LcNAC13 Physically Interacts with LcR1MYB1 to Coregulate Anthocyanin Biosynthesis-Related Genes during Litchi Fruit Ripening
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
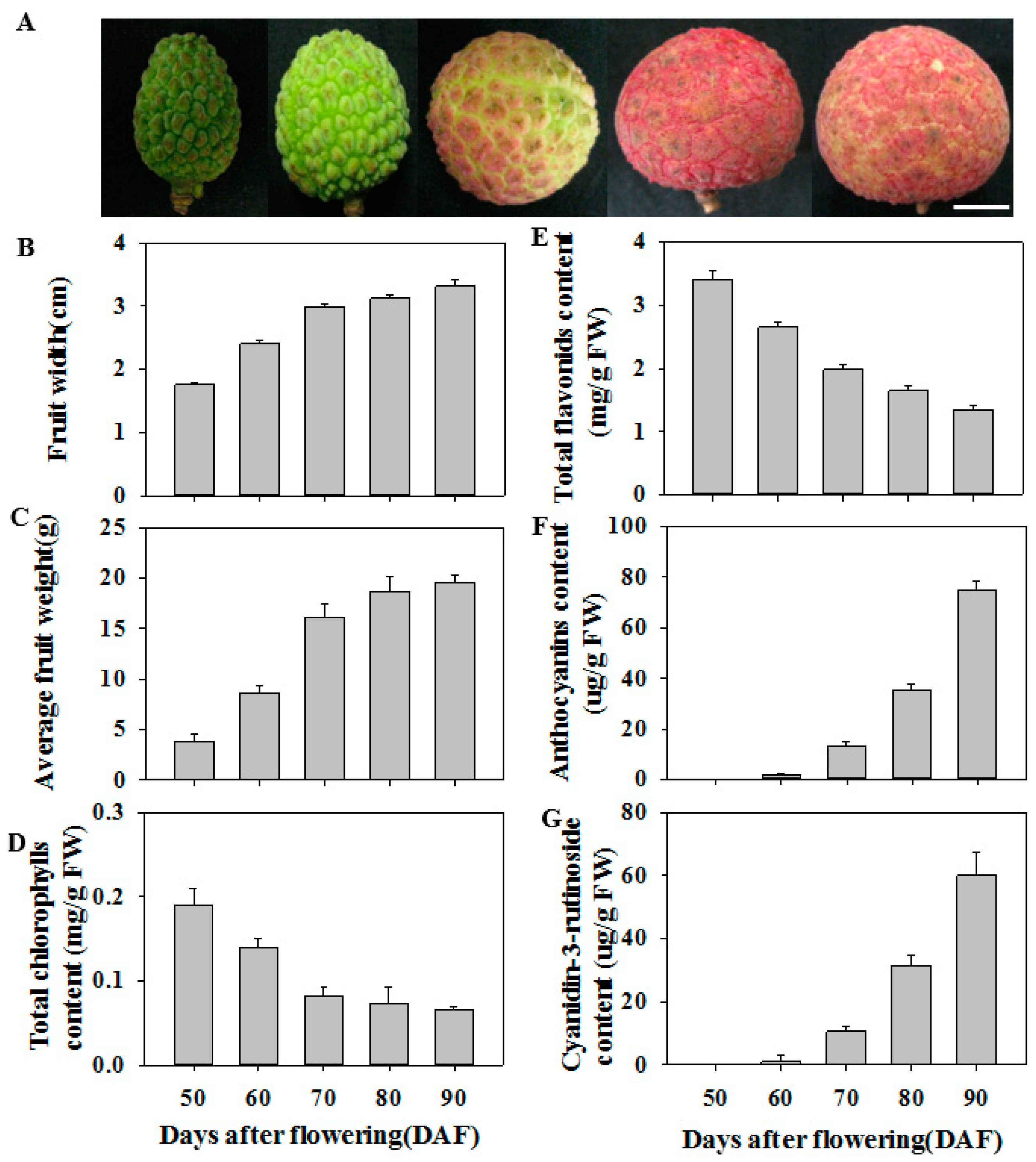
2.2. Flavonoid and Chlorophyll Measurement
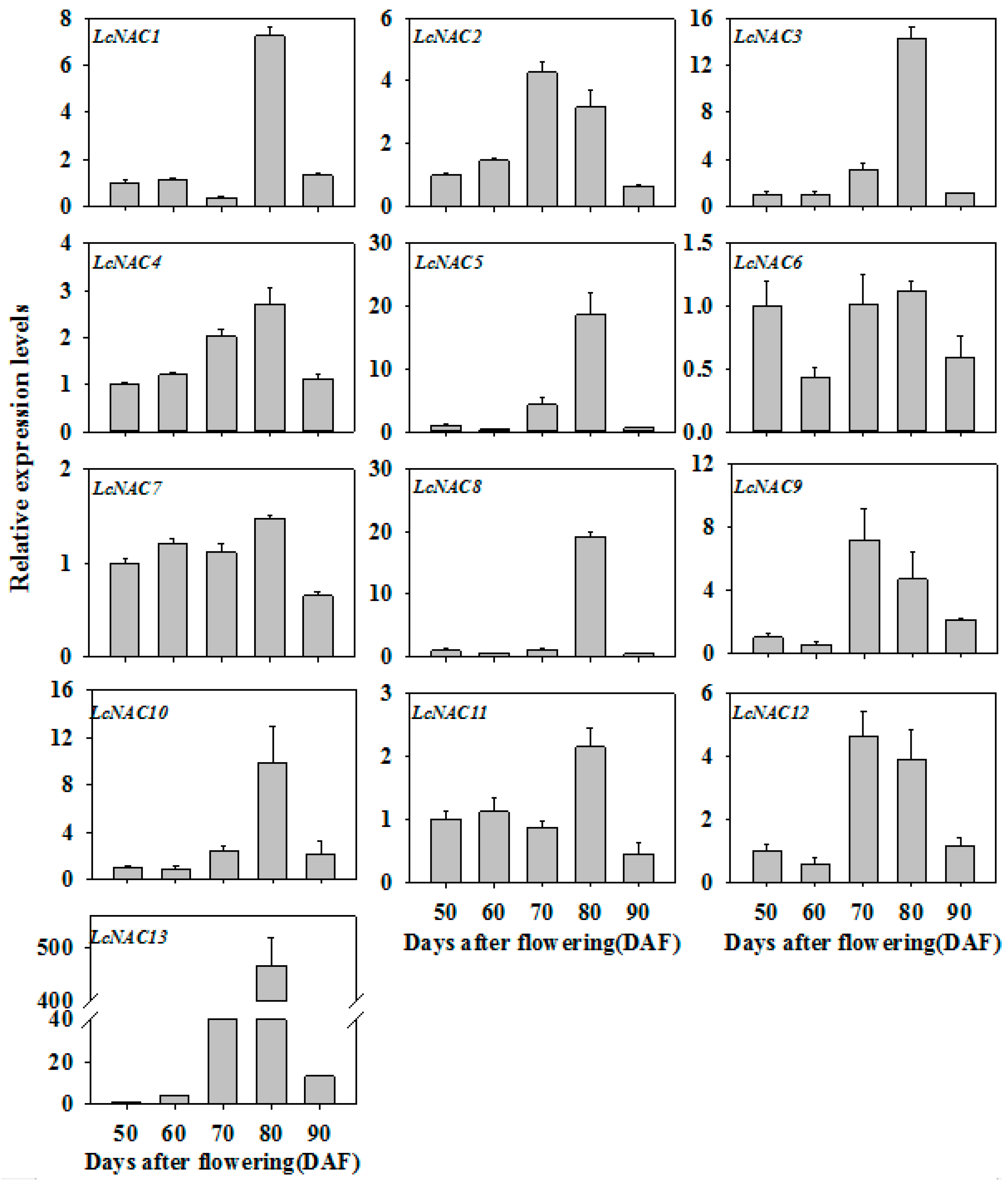
2.3. RNA Extraction and Quantitative Real-Time PCR Analysis
2.4. Promoter Isolation
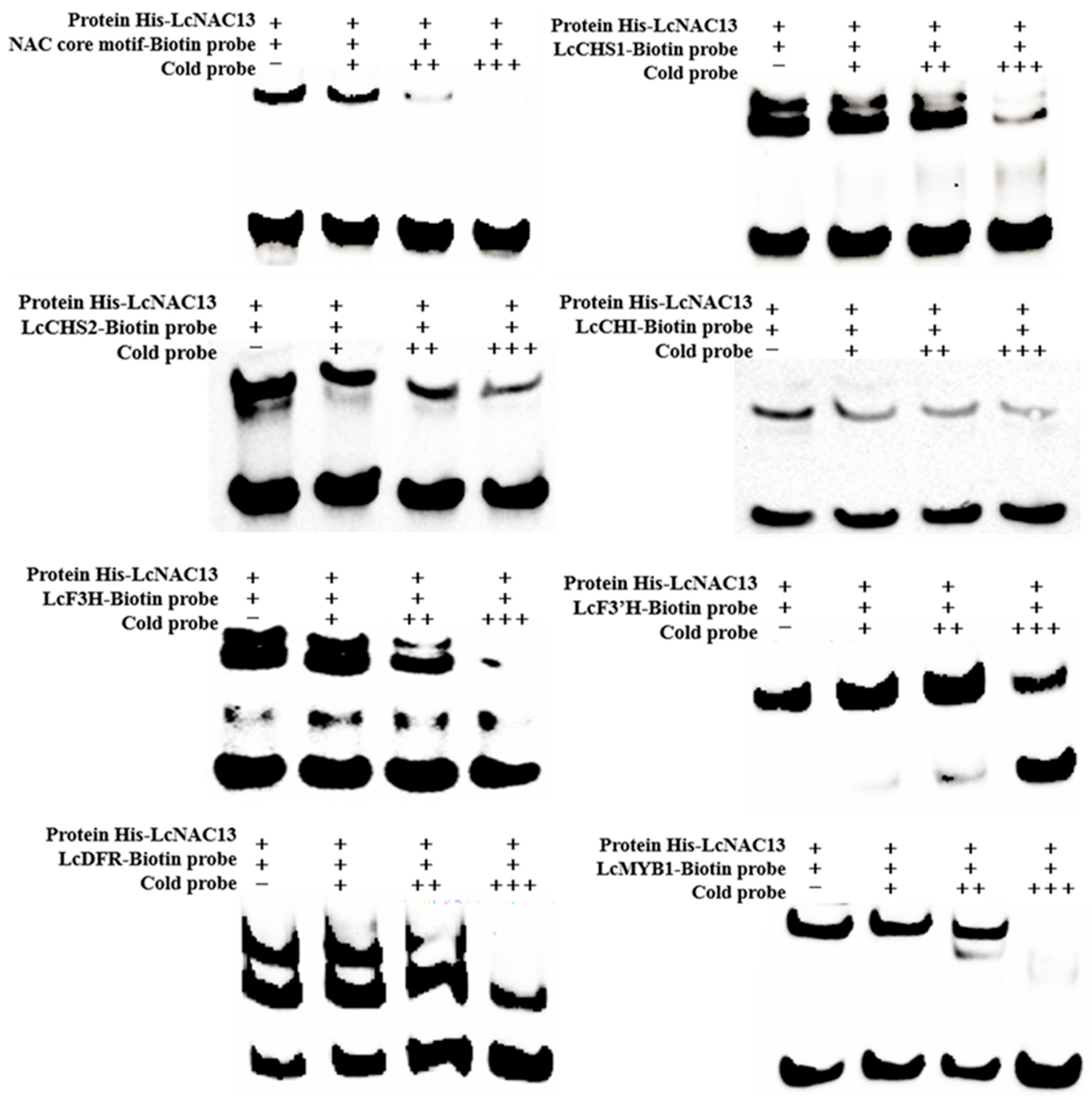
2.5. Electrophoretic Mobility Shift Assay (EMSA)
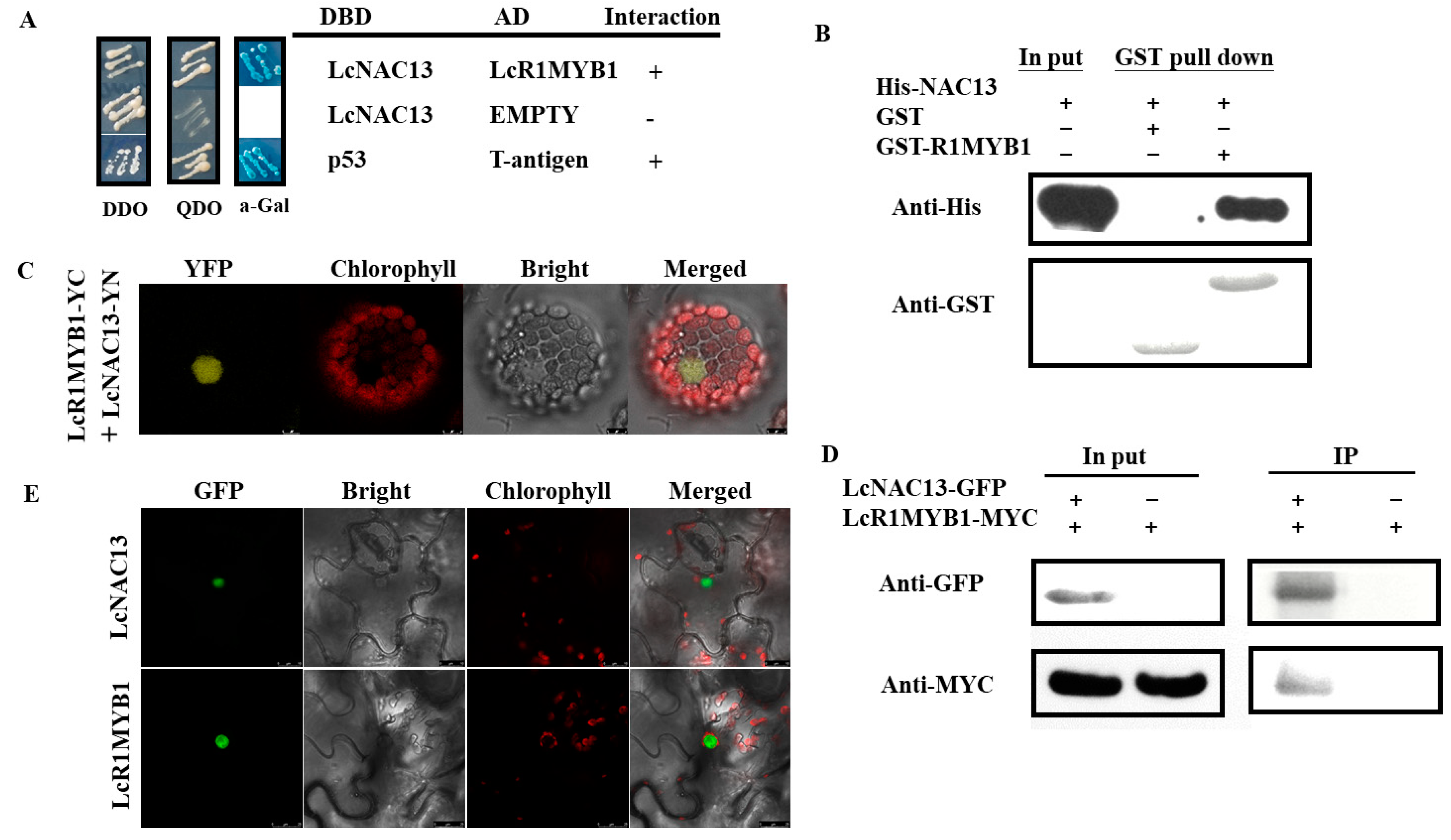
2.6. Yeast Two-Hybrid (Y2H) Assay
2.7. Expression and Purification of Recombinant LcNAC13 and LcR1MYB1
2.8. GST Pull-Down Assay
2.9. Bimolecular Fluorescence Complementation (BiFC) Assay
2.10. Co-IP Assay
2.11. Subcellular Localization Analysis
2.12. Dual-Luciferase Reporter Assay (DLR)
2.13. Data Handling
3. Results
3.1. Physiological Characteristics of Litchi Fruit during Ripening
3.2. Expression Profiles of LcNAC Genes during Litchi Fruit Ripening
3.3. LcNAC13 Binds to the Promoters of Anthocyanin Biosynthesis-Related Genes
3.4. LcNAC13 Physically Interacts with LcR1MYB1
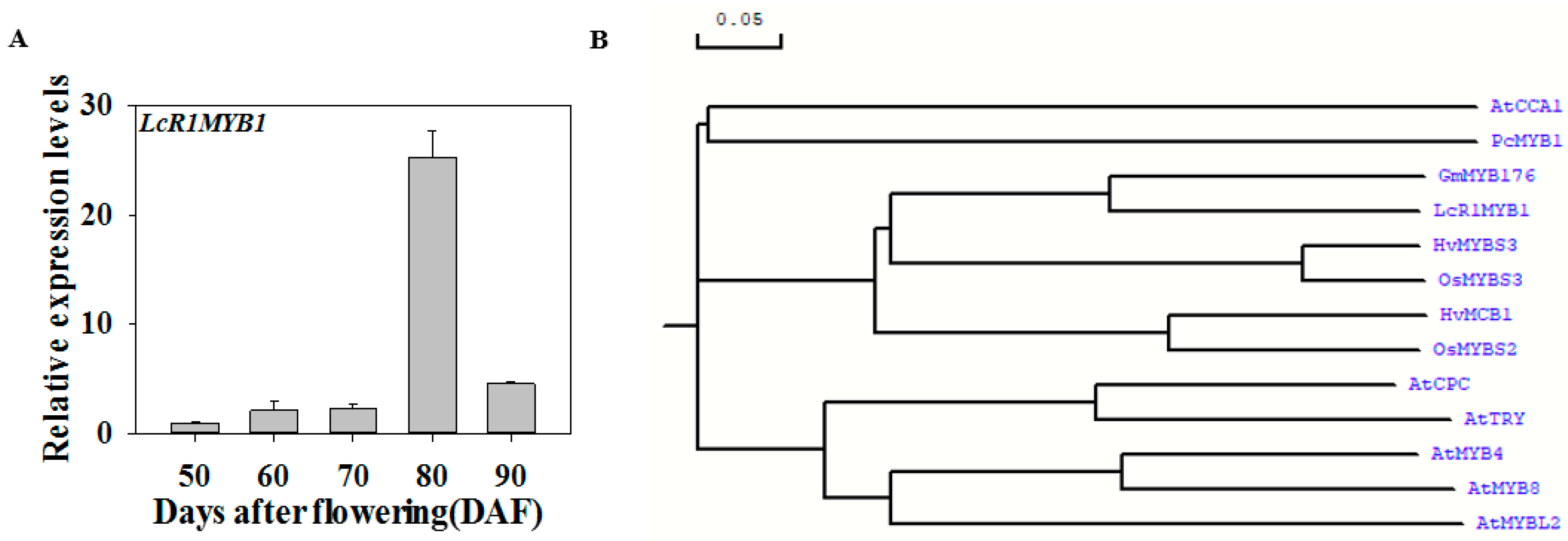
3.5. Expression Profiles of LcR1MYB1 during Litchi Fruit Ripening
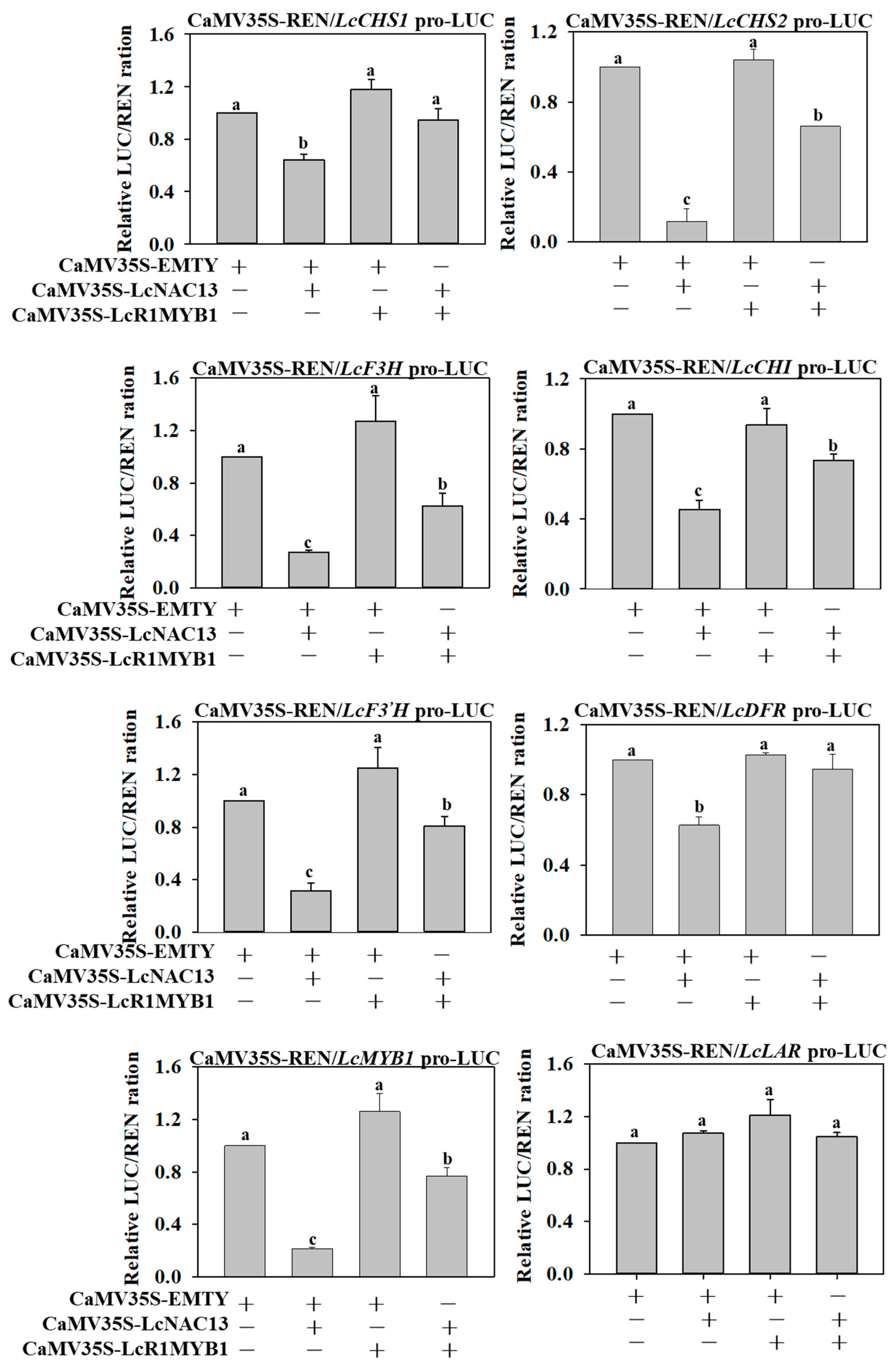
3.6. The Repressive Effect of LcNAC13 on Anthocyanin Biosynthesis-Related Genes is Reversed by LcR1MYB1 in Transient Expression Assays
3.7. Expression Patterns of LcNAC13-Targeted Genes are Correlated with the Change in Pericarp Color during Fruit Ripening
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.B.; Choi, G.; Park, Y.I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef] [PubMed]
- Barrett, D.M.; Beaulieu, J.C.; Shewfelt, R. Color, flavor, texture, and nutritional quality of fresh-cut fruits and vegetables: Desirable levels, instrumental and sensory measurement, and the effects of processing. Crit. Rev. Food Sci. Nutr. 2010, 50, 369–389. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Buer, C.S.; Imin, N.; Djordjevic, M.A. Flavonoids: New roles for old molecules. J. Integr. Plant Biol. 2010, 52, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Irving, L.J.; Jameson, P.E.; Davies, K.M. Light-induced vegetative anthocyanin pigmentation in Petunia. J. Exp. Bot. 2009, 60, 2191–2202. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Olsen, K.M.; Slimestad, R.; Lea, U.S.; Brede, C.; Lovdal, T.; Ruoff, P.; Verheul, M.; Lillo, C. Temperature and nitrogen effects on regulators and products of the flavonoid pathway: Experimental and kinetic model studies. Plant Cell Environ. 2009, 32, 286–299. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; De Stefano, R.; Schoonbeek, H.J.; Magusin, A.; Pagliarani, C.; Wellner, N.; Hill, L.; Orzaez, D.; Granell, A.; et al. Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr. Biol. 2013, 23, 1094–1100. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.X.; Lin, S.; Wen, L.R.; Jiang, Y.M.; Zhao, M.M.; Chen, F.; Prasad, K.N.; Duan, X.W.; Yang, B. Identification of a novel phenolic compound in litchi (Litchi chinensis Sonn.) pericarp and bioactivity evaluation. Food Chem. 2013, 136, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, R.; Vijayakumar, M.; Pushpangadan, P. Antioxidant approach to disease management and the role of ‘Rasayana’ herbs of Ayurveda. J. Ethnopharmacol. 2005, 99, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Khatun, H.; Rahman, A.; Biswas, M.; Islam, A.U. Water-soluble fraction of Abelmoschus esculentus L interacts with glucose and metformin hydrochloride and alters their absorption kinetics after coadministration in Rats. ISRN Pharm. 2011, 2011, 260537. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Springob, K.; Nakajima, J.; Yamazaki, M.; Saito, K. Recent advances in the biosynthesis and accumulation of anthocyanins. Nat. Prod. Rep. 2003, 20, 288–303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.B.; Abrahan, C.; Colquhoun, T.A.; Liu, C.J. A proteolytic regulator controlling chalcone synthase stability and flavonoid biosynthesis in Arabidopsis. Plant Cell 2017, 29, 1157–1174. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Avramenko, T.V.; Tsitsiashvili, G.S. Critical analysis of protein signaling networks involved in the regulation of plant secondary metabolism: Focus on anthocyanins. Crit. Rev. Biotechnol. 2017, 37, 685–700. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Wang, X.F.; Li, Y.Y.; Song, L.Q.; Zhao, L.L.; You, C.X.; Hao, Y.J. EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 2018. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Liu, C.H.; Yan, D.; Wen, X.H.; Liu, Y.L.; Wang, H.J.; Dai, J.Y.; Zhang, Y.J.; Liu, Y.F.; Zhou, B.; et al. MdHB1 down-regulation activates anthocyanin biosynthesis in the white-fleshed apple cultivar ‘Granny Smith’. J. Exp. Bot. 2017, 68, 1055–1069. [Google Scholar] [CrossRef] [PubMed]
- Morishita, T.; Kojima, Y.; Maruta, T.; Nishizawa-Yokoi, A.; Yabuta, Y.; Shigeoka, S. Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 2009, 50, 2210–2222. [Google Scholar] [CrossRef]
- Mahmood, K.; Xu, Z.; El-Kereamy, A.; Casaretto, J.A.; Rothstein, S.J. The Arabidopsis transcription factor ANAC032 represses anthocyanin biosynthesis in response to high sucrose and oxidative and abiotic stresses. Front. Plant Sci. 2016, 7, 1548. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Kui, L.W.; Wang, H.L.; Gu, C.; Dare, A.P.; Espley, R.V.; He, H.P.; Allan, A.C.; Han, Y.P. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015, 82, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.M.; Duan, X.W.; Joyce, D.; Zhang, Z.Q.; Li, J.R. Advances in understanding of enzymatic browning in harvested litchi fruit. Food Chem. 2004, 88, 443–446. [Google Scholar] [CrossRef]
- Lee, H.S.; Wicker, L. Anthocyanin pigments in the skin of lychee fruit. J. Food Sci. 1991, 56, 466–468. [Google Scholar] [CrossRef]
- Lai, B.; Li, X.J.; Hu, B.; Qin, Y.H.; Huang, X.M.; Wang, H.C.; Hu, G.B. LcMYB1 is a key determinant of differential anthocyanin accumulation among genotypes, tissues, developmental phases and ABA and light stimuli in Litchi chinensis. PLoS ONE 2014, 9, e86293. [Google Scholar] [CrossRef] [PubMed]
- Lai, B.; Du, L.N.; Liu, R.; Hu, B.; Su, W.B.; Qin, Y.H.; Zhao, J.T.; Wang, H.C.; Hu, G.B. Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in Nicotiana and Litchi chinensis during anthocyanin accumulation. Front. Plant Sci. 2016, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Lai, B.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Identification of MicroRNAs and their target genes related to the accumulation of anthocyanins in Litchi chinensis by high-throughput sequencing and degradome analysis. Front. Plant Sci. 2016, 7, 2059. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- Zhang, Z.Q.; Pang, X.Q.; Ji, Z.L.; Jiang, Y.M. Role of anthocyanin degradation in litchi pericarp browning. Food Chem. 2001, 75, 217–221. [Google Scholar] [CrossRef]
- Wang, H.C.; Hu, Z.Q.; Wang, Y.; Chen, H.B.; Huang, X.M. Phenolic compounds and the antioxidant activities in litchi pericarp: Difference among cultivars. Sci. Hortic. 2011, 129, 784–789. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Huber, D.J.; Qu, H.X.; Yun, Z.; Wang, H.; Huang, Z.H.; Huang, H.; Jiang, Y.M. Enzymatic browning and antioxidant activities in harvested litchi fruit as influenced by apple polyphenols. Food Chem. 2015, 171, 191–199. [Google Scholar] [CrossRef]
- Wan, C.Y.; Wilkins, T.A. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal. Biochem. 1994, 223, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.Y.; Chen, J.W.; Li, C.Q.; Chen, L.; Wu, J.Y.; Chen, J.Y.; Lu, W.J.; Li, J.G. Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep. 2011, 30, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Lai, B.; Hu, B.; Qin, Y.H.; Zhao, J.T.; Wang, H.C.; Hu, G.B. Transcriptomic analysis of Litchi chinensis pericarp during maturation with a focus on chlorophyll degradation and flavonoid biosynthesis. BMC Genom. 2015, 16, 225. [Google Scholar] [CrossRef]
- Jiang, G.; Yan, H.; Wu, F.; Zhang, D.; Zeng, W.; Qu, H.; Chen, F.; Tan, L.; Duan, X.; Jiang, Y. Litchi fruit LcNAC1 is a target of LcMYC2 and regulator of fruit senescence through its interaction with LcWRKY1. Plant Cell Physiol. 2017, 58, 1075–1089. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Chen, L.; Dhaubhadel, S. 14-3-3 proteins regulate the intracellular localization of the transcriptional activator GmMYB176 and affect isoflavonoid synthesis in soybean. Plant J. 2012, 71, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.X.; Derynck, M.R.; Li, X.Y.; Telmer, P.; Marsolais, F.; Dhaubhadel, S. A single-repeat MYB transcription factor, GmMYB176, regulates CHS8 gene expression and affects isoflavonoid biosynthesis in soybean. Plant J. 2010, 62, 1019–1034. [Google Scholar] [CrossRef]
- Zhang, Z.Q.; Pang, X.Q.; Yang, C.; Ji, Z.L.; Jiang, Y.M. Purification and structural analysis of anthocyanins from litchi pericarp. Food Chem. 2004, 84, 601–604. [Google Scholar] [CrossRef]
- He, X.J.; Mu, R.L.; Cao, W.H.; Zhang, Z.G.; Zhang, J.S.; Chen, S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005, 44, 903–916. [Google Scholar] [CrossRef]
- Hibara, K.; Takada, S.; Tasaka, M. CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 2003, 36, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.Z.; Ding, W.N.; Wu, Y.R.; Yu, J.; He, X.W.; Shou, H.X.; Wu, P. Overexpression of a NAC-domain protein promotes shoot branching in rice. New Phytol. 2007, 176, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, H.; Maruyama, K.; Takahashi, F.; Fujita, M.; Yoshida, T.; Nakashima, K.; Myouga, F.; Toyooka, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 2015, 84, 1114–1123. [Google Scholar] [CrossRef] [PubMed]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.S.; Penfold, C.A.; Jenkins, D.; et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef] [PubMed]
- Allan, A.C.; Espley, R.V. MYBs drive novel consumer traits in fruits and vegetables. Trends Plant Sci. 2018, 23, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Colinas, M.; Goossens, A. Combinatorial transcriptional control of plant specialized metabolism. Trends Plant Sci. 2018, 23, 324–336. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Feng, S.Q.; Wang, Y.L.; Yang, S.; Xu, Y.T.; Chen, X.S. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef]
- Schaart, J.G.; Dubos, C.; De La Fuente, I.R.; van Houwelingen, A.M.M.L.; de Vos, R.C.H.; Jonker, H.H.; Xu, W.J.; Routaboul, J.M.; Lepiniec, L.; Bovy, A.G. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria x ananassa) fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K.; Umemura, Y.; Ohme-Takagi, M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008, 55, 954–967. [Google Scholar] [CrossRef] [PubMed]
- Colanero, S.; Perata, P.; Gonzali, S. The atroviolacea Gene Encodes an R3-MYB Protein Repressing Anthocyanin Synthesis in Tomato Plants. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Gates, D.J.; Olson, B.J.S.C.; Clemente, T.E.; Smith, S.D. A novel R3 MYB transcriptional repressor associated with the loss of floral pigmentation in Iochroma. New Phytologist. 2018, 217, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Maier, A.; Hoecker, U. COP1/SPA ubiquitin ligase complexes repress anthocyanin accumulation under low light and high light conditions. Plant Signal. Behav. 2015, 10, e970440. [Google Scholar] [CrossRef] [PubMed]
- Maier, A.; Schrader, A.; Kokkelink, L.; Falke, C.; Welter, B.; Iniesto, E.; Rubio, V.; Uhrig, J.F.; Hulskamp, M.; Hoecker, U. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013, 74, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S.; et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yin, X.; Xiao, Y.; Zhang, Z.; Li, S.; Liu, X.; Zhang, B.; Yang, X.; Grierson, D.; Jiang, G.; et al. An ETHYLENE RESPONSE FACTOR-MYB transcription complex regulates furaneol biosynthesis by activating QUINONE OXIDOREDUCTASE expression in Strawberry. Plant. Physiol. 2018. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, G.; Li, Z.; Song, Y.; Zhu, H.; Lin, S.; Huang, R.; Jiang, Y.; Duan, X. LcNAC13 Physically Interacts with LcR1MYB1 to Coregulate Anthocyanin Biosynthesis-Related Genes during Litchi Fruit Ripening. Biomolecules 2019, 9, 135. https://doi.org/10.3390/biom9040135
Jiang G, Li Z, Song Y, Zhu H, Lin S, Huang R, Jiang Y, Duan X. LcNAC13 Physically Interacts with LcR1MYB1 to Coregulate Anthocyanin Biosynthesis-Related Genes during Litchi Fruit Ripening. Biomolecules. 2019; 9(4):135. https://doi.org/10.3390/biom9040135
Chicago/Turabian StyleJiang, Guoxiang, Zhiwei Li, Yunbo Song, Hong Zhu, Sen Lin, Riming Huang, Yueming Jiang, and Xuewu Duan. 2019. "LcNAC13 Physically Interacts with LcR1MYB1 to Coregulate Anthocyanin Biosynthesis-Related Genes during Litchi Fruit Ripening" Biomolecules 9, no. 4: 135. https://doi.org/10.3390/biom9040135
APA StyleJiang, G., Li, Z., Song, Y., Zhu, H., Lin, S., Huang, R., Jiang, Y., & Duan, X. (2019). LcNAC13 Physically Interacts with LcR1MYB1 to Coregulate Anthocyanin Biosynthesis-Related Genes during Litchi Fruit Ripening. Biomolecules, 9(4), 135. https://doi.org/10.3390/biom9040135








