Dose-Dependent Alterations to In Vitro Human Microbiota Composition and Butyrate Inhibition by a Supercritical Carbon Dioxide Hops Extract
Abstract
1. Introduction
2. Materials and Methods
2.1. Hops Composition
2.2. In Vitro Fermentations
2.3. Characterisation of the Microbiome
2.4. Quantification of Organic Acid Metabolites
2.5. Bioinformatics
2.6. Statistical Analysis
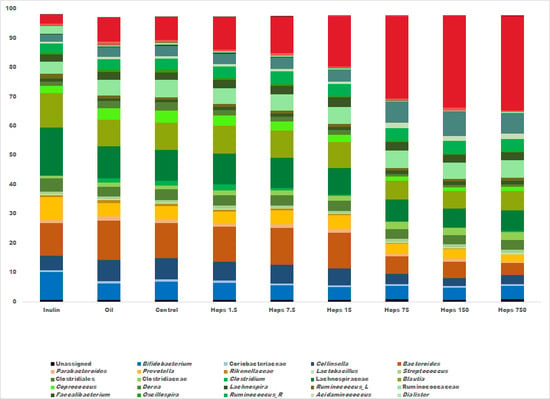
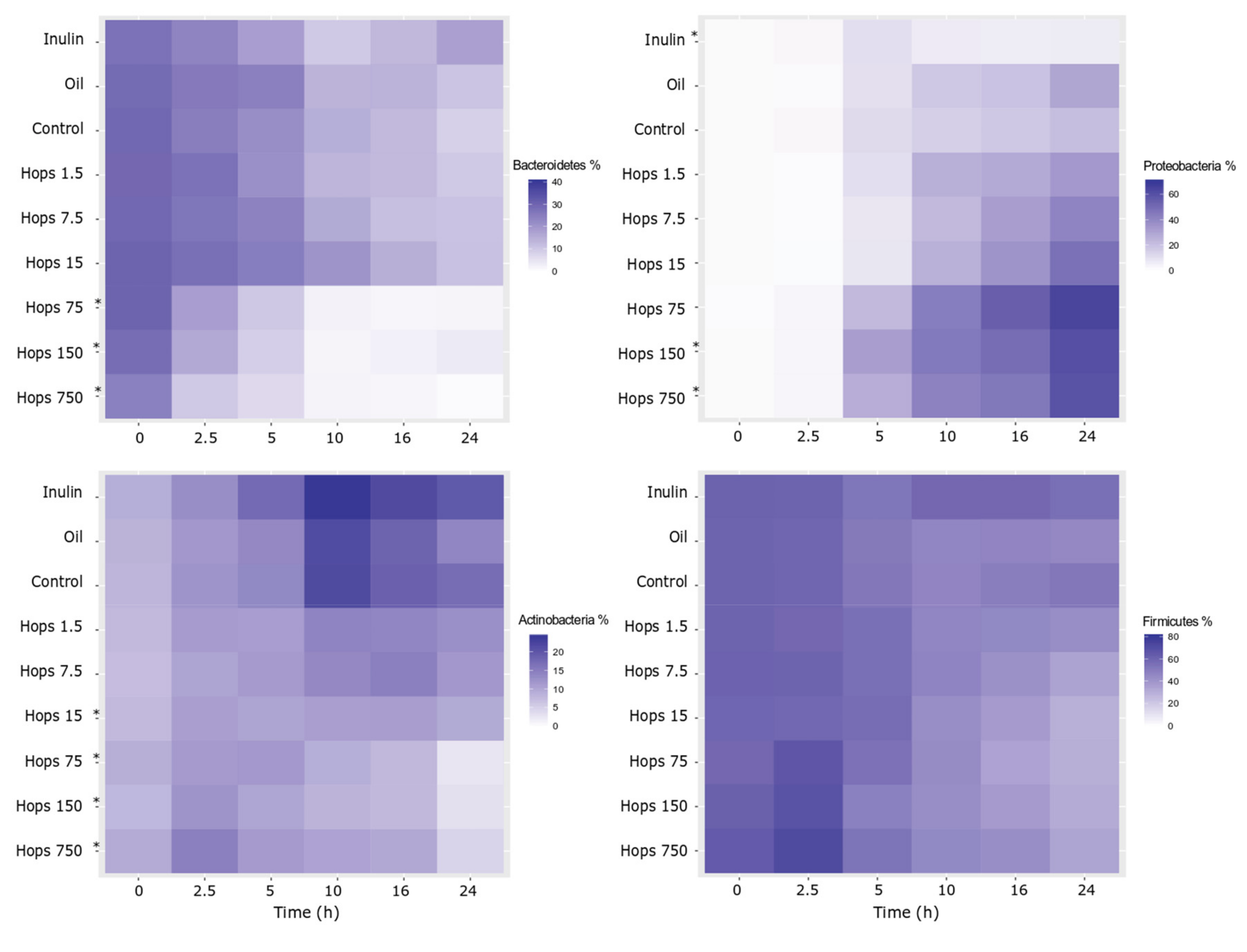
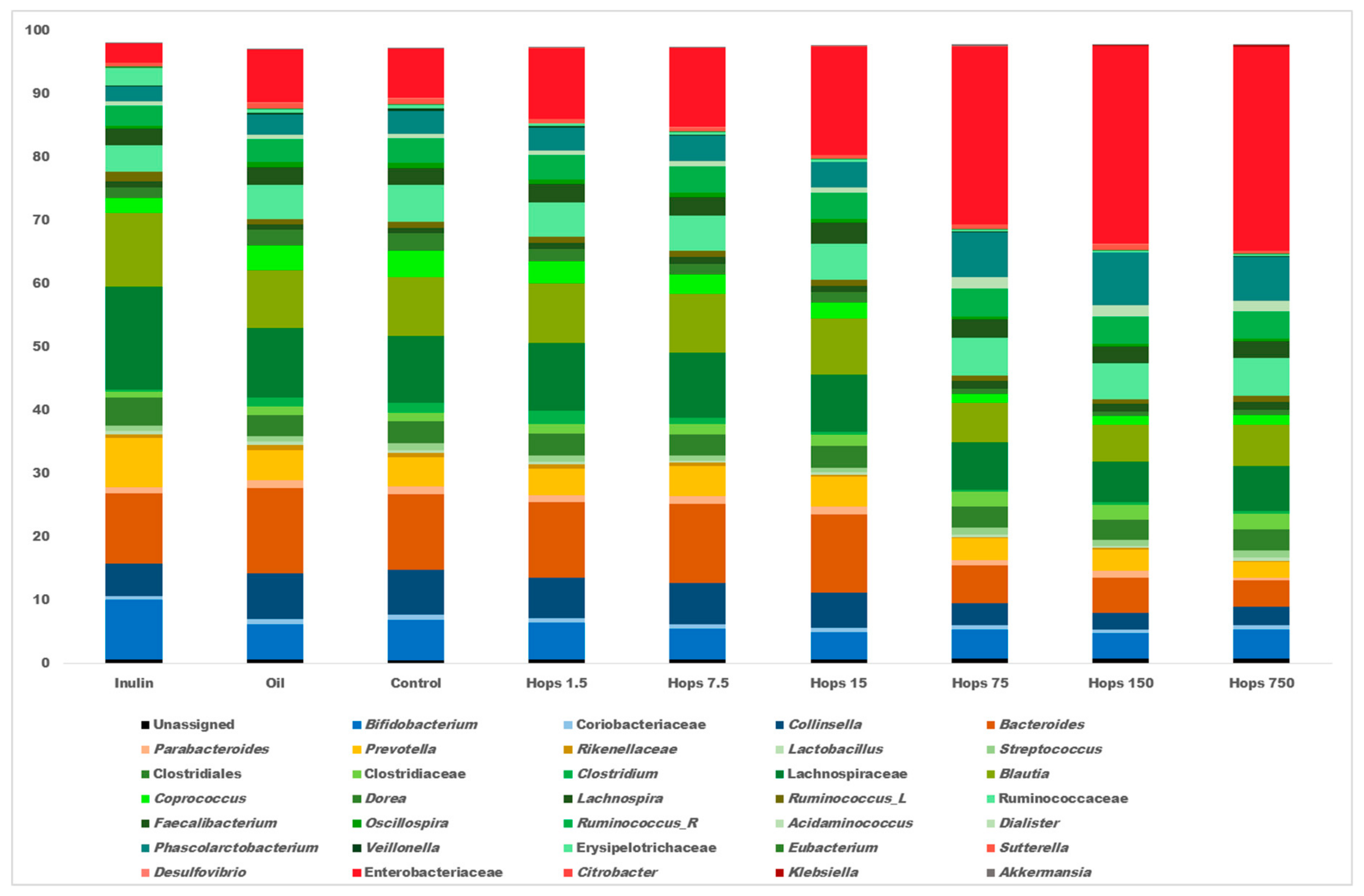
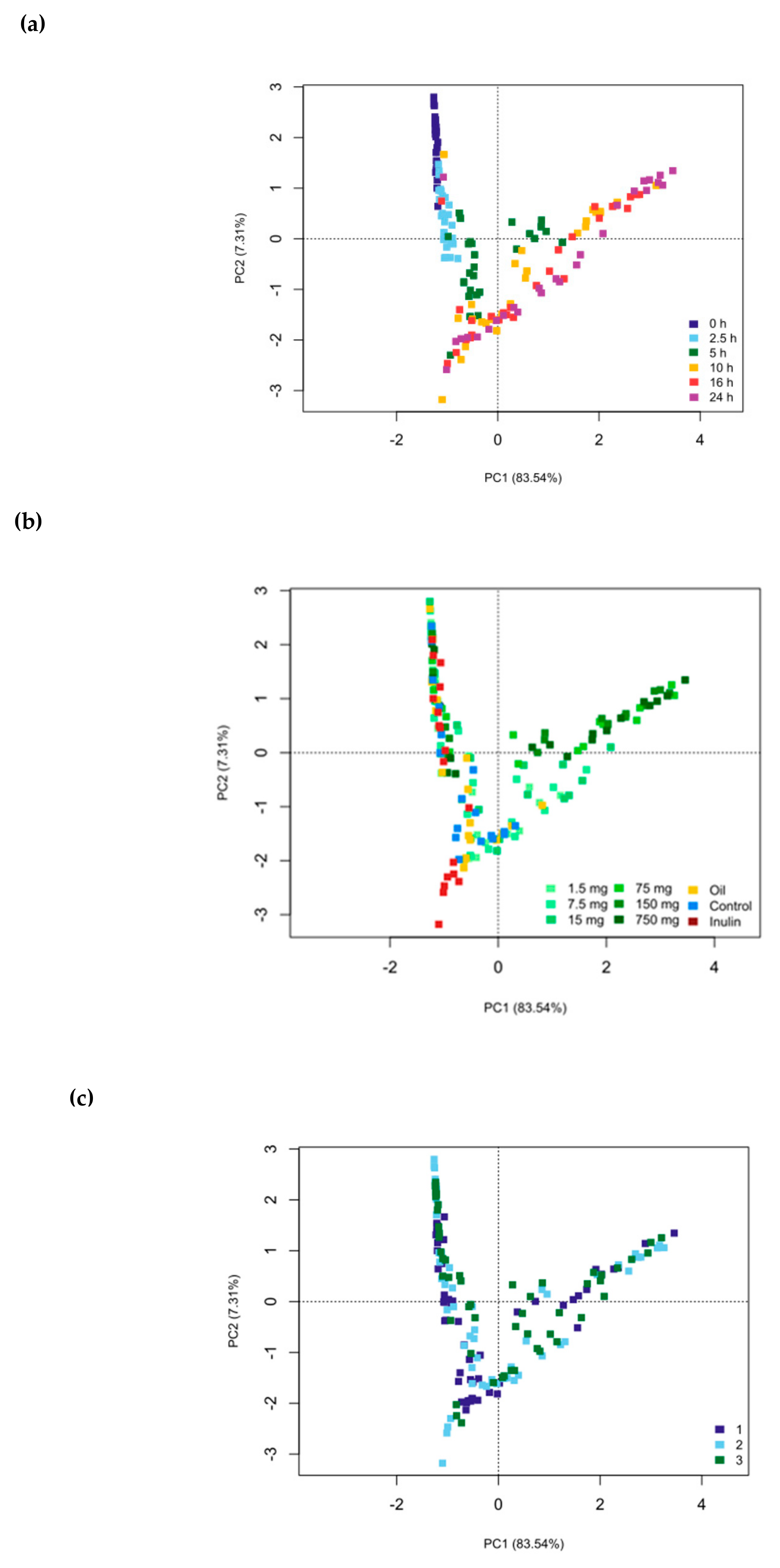
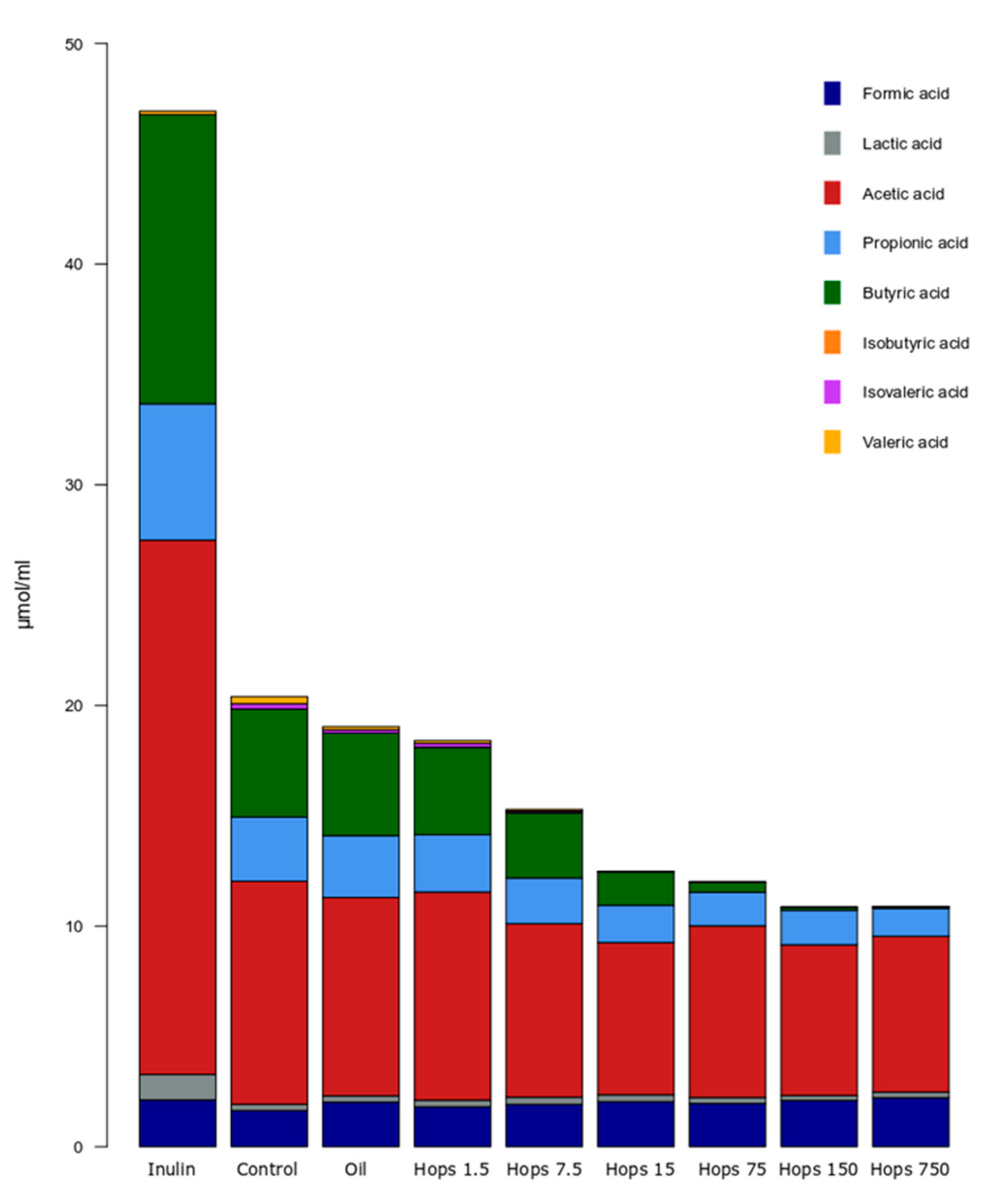
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Zhang, S.; Zheng, X.; Jia, W. Metabolomics approaches for characterizing metabolic interactions between host and its commensal microbes. Electrophoresis 2013, 34, 2787–2798. [Google Scholar] [CrossRef]
- Krumbeck, J.A.; Maldonado-Gomez, M.X.; Ramer-Tait, A.E.; Hutkins, R.W. Prebiotics and synbiotics: dietary strategies for improving gut health. Curr. Opin. Gastroenterol. 2016, 32, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Ince, J.; Duncan, S.H.; Webster, L.M.; Holtrop, G.; Ze, X.; Brown, D.; Stares, M.D.; Scott, P.; Bergerat, A.; et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011, 5, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Lamothe, R.; Mitchell, G.; Gattuso, M.; Diarra, M.S.; Malouin, F.; Bouarab, K. Plant antimicrobial agents and their effects on plant and human pathogens. Int. J. Mol. Sci. 2009, 10, 3400–3419. [Google Scholar] [CrossRef]
- Scalbert, A.; Morand, C.; Manach, C.; Remesy, C. Absorption and metabolism of polyphenols in the gut and impact on health. Biomed. Pharmacother. 2002, 56, 276–282. [Google Scholar] [CrossRef]
- Karabin, M.; Hudcova, T.; Jelinek, L.; Dostalek, P. Biologically active compounds from hops and prospects for their use. Compr. Rev. Food Sci. Food Saf. 2016, 15, 542–567. [Google Scholar] [CrossRef]
- Kramer, B.; Thielmann, J.; Hickisch, A.; Muranyi, P.; Wunderlich, J.; Hauser, C. Antimicrobial activity of hop extracts against foodborne pathogens for meat applications. J. Appl. Microbiol. 2015, 118, 648–657. [Google Scholar] [CrossRef]
- Sakamoto, K.; Konings, W.N. Beer spoilage bacteria and hop resistance. Int. J. Food Microbiol. 2003, 89, 105–124. [Google Scholar] [CrossRef]
- Possemiers, S.; Bolca, S.; Grootaert, C.; Heyerick, A.; Decroos, K.; Dhooge, W.; De Keukeleire, D.; Rabot, S.; Verstraete, W.; Van de Wiele, T. The prenylflavonoid isoxanthohumol from hops (Humulus lupulus L.) is activated into the potent phytoestrogen 8-prenylnaringenin in vitro and in the human intestine. J. Nutr. 2006, 136, 1862–1867. [Google Scholar] [CrossRef]
- Langezaal, C.R.; Chandra, A.; Katsiotis, S.T.; Scheffer, J.J.; De Haan, A.B. Analysis of supercritical carbon dioxide extracts from cones and leaves of a Humulus lupulus L. cultivar. J. Sci. Food Agric. 1990, 53, 455–463. [Google Scholar] [CrossRef]
- Rój, E.; Tadić, V.M.; Mišić, D.; Žižović, I.; Arsić, I.; Dobrzyńska-Inger, A.; Kostrzewa, D. Supercritical carbon dioxide hops extracts with antimicrobial properties. Open Chem. 2015, 13, 1157–1171. [Google Scholar] [CrossRef]
- Bruck, W.M.; Graverholt, G.; Gibson, G.R. Use of batch culture and a two-stage continuous culture system to study the effect of supplemental alpha-lactalbumin and glycomacropeptide on mixed populations of human gut bacteria. FEMS Microbiol. Ecol. 2002, 41, 231–237. [Google Scholar] [CrossRef]
- Bortoluzzi, C.; Menten, J.F.M.; Pereira, R.; Fagundes, N.S.; Napty, G.S.; Pedroso, A.A.; Bigaton, A.D.; Andreote, F.D. Hops beta-acids and zinc bacitracin affect the performance and intestinal microbiota of broilers challenged with Eimeria acervulina and Eimeria tenella. Anim. Feed Sci. Technol. 2015, 207, 181–189. [Google Scholar] [CrossRef]
- Tillman, G.E.; Haas, G.J.; Wise, M.G.; Oakley, B.; Smith, M.A.; Siragusa, G.R. Chicken intestine microbiota following the administration of lupulone, a hop-based antimicrobial. FEMS Microbiol. Ecol. 2011, 77, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Gerhauser, C. Broad spectrum antiinfective potential of xanthohumol from hop (Humulus lupulus L.) in comparison with activities of other hop constituents and xanthohumol metabolites. Mol. Nutr. Food Res. 2005, 49, 827–831. [Google Scholar] [CrossRef]
- Possemiers, S.; Heyerick, A.; Robbens, V.; De Keukeleire, D.; Verstraete, W. Activation of proestrogens from hops (Humulus lupulus L.) by intestinal microbiota; Conversion of isoxanthohumol into 8-prenylnaringenin. J. Agric. Food Chem. 2005, 53, 6281–6288. [Google Scholar] [CrossRef]
- Possemiers, S.; Rabot, S.; Espin, J.C.; Bruneau, A.; Philippe, C.; Gonzalez-Sarrias, A.; Heyerick, A.; Tomas-Barberan, F.A.; De Keukeleire, D.; Verstraete, W. Eubacterium limosum activates isoxanthohumol from hops (Humulus lupulus L.) into the potent phytoestrogen 8-prenylnaringenin in vitro and in rat intestine. J. Nutr. 2008, 138, 1310–1316. [Google Scholar] [CrossRef]
- Blatchford, P.K.; Bentley-Hewitt, H.S.; McGhie, T.; Gearry, R.; Gibson, G.; Ansell, J. In vitro characterisation of the fermentation profile and prebiotic capacity of gold-fleshed kiwifruit. Benef. Microbes 2015, 6, 829–839. [Google Scholar] [CrossRef]
- Gietl, E.; Mengerink, W.; De Slegte, J.; Gibson, G.; Rastall, R.; Van den Heuvel, E. Factors involved in the in vitro fermentability of short carbohydrates in static faecal batch cultures. Int. J. Carbohydr. Chem. 2012, 2012. [Google Scholar] [CrossRef]
- Parkar, S.G.; Davis, P.; Trower, T.M.; Hedderley, D.I.; Sutton, K.H.; Ingram, J.R. Factors affecting microbial metabolism in a human fecal fermentation model to evaluate prebiotics. J. Nutr. Intermed. Metab. 2019, 15, 55–57. [Google Scholar] [CrossRef]
- Nadkarni, M.A.; Martin, F.E.; Jacques, N.A.; Hunter, N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002, 148, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Rinttila, T.; Kassinen, A.; Malinen, E.; Krogius, L.; Palva, A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 2004, 97, 1166–1177. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.J.; Carter, J.N.; Li, Y.; Porter, J.H.; Kerley, M.S. Comparison of agar plate and real-time PCR on enumeration of Lactobacillus, Clostridium perfringens and total anaerobic bacteria in dog faeces. Lett. Appl. Microbiol. 2006, 42, 490–494. [Google Scholar] [CrossRef]
- Kok, R.G.; DeWaal, A.; Schut, F.; Welling, G.W.; Weenk, G.; Hellingwerf, K.J. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl. Environ. Microbiol. 1996, 62, 3668–3672. [Google Scholar] [PubMed]
- Paturi, G.; Butts, C.A.; Bentley-Hewitt, K.L.; Ansell, J. Influence of green and gold kiwifruit on indices of large bowel function in healthy rats. J. Food Sci. 2014, 79, H1611–H1620. [Google Scholar] [CrossRef] [PubMed]
- Leahy, S.C.; Kelly, W.J.; Altermann, E.; Ronimus, R.S.; Yeoman, C.J.; Pacheco, D.M.; Li, D.; Kong, Z.H.; McTavish, S.; Sang, C.; et al. The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS ONE 2010, 5, e8926. [Google Scholar] [CrossRef]
- Kurakawa, T.; Kubota, H.; Tsuji, H.; Matsuda, K.; Takahashi, T.; Ramamurthy, T.; Nair, G.B.; Takeda, Y.; Nomoto, K. Intestinal Enterobacteriaceae and Escherichia coli populations in Japanese adults demonstrated by the reverse transcription-quantitative PCR and the clone library analyses. J. Microbiol. Methods 2013, 92, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Parkar, S.G.; Trower, T.M.; Stevenson, D.E. Fecal microbial metabolism of polyphenols and its effects on human gut microbiota. Anaerobe 2013, 23, 12–19. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Oksanen, J.; Blanchet, F.; Kindt, R.; Minchin, P.; O’Hara, P.; Simpson, G.; Simpson, G.; Stevens, M.; Wagner, H. Package ‘Vegan’: Community ecology package, version 2.0 10. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 10 January 2017).
- Wickham, H. ggplot2—Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016. [Google Scholar]
- Leinonen, R.; Sugawara, H.; Shumway, M.; International Nucleotide Sequence Database Collaboration. The sequence read archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef]
- Dostalek, P.; Karabin, M.; Jelinek, L. Hop phytochemicals and their potential role in metabolic syndrome prevention and therapy. Molecules 2017, 22, 1761. [Google Scholar] [CrossRef]
- Haas, G.J.; Barsoumian, R. Antimicrobial activity of hop resins. J. Food Prot. 1994, 57, 59–61. [Google Scholar] [CrossRef]
- Schonberger, C.; Kostelecky, T. 125th Anniversary Review: The Role of Hops in Brewing. J. Inst. Brew. 2011, 117, 259–267. [Google Scholar] [CrossRef]
- Chadwick, L.R.; Pauli, G.F.; Farnsworth, N.R. The pharmacognosy of Humulus lupulus L. (hops) with an emphasis on estrogenic properties. Phytomedicine 2006, 13, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Cattoor, K. Bioavailability of hop-derived bitter acids: Key Factors for the Health-Beneficial Properties of Beer. PhD Thesis, Ghent University, Gent, Belgium, 2013. [Google Scholar]
- Hofte, A.; Van der Hoeven, R.; Fung, S.; Verpoorte, R.; Tjaden, U.; Van der Greef, J. Characterization of hop acids by liquid chromatography with negative electrospray ionization mass spectrometry. J. Am. Soc. Brew. Chem. 1998, 56, 118–122. [Google Scholar] [CrossRef]
- Aguirre, M.; Ramiro-Garcia, J.; Koenen, M.E.; Venema, K. To pool or not to pool? Impact of the use of individual and pooled fecal samples for in vitro fermentation studies. J. Microbiol. Methods 2014, 107, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Rosendale, D.; Butts, C.; De Guzman, E.; Maddox, I.; Martell, S.; McIntyre, L.; Skinner, M.; Smith, H.; Ansell, J. The contribution of dietary broccoli sprouts towards the microbial metabolite profile in the hind gut of mice. Int. J. Food Sci. Technol. 2012, 47, 1328–1332. [Google Scholar] [CrossRef]
- Cummings, J.H.; Englyst, H.N. Fermentation in the human large-intestine and the available substrates. Am. J. Clin. Nutr. 1987, 45, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef] [PubMed]
- McNabney, S.M.; Henagan, T.M. Short chain fatty acids in the colon and peripheral tissues: A focus on butyrate, colon cancer, obesity and insulin resistance. Nutrients 2017, 9, 1348. [Google Scholar] [CrossRef] [PubMed]
- Yadav, H.; Lee, J.-H.; Lloyd, J.; Walter, P.; Rane, S.G. Beneficial metabolic effects of a probiotic via butyrate-induced GLP-1 hormone secretion. J. Biol. Chem. 2013, 288, 25088–25097. [Google Scholar] [CrossRef] [PubMed]
- Canani, R.B.; Di Costanzo, M.; Leone, L.; Pedata, M.; Meli, R.; Calignano, A. Potential beneficial effects of butyrate in intestinal and extrainitestinal diseases. World J. Gastroenterol. 2011, 17, 1519–1528. [Google Scholar] [CrossRef]
- Vital, M.; Howe, A.C.; Tiedje, J.M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio 2014, 5, e00889. [Google Scholar] [CrossRef]
- Barcenilla, A.; Pryde, S.E.; Martin, J.C.; Duncan, S.H.; Stewart, C.S.; Henderson, C.; Flint, H.J. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 2000, 66, 1654–1661. [Google Scholar] [CrossRef]
- Pryde, S.E.; Duncan, S.H.; Hold, G.L.; Stewart, C.S.; Flint, H.J. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 2002, 217, 133–139. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Leroy, F. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int. J. Food Microbiol. 2011, 149, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.R.; Gratz, S.W.; Duncan, S.H.; Holtrop, G.; Ince, J.; Scobbie, L.; Duncan, G.; Johnstone, A.M.; Lobley, G.E.; Wallace, R.J.; et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am. J. Clin. Nutr. 2011, 93, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Behr, J.; Ganzle, M.G.; Vogel, R.F. Characterization of a highly hop-resistant Lactobacillus brevis strain lacking hop transport. Appl. Environ. Microbiol. 2006, 72, 6483–6492. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Iijima, K.; Sakamoto, K.; Sami, M.; Yamashita, H. A review of hop resistance in beer spoilage lactic acid bacteria. J. Inst. Brew. 2006, 112, 173–191. [Google Scholar] [CrossRef]
- Cermak, P.; Olsovska, J.; Mikyska, A.; Dusek, M.; Kadleckova, Z.; Vanicek, J.; Nyc, O.; Sigler, K.; Bostikova, V.; Bostik, P. Strong antimicrobial activity of xanthohumol and other derivatives from hops (Humulus lupulus L.) on gut anaerobic bacteria. APMIS 2017, 125, 1033–1038. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Everard, A.; Geurts, L.; Van Roye, M.; Delzenne, N.M.; Cani, P.D. Tetrahydro iso-alpha acids from hops improve glucose homeostasis and reduce body weight gain and metabolic endotoxemia in high-fat diet-fed mice. PLoS ONE 2012, 7, e33858. [Google Scholar] [CrossRef]
- Kemperman, R.A.; Gross, G.; Mondot, S.; Possemiers, S.; Marzorati, M.; Van de Wiele, T.; Dore, J.; Vaughan, E.E. Impact of polyphenols from black tea and red wine/grape juice on a gut model microbiome. Food Res. Int. 2013, 53, 659–669. [Google Scholar] [CrossRef]
- Blatchford, P.; Stoklosinski, H.; Walton, G.; Swann, J.; Gibson, G.; Gearry, R.; Ansell, J. Kiwifruit fermentation drives positive gut microbial and metabolic changes irrespective of initial microbiota composition. Bioact. Carbohydr. Diet. Fibre 2015, 6, 37–45. [Google Scholar] [CrossRef]
- Long, W.; Xue, Z.; Zhang, Q.; Feng, Z.; Bridgewater, L.; Wang, L.; Zhao, L.; Pang, X. Differential responses of gut microbiota to the same prebiotic formula in oligotrophic and eutrophic batch fermentation systems. Sci. Rep. 2015, 5, 13469. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Macfarlane, S.; Gibson, G.R. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb. Ecol. 1998, 35, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Marianelli, C.; Cifani, N.; Pasquali, P. Evaluation of antimicrobial activity of probiotic bacteria against Salmonella enterica subsp. enterica serovar typhimurium 1344 in a common medium under different environmental conditions. Res. Microbiol. 2010, 161, 673–680. [Google Scholar]
- Beards, E.; Tuohy, K.; Gibson, G. Bacterial, SCFA and gas profiles of a range of food ingredients following in vitro fermentation by human colonic microbiota. Anaerobe 2010, 16, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Rycroft, C.E.; Jones, M.R.; Gibson, G.R.; Rastall, R.A. A comparative in vitro evaluation of the fermentation properties of prebiotic oligosaccharides. J. Appl. Microbiol. 2001, 91, 878–887. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-F.; Cao, W.-W.; Cerniglia, C.E. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl. Environ. Microbiol. 1996, 62, 1242–1247. [Google Scholar] [PubMed]
- Respondek, F.; Gerard, P.; Bossis, M.; Boschat, L.; Bruneau, A.; Rabot, S.; Wagner, A.; Martin, J.-C. Short-chain fructo-oligosaccharides modulate intestinal microbiota and metabolic parameters of humanized gnotobiotic diet induced obesity mice. PLoS ONE 2013, 8, e71026. [Google Scholar] [CrossRef] [PubMed]
| Substrate | Sample Name | Hops Extract (mg) Added to Each Vessel | Final Hops Extract Concentration (µg/mL) | Inulin (mg) | Canola Oil (mg) |
|---|---|---|---|---|---|
| Hops | Hops 750 | 750 | 5000 | 0 | 375 |
| Hops | Hops 150 | 150 | 1000 | 0 | 375 |
| Hops | Hops 75 | 75 | 500 | 0 | 375 |
| Hops | Hops 15 | 15 | 100 | 0 | 375 |
| Hops | Hops 7.5 | 7.5 | 50 | 0 | 375 |
| Hops | Hops 1.5 | 1.5 | 10 | 0 | 375 |
| Canola Oil | Oil | 0 | 0 | 0 | 375 |
| Unsupplemented | Control | 0 | 0 | 0 | 0 |
| Inulin | Inulin | 0 | 0 | 1500 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blatchford, P.A.; Parkar, S.G.; Hopkins, W.; Ingram, J.R.; Sutton, K.H. Dose-Dependent Alterations to In Vitro Human Microbiota Composition and Butyrate Inhibition by a Supercritical Carbon Dioxide Hops Extract. Biomolecules 2019, 9, 390. https://doi.org/10.3390/biom9090390
Blatchford PA, Parkar SG, Hopkins W, Ingram JR, Sutton KH. Dose-Dependent Alterations to In Vitro Human Microbiota Composition and Butyrate Inhibition by a Supercritical Carbon Dioxide Hops Extract. Biomolecules. 2019; 9(9):390. https://doi.org/10.3390/biom9090390
Chicago/Turabian StyleBlatchford, Paul A., Shanthi G. Parkar, Wendy Hopkins, John R. Ingram, and Kevin H. Sutton. 2019. "Dose-Dependent Alterations to In Vitro Human Microbiota Composition and Butyrate Inhibition by a Supercritical Carbon Dioxide Hops Extract" Biomolecules 9, no. 9: 390. https://doi.org/10.3390/biom9090390
APA StyleBlatchford, P. A., Parkar, S. G., Hopkins, W., Ingram, J. R., & Sutton, K. H. (2019). Dose-Dependent Alterations to In Vitro Human Microbiota Composition and Butyrate Inhibition by a Supercritical Carbon Dioxide Hops Extract. Biomolecules, 9(9), 390. https://doi.org/10.3390/biom9090390