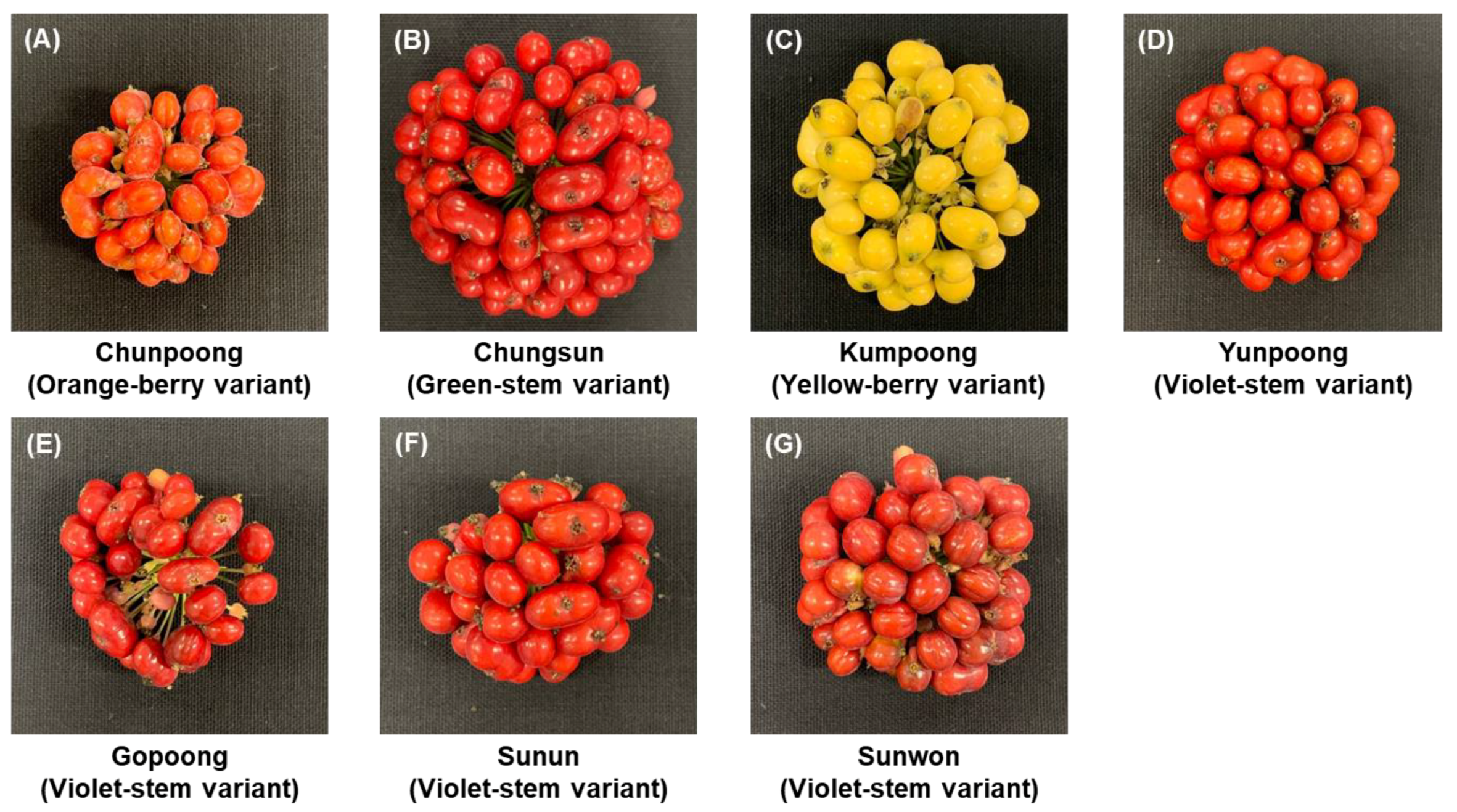
2.1. Ginseng Berry Samples
2.2. Standard Constituents and Reagents
2.3. Sample Preparation
2.4. UPLC-QTOF/MS Analysis
2.5. HR-MAS NMR Analysis
3. Results
3.1. Construction of LC-MS Conditions to Profile Various Ginsenosides
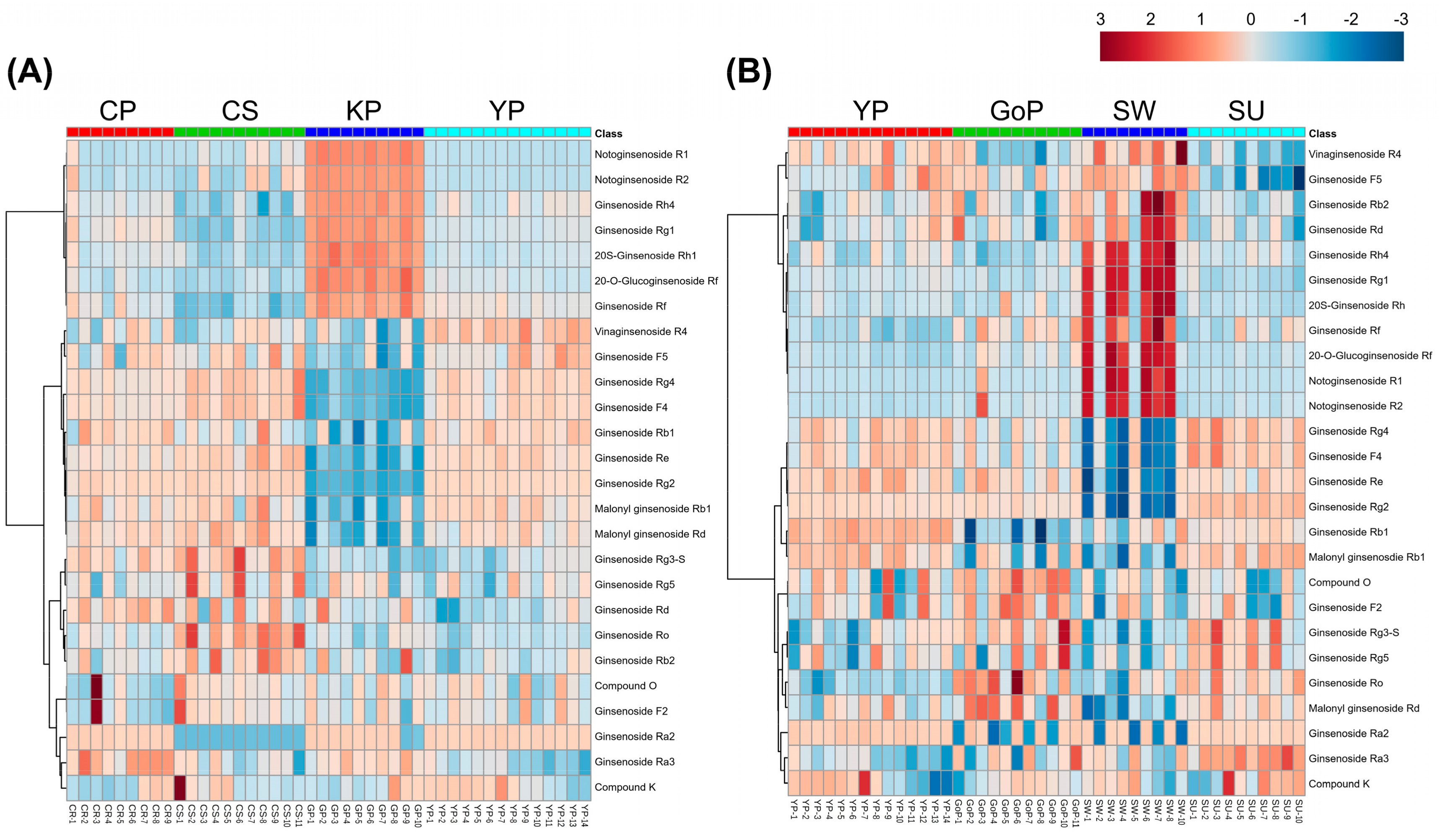
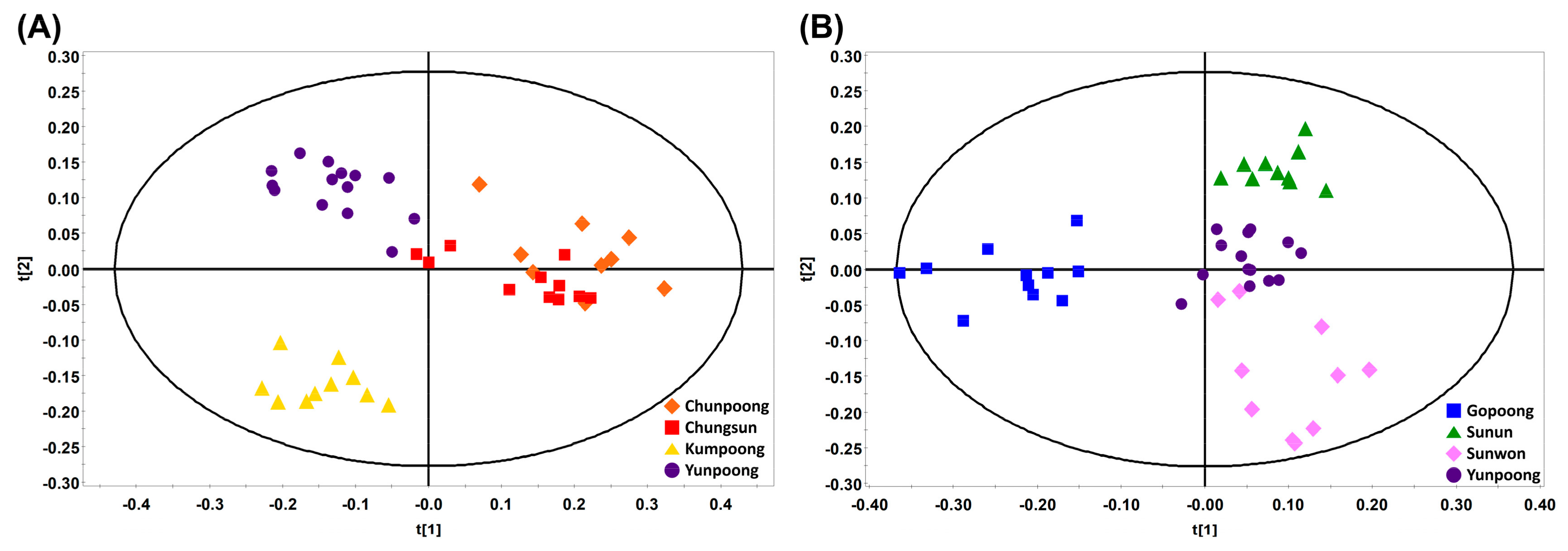
3.2. Profiling of Various Ginsenosides from Seven Different Cultivars of Ginseng Berries
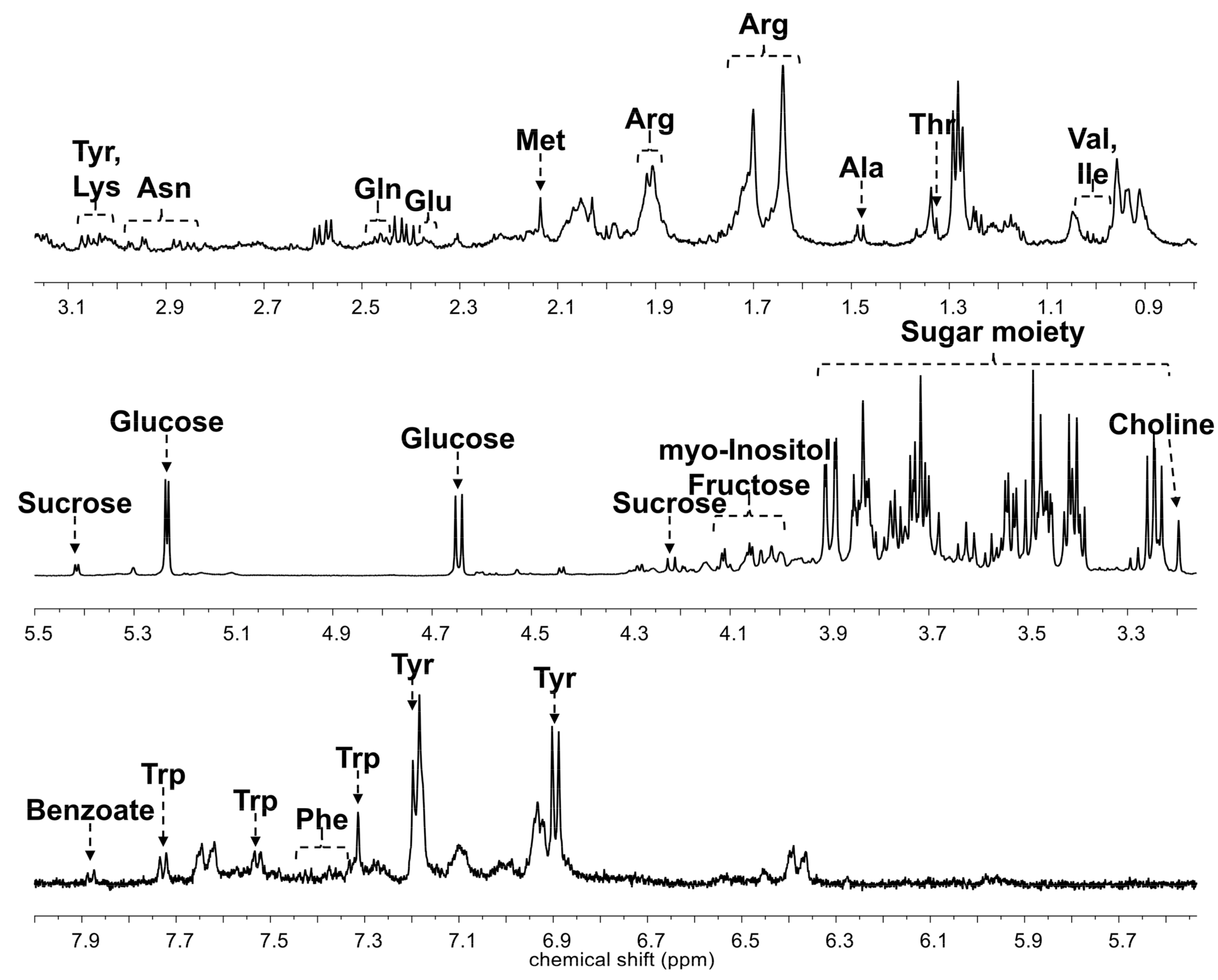
3.3. Profiling of Primary Metabolites from Different Cultivars of Ginseng Berries Using HR-MAS NMR
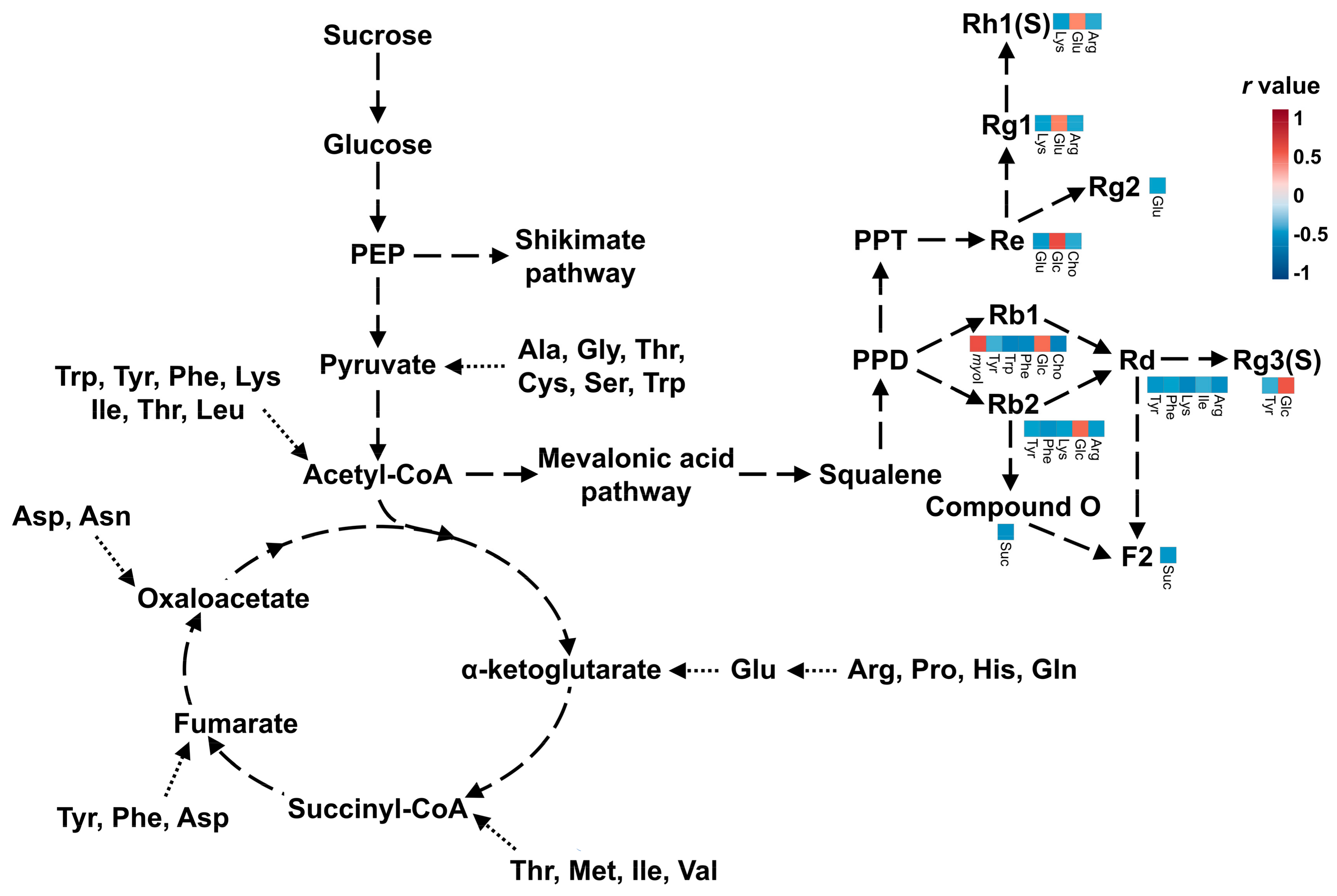
3.4. Pearson Correlation Analysis of Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ANOVA | analysis of variance |
| ELSD | evaporative light-scattering detector |
| ESI | electrospray ionization |
| HPLC | high-performance liquid chromatography |
| HR-MAS | high-resolution magic angle spinning |
| HR-MAS | high-resolution magic angle spinning |
| LODs | limits of detection |
| MS | mass spectrometry |
| NMR | nuclear magnetic resonance |
| OPLS-DA | orthogonal partial least squares discriminant analysis |
| PCA | principal component analysis |
| PLS-DA | partial least squares discriminant analysis |
| PPD | protopanaxadiol |
| PPT | protopanaxatriol |
| QTOF | quadrupole time of flight |
| RT | retention time |
References
- Wang, Y.; Pan, J.Y.; Xiao, X.Y.; Lin, R.C.; Cheng, Y.Y. Simultaneous determination of ginsenosides in Panax ginseng with different growth ages using high-performance liquid chromatography–mass spectrometry. Phytochem. Anal. 2006, 17, 424–430. [Google Scholar] [CrossRef]
- Xie, J.-T.; Mehendale, S.R.; Li, X.; Quigg, R.; Wang, X.; Wang, C.-Z.; Wu, J.A.; Aung, H.H.; Rue, P.A.; Bell, G.I. Anti-diabetic effect of ginsenoside Re in ob/ob mice. BBA-Mol. Basis Dis. 2005, 1740, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Kenarova, B.; Neychev, H.; Hadjiivanova, C.; Petkov, V.D. Immunomodulating activity of ginsenoside Rg1 from Panax ginseng. Jpn. J. Pharmacol. 1990, 54, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.Y.K.; Mak, N.K.; Cheng, Y.K.; Leung, K.W.; Ng, T.B.; Fan, D.T.P.; Yeung, H.W.; Wong, R.N.S. Pharmacogenomics and the Yin/Yang actions of ginseng: Anti-tumor, angiomodulating and steroid-like activities of ginsenosides. Chin. Med.-UK 2007, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Zhang, Y.; Gao, J.; Ding, X.; Gao, S. The anti-fatigue effect of 20 (R)-ginsenoside Rg3 in mice by intranasally administration. Biol. Pharm. Bull. 2008, 31, 2024–2027. [Google Scholar] [CrossRef] [PubMed]
- Kang, O.-J.; Kim, J.-S. Comparison of Ginsenoside Contents in Different Parts of Korean Ginseng (Panax ginseng C.A. Meyer). Prev. Nutr. Food Sci. 2016, 21, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Wang, Y.T.; Li, J.; Zhang, H.Q.; Ding, L. Investigation of ginsenosides in different parts and ages of Panax ginseng. Food Chem. 2007, 102, 664–668. [Google Scholar] [CrossRef]
- Lee, J.W.; Choi, B.-R.; Kim, Y.-C.; Choi, D.J.; Lee, Y.-S.; Kim, G.-S.; Baek, N.-I.; Kim, S.-Y.; Lee, D.Y. Comprehensive Profiling and Quantification of Ginsenosides in the Root, Stem, Leaf, and Berry of Panax ginseng by UPLC-QTOF/MS. Molecules 2017, 22, 2147. [Google Scholar] [CrossRef]
- Attele, A.S.; Zhou, Y.P.; Xie, J.T.; Wu, J.A.; Zhang, L.; Dey, L.; Pugh, W.; Rue, P.A.; Polonsky, K.S.; Yuan, C.S. Antidiabetic effects of Panax ginseng berry extract and the identification of an effective component. Diabetes 2002, 51, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.J.; Han, I.H.; Kim, Y.J.; Yamabe, N.; Lee, D.; Hwang, G.S.; Oh, M.; Choi, K.C.; Kim, S.N.; Ham, J.; et al. Anticarcinogenic Effects of Products of Heat-Processed Ginsenoside Re, a Major Constituent of Ginseng Berry, on Human Gastric Cancer Cells. J. Agric. Food Chem. 2014, 62, 2830–2836. [Google Scholar] [CrossRef]
- Mehendale, S.; Aung, H.; Wang, A.; Yin, J.-J.; Wang, C.-Z.; Xie, J.-T.; Yuan, C.-S. American ginseng berry extract and ginsenoside Re attenuate cisplatin-induced kaolin intake in rats. Cancer Chemother. Pharmacol. 2005, 56, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.-T.; Shao, Z.-H.; Vanden Hoek, T.L.; Chang, W.-T.; Li, J.; Mehendale, S.; Wang, C.-Z.; Hsu, C.-W.; Becker, L.B.; Yin, J.-J.; et al. Antioxidant effects of ginsenoside Re in cardiomyocytes. Eur. J. Pharmacol. 2006, 532, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Dey, L.; Xie, J.T.; Wang, A.; Wu, J.; Maleckar, S.A.; Yuan, C.-S. Anti-hyperglycemic effects of ginseng: Comparison between root and berry. Phytomedicine 2003, 10, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.-T.; Du, G.-J.; McEntee, E.; Aung, H.H.; He, H.; Mehendale, S.R.; Wang, C.-Z.; Yuan, C.-S. Effects of Triterpenoid Glycosides from Fresh Ginseng Berry on SW480 Human Colorectal Cancer Cell Line. Cancer Res. Treat. 2011, 43, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.-H.; Xie, J.-T.; Vanden Hoek, T.L.; Mehendale, S.; Aung, H.; Li, C.-Q.; Qin, Y.; Schumacker, P.T.; Becker, L.B.; Yuan, C.-S. Antioxidant effects of American ginseng berry extract in cardiomyocytes exposed to acute oxidant stress. BBA-Gen. Subj. 2004, 1670, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, Y.K.; Park, N.I.; Kim, C.S.; Lee, C.Y.; Park, S.U. Chemical constituents and biological activities of the berry of Panax ginseng. J. Med. Plants Res. 2010, 4, 349–353. [Google Scholar]
- Wang, C.-Z.; Wu, J.A.; MCENTEE, E.; Yuan, C.-S. Saponins Composition in American Ginseng Leaf and Berry Assayed by High-Performance Liquid Chromatography. J. Agric. Food Chem. 2006, 54, 2261–2266. [Google Scholar] [CrossRef]
- Kim, S.N.; Ha, Y.W.; Shin, H.; Son, S.H.; Wu, S.J.; Kim, Y.S. Simultaneous quantification of 14 ginsenosides in Panax ginseng CA Meyer (Korean red ginseng) by HPLC-ELSD and its application to quality control. J. Pharm. Biomed. 2007, 45, 164–170. [Google Scholar] [CrossRef]
- Mao, Q.; Bai, M.; Xu, J.-D.; Kong, M.; Zhu, L.-Y.; Zhu, H.; Wang, Q.; Li, S.-L. Discrimination of leaves of Panax ginseng and P. quinquefolius by ultra-high performance liquid chromatography quadrupole/time-of-flight mass spectrometry based metabolomics approach. J. Pharm. Biomed. Anal. 2014, 97, 129–140. [Google Scholar] [CrossRef]
- Xie, G.X.; Ni, Y.; Su, M.M.; Zhang, Y.Y.; Zhao, A.H.; Gao, X.F.; Liu, Z.; Xiao, P.G.; Jia, W. Application of ultra-performance LC-TOF MS metabolite profiling techniques to the analysis of medicinal Panax herbs. Metabolomics 2008, 4, 248–260. [Google Scholar] [CrossRef]
- Alam, T.M.; Jenkins, J.E. HR-MAS NMR spectroscopy in material science. In Advanced Aspects of Spectroscopy; IntechOpen: London, UK, 2012; pp. 279–306. [Google Scholar]
- Mun, J.-H.; Lee, H.; Yoon, D.; Kim, B.-S.; Kim, M.-B.; Kim, S. Discrimination of Basal Cell Carcinoma from Normal Skin Tissue Using High-Resolution Magic Angle Spinning 1H NMR Spectroscopy. PLoS ONE 2016, 11, e0150328. [Google Scholar] [CrossRef] [PubMed]
- Raja, G.; Kim, S.; Yoon, D.; Yoon, C.; Kim, S. 1H-NMR-based Metabolomics Studies of the Toxicity of Mesoporous Carbon Nanoparticles in Zebrafish (Danio rerio). Bull. Korean. Chem. Soc. 2017, 38, 271–277. [Google Scholar] [CrossRef]
- Sedgwick, P. Pearson’s correlation coefficient. BMJ 2012, 345, e4483. [Google Scholar] [CrossRef]
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation coefficients: Appropriate use and interpretation. Anesth. Analg. 2018, 126, 1763–1768. [Google Scholar] [CrossRef] [PubMed]
- Vickers, C.E.; Williams, T.C.; Peng, B.; Cherry, J. Recent advances in synthetic biology for engineering isoprenoid production in yeast. Curr. Opin. Chem. Biol. 2017, 40, 47–56. [Google Scholar] [CrossRef]
- Ward, J.M.; Kühn, C.; Tegeder, M.; Frommer, W.B. Sucrose transport in higher plants. Int. Rev. Cytol. 1997, 178, 41–71. [Google Scholar]
- Falasca, A.; Melck, D.; Paris, D.; Saviano, G.; Motta, A.; Iorizzi, M. Seasonal changes in the metabolic fingerprint of Juniperus communis L. berry extracts by 1H NMR-based metabolomics. Metabolomics 2014, 10, 165–174. [Google Scholar] [CrossRef]
- Loewus, F.A.; Murthy, P.P. myo-Inositol metabolism in plants. Plant Sci. 2000, 150, 1–19. [Google Scholar] [CrossRef]
- Cho, W.C.; Chung, W.S.; Lee, S.K.; Leung, A.W.; Cheng, C.H.; Yue, K.K. Ginsenoside Re of Panax ginseng possesses significant antioxidant and antihyperlipidemic efficacies in streptozotocin-induced diabetic rats. Eur. J. Pharmacol. 2006, 550, 173–179. [Google Scholar] [CrossRef]
- Liu, Y.W.; Zhu, X.; Li, W.; Lu, Q.; Wang, J.Y.; Wei, Y.Q.; Yin, X.X. Ginsenoside Re attenuates diabetes-associated cognitive deficits in rats. Pharmacol. Biochem. Behav. 2012, 101, 93–98. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X.; Lv, W.; Yang, Y.; Gao, H.; Yang, J.; Gao, H.; Yang, J.; Shen, Y.; Ning, G. Ginsenoside Re reduces insulin resistance through inhibition of c-Jun NH2-terminal kinase and nuclear factor-κB. Mol. Endocrinol. 2008, 22, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Wang, M.; Wang, X.; Sun, G.; Ye, J.; Xu, H.; Sun, X. Suppression of NADPH oxidase-and mitochondrion-derived superoxide by Notoginsenoside R1 protects against cerebral ischemia?reperfusion injury through estrogen receptor-dependent activation of Akt/Nrf2 pathways. Free Radic. Res. 2014, 48, 823–838. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Sun, G.; Ye, J.; Xu, H.; Wang, H.; Sun, X. Notoginsenoside R1-mediated neuroprotection involves estrogen receptor-dependent crosstalk between Akt and ERK1/2 pathways: A novel mechanism of Nrf2/ARE signaling activation. Free Radic. Res. 2014, 48, 445–460. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Meng, X.; Luo, Y.; Wu, Y.; Ye, T.; Zhou, P.; Ding, S.; Wang, M.; Lu, S.; Zhu, L.; et al. Notoginsenoside R1 ameliorates diabetic encephalopathy by activating the Nrf2 pathway and inhibiting NLRP3 inflammasome activation. Oncotarget 2018, 9, 9344. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.B.; Sun, G.B.; Wang, M.; Sun, J.; Qin, M.; Sun, X.B. P90RSK and Nrf2 activation via MEK1/2-ERK1/2 pathways mediated by notoginsenoside R2 to prevent 6-hydroxydopamine-induced apoptotic death in SH-SY5Y cells. Evid. Based Complement. Altern. 2013, 2013, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.C.; Zhu, Y.G.; Zhu, L.A.; Huang, C.; Chen, Y.; Chen, L.M.; Fang, F.; Zhou, Y.C.; Zhao, C.H. Ginsenoside Rg1 attenuates dopamine-induced apoptosis in PC12 cells by suppressing oxidative stress. Eur. J. Pharmacol. 2003, 473, 1–7. [Google Scholar] [CrossRef]
- Chen, X.C.; Zhou, Y.C.; Chen, Y.; Zhu, Y.G.; Fang, F.; Chen, L.M. Ginsenoside Rg1 reduces MPTP-induced substantia nigra neuron loss by suppressing oxidative stress 1. Acta Pharmacol. Sin. 2005, 26, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.S.; Ahn, J.H.; Le, T.K.; Kim, D.H.; Kim, H.S. Protopanaxatriol ginsenoside Rh1 inhibits the expression of matrix metalloproteinases and the in vitro invasion/migration of human astroglioma cells. Neurochem. Int. 2013, 63, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.S.; Kim, D.H.; Kim, H.S. Ginsenoside Rh1 suppresses inducible nitric oxide synthase gene expression in IFN-γ-stimulated microglia via modulation of JAK/STAT and ERK signaling pathways. Biochem. Biophys. Res. Commun. 2010, 397, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.; Ma, Z.; Yu, X.; Qu, S.; Sui, D. Effect of ginsenoside Rh1 on myocardial injury and heart function in isoproterenol-induced cardiotoxicity in rats. Toxicol. Mech. Method 2012, 22, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Sun, G.; Luo, Y.; Wang, M.; Chen, R.; Zhang, J.; Ai, Q.; Xing, N.; Sun, X. Cardioprotective effects of Notoginsenoside R1 against ischemia/reperfusion injuries by regulating oxidative stress-and endoplasmic reticulum stress-related signaling pathways. Sci. Rep.-UK 2016, 6, 21730. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Zhou, X.L.; Liu, Y.S.; Wang, Y.M.; Ma, F.; Guo, B.L.; Yan, Z.Q.; Zhang, Q.Y. Estrogen receptor α mediates the effects of notoginsenoside R1 on endotoxin-induced inflammatory and apoptotic responses in H9c2 cardiomyocytes. Mol. Med. Rep. 2015, 12, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.R.; Xu, M.C.; Huang, Y.U.; Zhang, C.J.; Lin, J.E.; Ruan, C.W. Cardioprotective effect of notoginsenoside R1 in a rabbit lung remote ischemic postconditioning model via activation of the TGF? β1/TAK1 signaling pathway. Exp. Ther. Med. 2016, 11, 2341–2348. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Gonzalez, F.J.; Yoon, M. Ginsenoside Rf, a component of ginseng, regulates lipoprotein metabolism through peroxisome proliferator-activated receptor α. Biochem. Biophys. Res. Commun. 2006, 339, 196–203. [Google Scholar] [CrossRef] [PubMed]


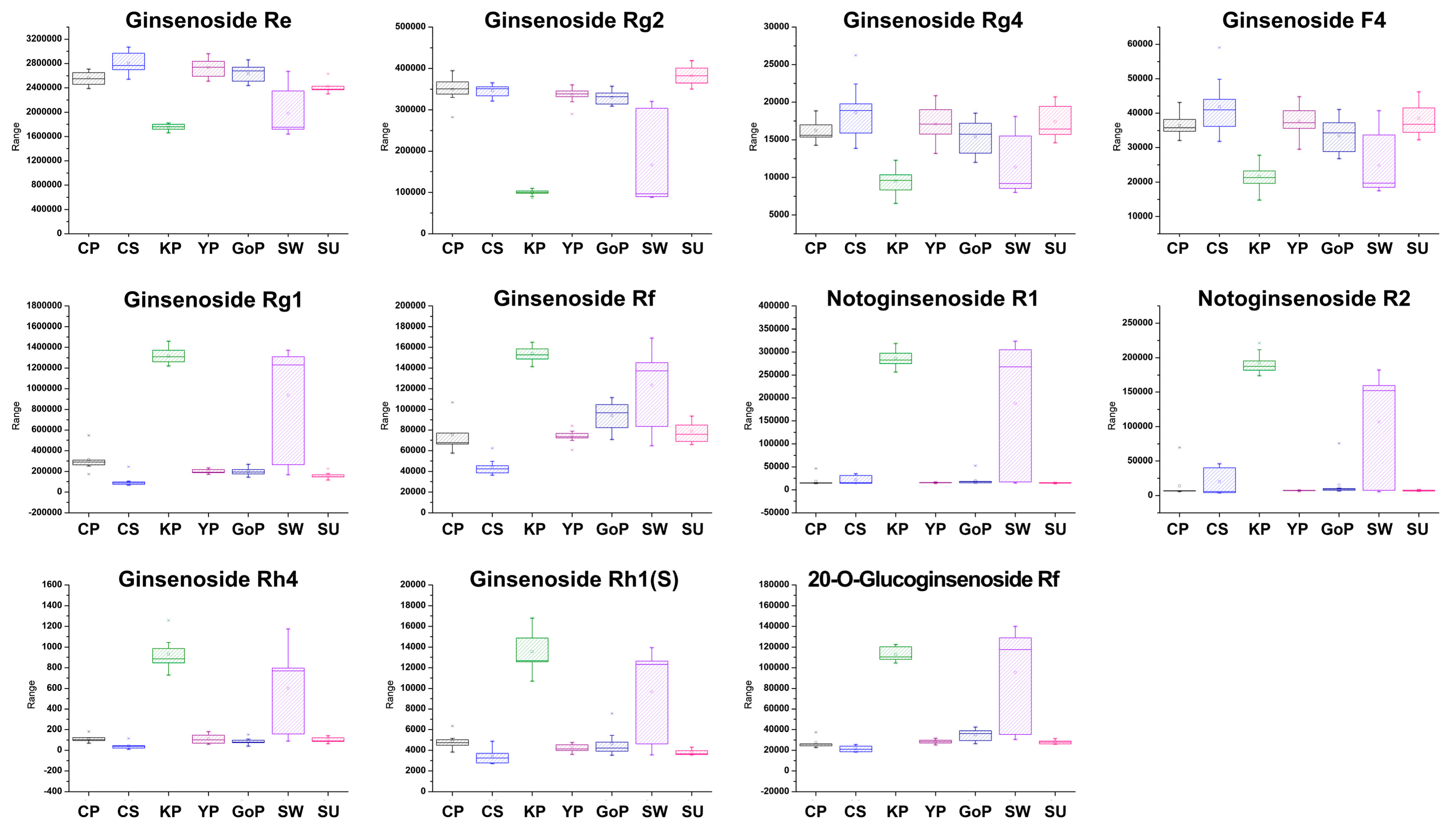
| No. | Expected RT (min) | Component Name | Formula | Observed m/z | Observed RT (min) | Adducts | Average of Relative Percentage (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CP | CS | KP | YP | GoP | SW | SU | |||||||
| 1 | 3.6 | 20-O-Glucoginsenoside Rf | C48H82O19 | 1007.5265 | 3.62 | +HCOO, -H | 0.0732 | 0.0563 | 0.2943 | 0.0764 | 0.0899 | 0.2539 | 0.0787 |
| 2 | 4.07 | Notoginsenoside R1 | C47H80O18 | 977.5161 | 4.09 | +HCOO, -H | 0.0478 | 0.0570 | 0.7476 | 0.0409 | 0.0511 | 0.4965 | 0.0406 |
| 3 | 5.35 | Ginsenoside Rg1 | C42H72O14 | 845.4773 | 5.45 | +HCOO, -H | 0.8297 | 0.2613 | 3.4380 | 0.5507 | 0.5116 | 2.4831 | 0.4419 |
| 4 | 5.66 | Ginsenoside Re | C48H82O18 | 991.5347 | 5.71 | +HCOO, -H | 6.7781 | 7.3395 | 4.5799 | 7.3372 | 6.7865 | 5.3904 | 6.7817 |
| 5 | 11.2 | vinaginsenoside R4 | C48H82O19 | 1007.5265 | 11.30 | +HCOO, -H | 0.2449 | 0.2195 | 0.2489 | 0.2805 | 0.2415 | 0.3290 | 0.2317 |
| 6 | 12.11 | Ginsenoside Rf | C42H72O14 | 845.4770 | 12.17 | +HCOO, -H | 0.1992 | 0.1142 | 0.4025 | 0.1991 | 0.2414 | 0.3307 | 0.2202 |
| 7 | 12.81 | Notoginsenoside R2 | C41H70O13 | 815.4644 | 12.89 | +HCOO, -H | 0.0368 | 0.0528 | 0.5008 | 0.0193 | 0.0384 | 0.2823 | 0.0203 |
| 8 | 13.43 | Ginsenoside F5 | C41H70O13 | 815.4669 | 13.58 | +HCOO, -H | 0.4852 | 0.4637 | 0.4576 | 0.5050 | 0.4842 | 0.5796 | 0.4123 |
| 9 | 13.56 | 20(S)-Ginsenoside Rh1 | C36H62O9 | 683.4246 | 13.66 | +HCOO | 0.0128 | 0.0090 | 0.0354 | 0.0114 | 0.0119 | 0.0258 | 0.0107 |
| 10 | 13.7 | Ginsenoside Rg2 | C42H72O13 | 829.4824 | 13.79 | +HCOO, -H | 0.9250 | 0.9064 | 0.2598 | 0.9042 | 0.8504 | 0.4588 | 1.0673 |
| 11 | 14.51 | Ginsenoside Ra3 | C59H100O27 | 1285.6185 | 14.27 | +HCOO, -H | 0.0033 | 0.0023 | 0.0032 | 0.0022 | 0.0026 | 0.0027 | 0.0031 |
| 12 | 14.35 | Ginsenoside Ra2 | C58H98O26 | 1255.6049 | 14.52 | +HCOO, -H | 0.0054 | 0 | 0.0057 | 0.0051 | 0.0031 | 0.0027 | 0.0038 |
| 13 | 14.53 | Ginsenoside Rb1 | C54H92O23 | 1153.5806 | 14.72 | +HCOO, -H | 0.5071 | 0.4411 | 0.4727 | 0.4999 | 0.3281 | 0.4326 | 0.4280 |
| 14 | 15.12 | Malonyl ginsenoside Rb1 | C57H94O26 | 1193.5794 | 15.24 | -H | 3.0830 | 2.9241 | 2.6016 | 3.1462 | 2.5714 | 2.5060 | 3.1080 |
| 15 | 15.69 | Ginsenoside Ro | C48H76O19 | 955.4726 | 15.73 | -H | 0.0486 | 0.0705 | 0.0530 | 0.0437 | 0.0621 | 0.0506 | 0.0563 |
| 16 | 16.3 | Ginsenoside Rb2 | C53H90O22 | 1123.5710 | 16.48 | +HCOO, -H | 0.9968 | 1.0124 | 1.1320 | 0.9467 | 0.9372 | 1.1441 | 0.9034 |
| 17 | 18.34 | Ginsenoside Rd | C48H82O18 | 991.5318 | 18.58 | +HCOO, -H | 1.6839 | 1.4716 | 1.7702 | 1.4030 | 1.4115 | 1.7513 | 1.3188 |
| 18 | 19.11 | Malonyl ginsenoside Rd | C51H84O21 | 1031.5279 | 19.32 | -H | 5.0173 | 4.8832 | 4.3847 | 4.8414 | 5.1380 | 4.6440 | 4.9302 |
| 19 | 23.46 | Ginsenoside Rg4 | C42H70O12 | 811.4684 | 23.75 | +HCOO, -H | 0.0430 | 0.0489 | 0.0249 | 0.0459 | 0.0397 | 0.0310 | 0.0488 |
| 20 | 23.5 | Compound O | C47H80O17 | 961.5136 | 23.77 | +HCOO, -H | 0.0930 | 0.0498 | 0.0527 | 0.0439 | 0.0610 | 0.0338 | 0.0317 |
| 21 | 24.28 | Ginsenoside F4 | C42H70O12 | 811.4695 | 24.52 | +HCOO, -H | 0.0964 | 0.1100 | 0.0566 | 0.1013 | 0.0864 | 0.0678 | 0.1077 |
| 22 | 24.68 | Ginsenoside Rh4 | C36H60O8 | 665.4140 | 24.90 | +HCOO | 0.0003 | 0.0001 | 0.0024 | 0.0003 | 0.0002 | 0.0016 | 0.0003 |
| 23 | 25.19 | Ginsenoside F2 | C42H72O13 | 829.4626 | 25.44 | +HCOO, -H | 0.0655 | 0.0346 | 0.0276 | 0.0238 | 0.0333 | 0.0201 | 0.0185 |
| 24 | 26.01 | Ginsenoside Rg3-S | C42H72O13 | 829.4784 | 26.19 | +HCOO, -H | 0.0240 | 0.0252 | 0.0219 | 0.0198 | 0.0224 | 0.0199 | 0.0223 |
| 25 | 27.35 | Compound K | C36H62O8 | 667.4320 | 27.54 | +HCOO | 0.0028 | 0.0060 | 0.0038 | 0.0039 | 0.0034 | 0.0038 | 0.0043 |
| 26 | 27.55 | Ginsenoside Rg5 | C42H70O12 | 811.4685 | 27.75 | -H | 0.0135 | 0.0153 | 0.0157 | 0.0135 | 0.0140 | 0.0134 | 0.0148 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, D.; Choi, B.-R.; Kim, Y.-C.; Oh, S.M.; Kim, H.-G.; Kim, J.-U.; Baek, N.-I.; Kim, S.; Lee, D.Y. Comparative Analysis of Panax ginseng Berries from Seven Cultivars Using UPLC-QTOF/MS and NMR-Based Metabolic Profiling. Biomolecules 2019, 9, 424. https://doi.org/10.3390/biom9090424
Yoon D, Choi B-R, Kim Y-C, Oh SM, Kim H-G, Kim J-U, Baek N-I, Kim S, Lee DY. Comparative Analysis of Panax ginseng Berries from Seven Cultivars Using UPLC-QTOF/MS and NMR-Based Metabolic Profiling. Biomolecules. 2019; 9(9):424. https://doi.org/10.3390/biom9090424
Chicago/Turabian StyleYoon, Dahye, Bo-Ram Choi, Young-Chang Kim, Seon Min Oh, Hyoung-Geun Kim, Jang-Uk Kim, Nam-In Baek, Suhkmann Kim, and Dae Young Lee. 2019. "Comparative Analysis of Panax ginseng Berries from Seven Cultivars Using UPLC-QTOF/MS and NMR-Based Metabolic Profiling" Biomolecules 9, no. 9: 424. https://doi.org/10.3390/biom9090424
APA StyleYoon, D., Choi, B.-R., Kim, Y.-C., Oh, S. M., Kim, H.-G., Kim, J.-U., Baek, N.-I., Kim, S., & Lee, D. Y. (2019). Comparative Analysis of Panax ginseng Berries from Seven Cultivars Using UPLC-QTOF/MS and NMR-Based Metabolic Profiling. Biomolecules, 9(9), 424. https://doi.org/10.3390/biom9090424







