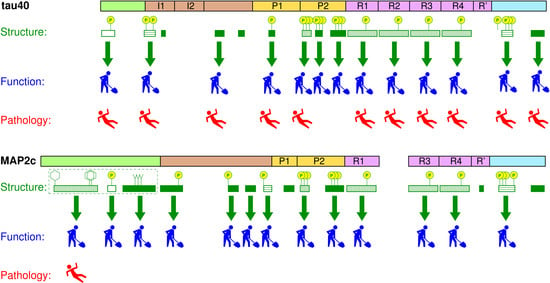
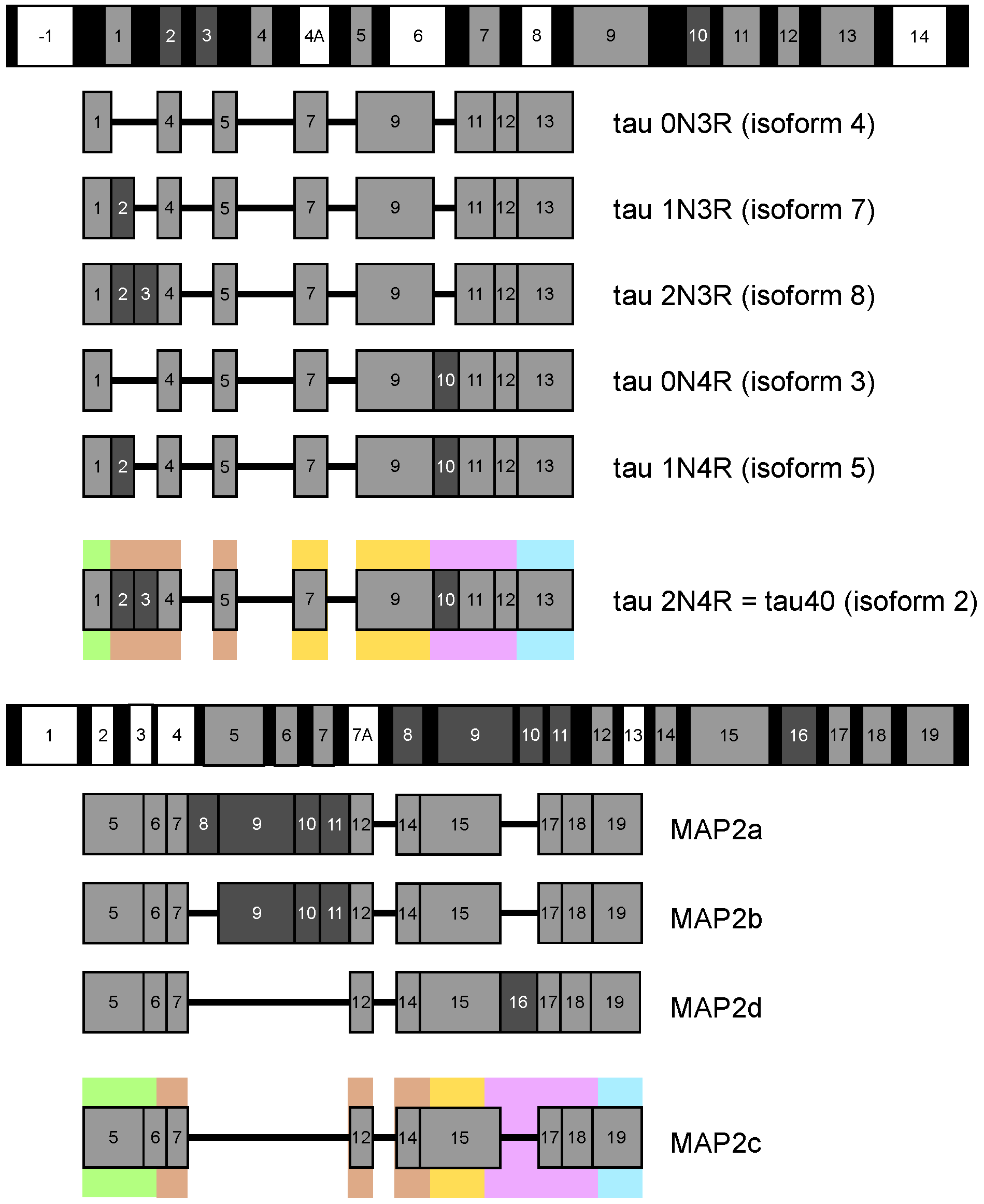
Structure and Functions of Microtubule Associated Proteins Tau and MAP2c: Similarities and Differences
Abstract
:1. Introduction
2. Measurement and Presentation of Transient Local Structures
3. N-Terminal Regions
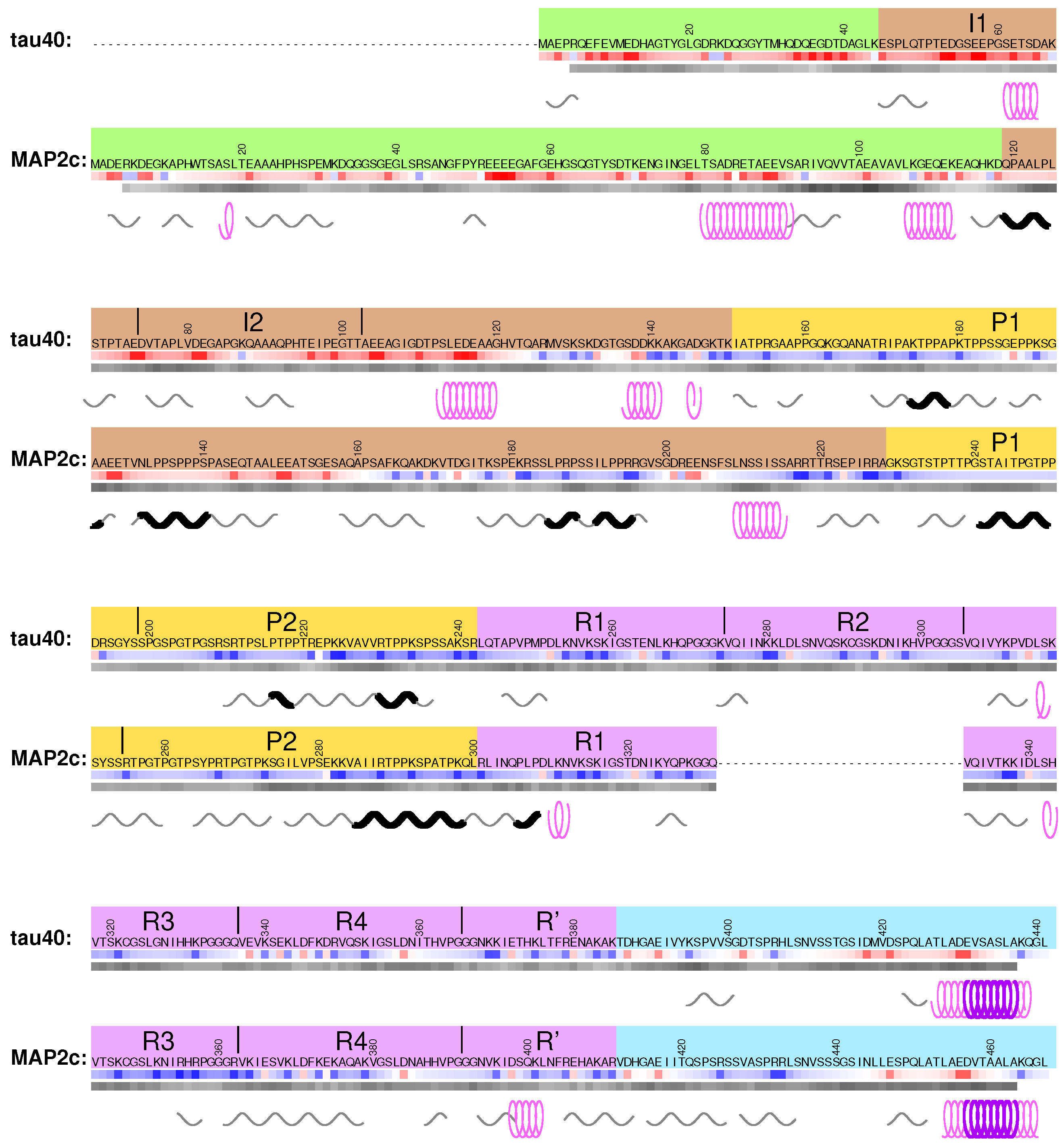
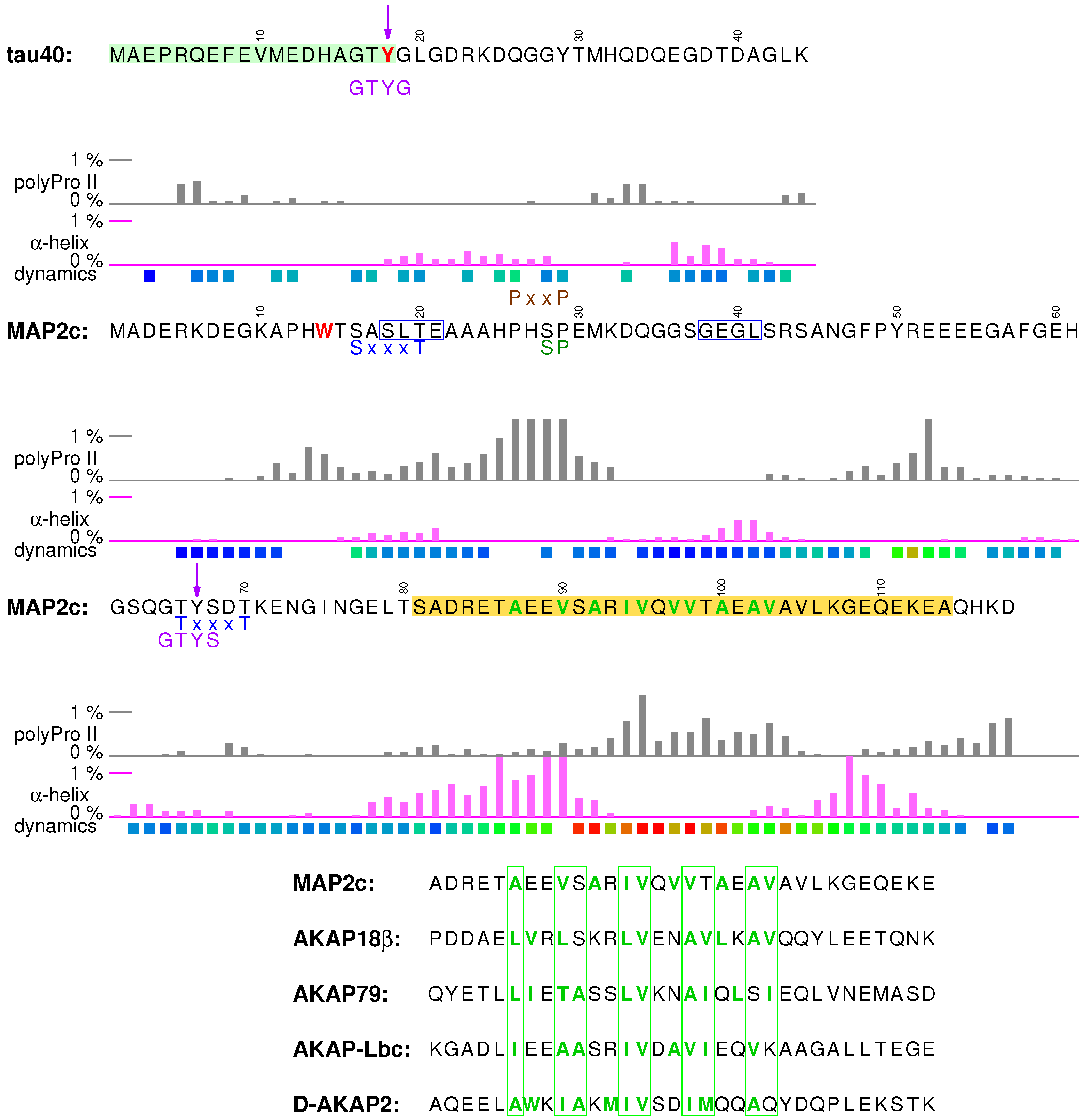
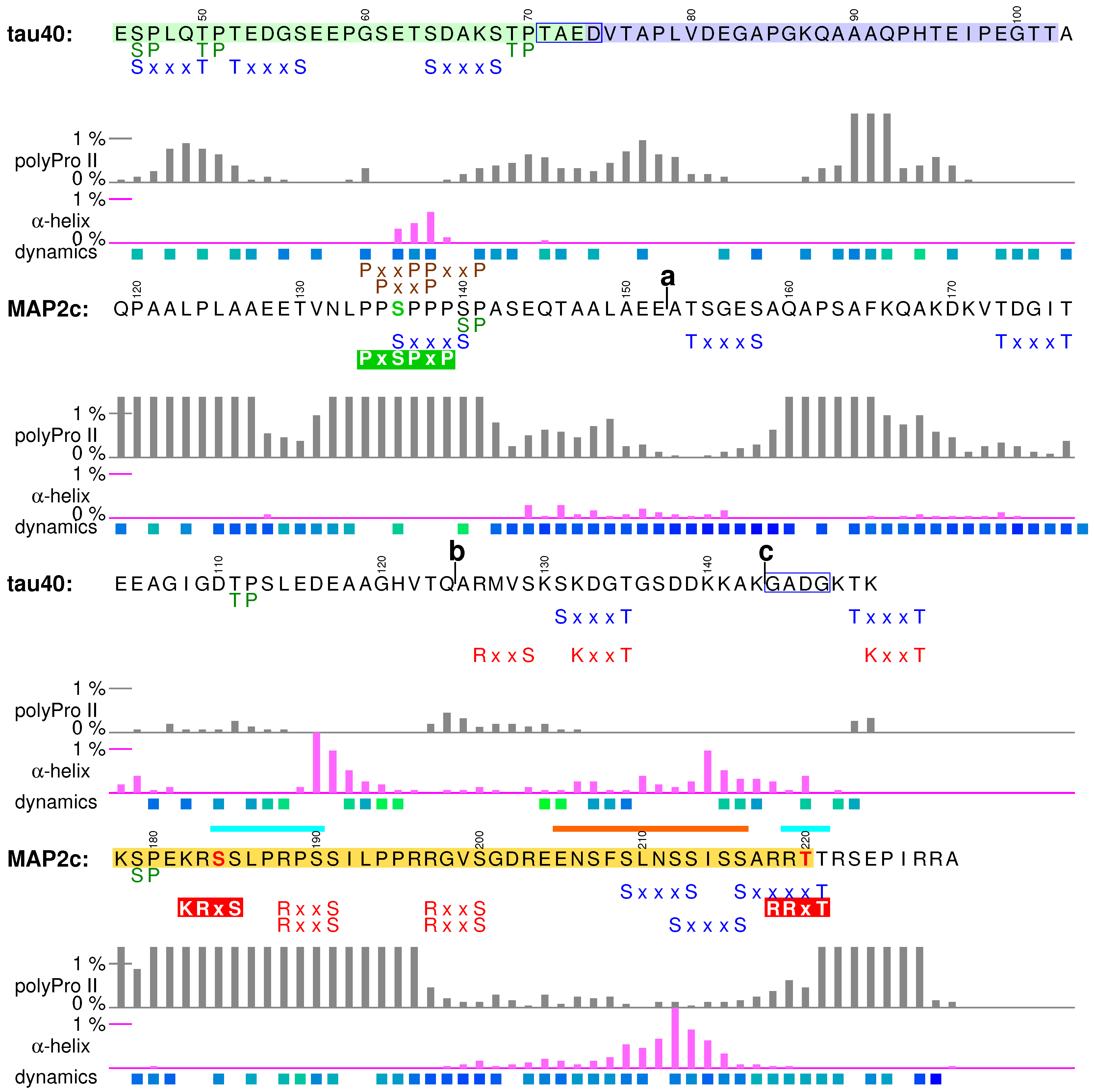
3.1. Structural Properties of N-Terminal Region of tau40
3.2. Phosphatase Activation by Tau N-Terminus and Axonal Transport
3.3. Interactions of Tau with Neuronal Membranes
3.4. Structural Properties of N-Terminal Region of MAP2c
3.5. Neurosteroid Binding to the N-Terminal Region of MAP2c
3.6. Binding Site for the RII Regulatory Subunit of PKA in the N-Terminal Region of MAP2c
3.7. Phosphorylation of MAP2c Tyr67 and SH2 Binding
3.8. Summary
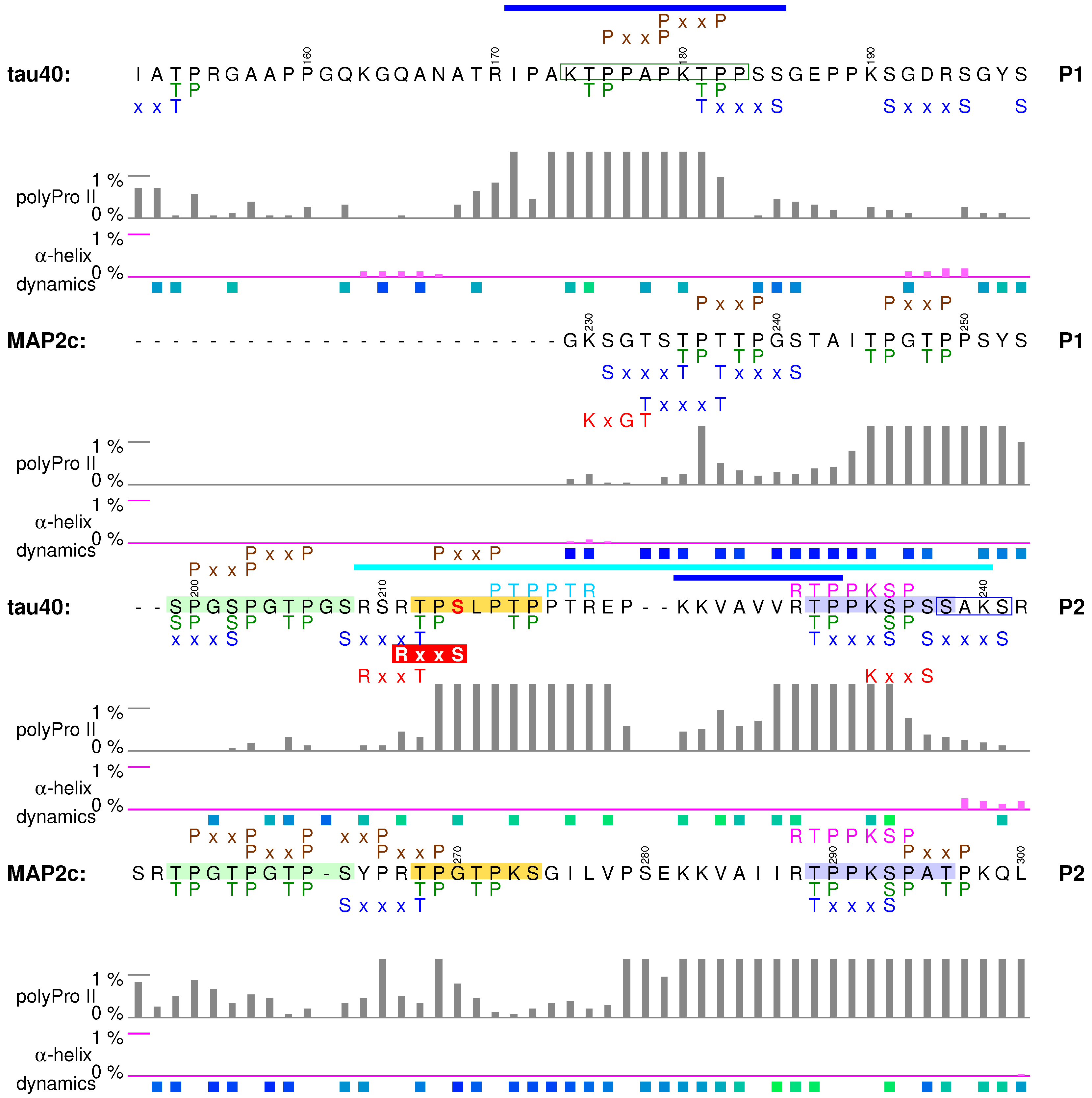
4. Variable Central Regions Preceding Proline-Rich Domains
4.1. Structural Properties of Variable Central Region of tau40
4.2. Regulatory Post-Translational Modifications: Phosphorylation of Tau Insert I1 and Truncation
4.3. Interactions of the Region Encoded by Tau Exon 4 with the Dynactin Complex
4.4. Structural Properties of Variable Central Region of MAP2c
4.5. Neural-Activity-Dependent Phosphorylation of MAP2c Epitope AP-18
4.6. Helical Motif of MAP2c Flanked by PKA Phosphorylation Sites and Involved in Interactions Interfering with MT Binding
4.7. Summary
5. Proline-Rich Domains
5.1. Structural Properties of Proline-Rich Domains of Tau and MAP2c
5.2. Tubulin-Binding Motif in Tau Region P1
5.3. Phosphorylation-Controlled Conformational Switch in Tau Epitope AT8
5.4. Formation of a Salt Bridge in Tau Epitope AT180 Interferes with MT Stabilization
5.5. Specific Phosphorylation of Ser214 by PKA and 14-3-3 Binding in the AT100 Epitope of Tau
5.6. Binding Sites for the SH3 Domains
5.7. Summary
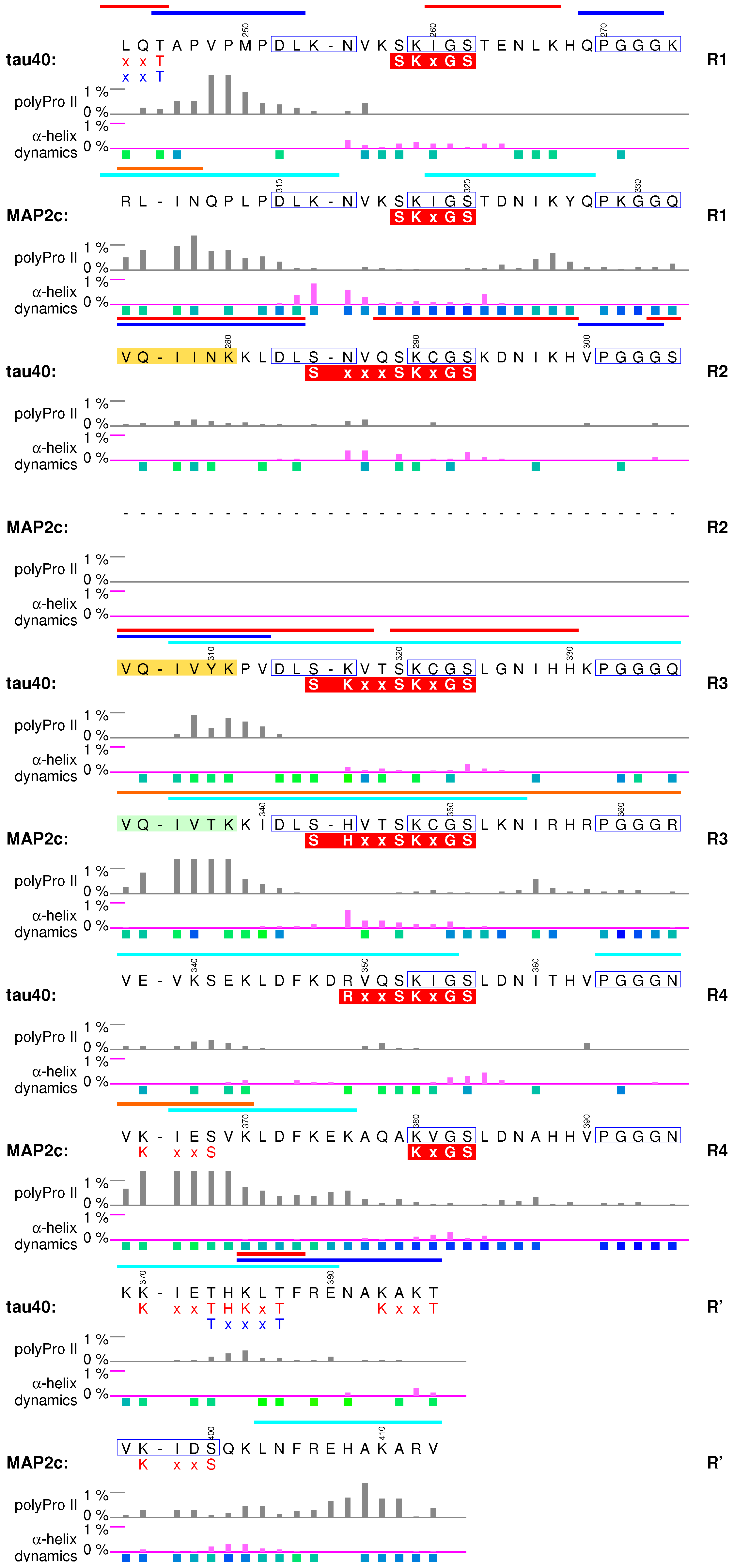
6. Microtubule-Binding Domains
6.1. Structural Properties of Microtubule-Binding Domains of tau40 and MAP2c
6.2. Interactions with Microtubules
6.3. Interactions with Actin and Other Proteins
6.4. Aggregation of tau40
6.5. Summary
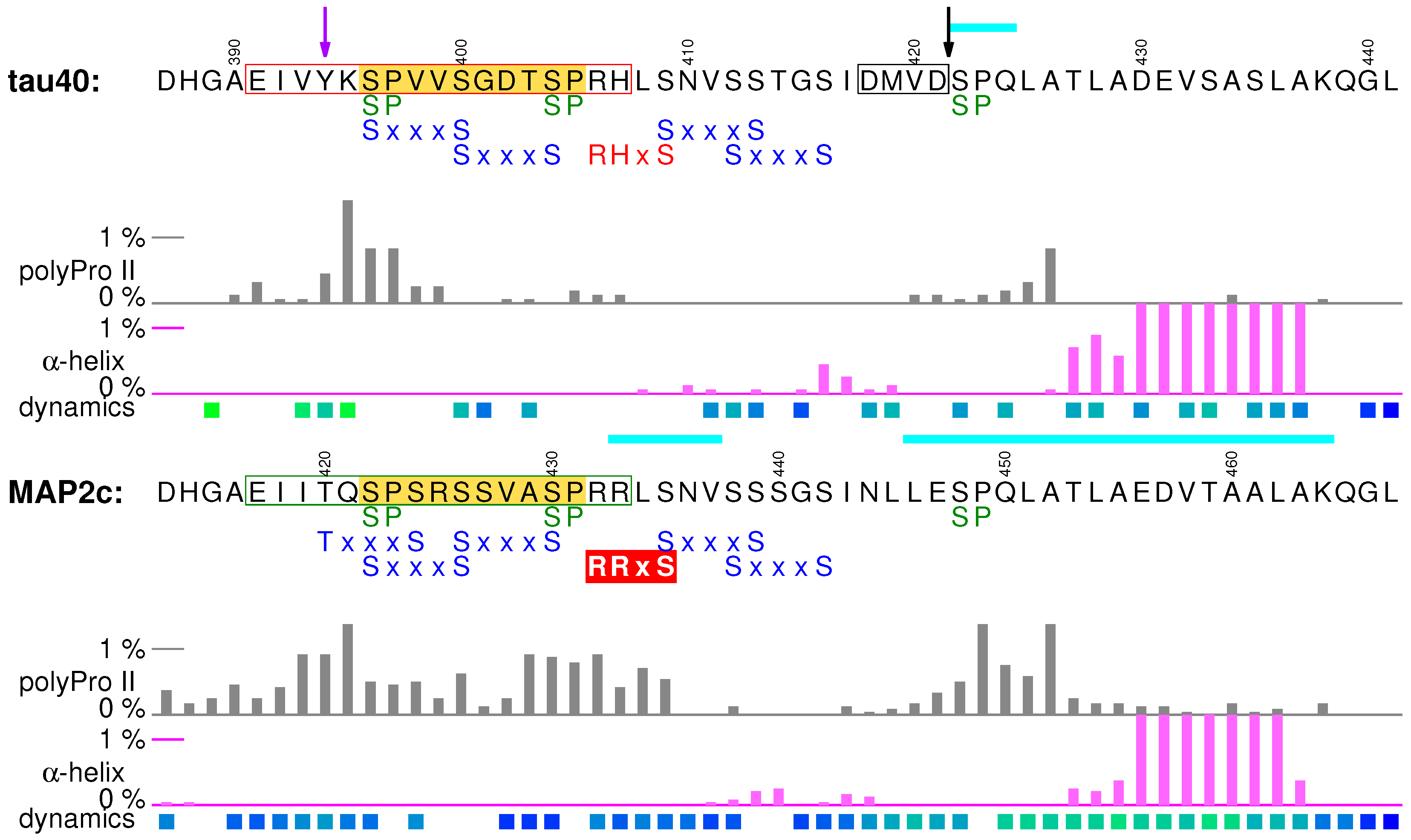
7. C-Terminal Regions
7.1. Structural Properties of C-Terminal Regions of tau40 and MAP2c
7.2. Muscarinic Receptor Activation and the PHF-1 Epitope of Tau
7.3. Rapid Phosphorylation at Ser435 and 14-3-3 Binding of MAP2c
7.4. Protective Role of the C-Terminal Helix
7.5. Summary
8. Global Structural Features of MAP2c and tau40
9. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
References
- Amos, L.A.; Schlieper, D. Microtubules and MAPs. Adv. Protein Chem. 2005, 71, 257–298. [Google Scholar] [PubMed]
- Dehmelt, L.; Halpain, S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 2004, 6, 204. [Google Scholar] [CrossRef]
- Arendt, T.; Stieler, J.T.; Holzer, M. Tau and tauopathies. Brain Res. Bull. 2016, 126, 238–292. [Google Scholar] [CrossRef] [PubMed]
- Caillet-Boudin, M.L.; Buée, L.; Sergeant, N.; Lefebvre, B. Regulation of human MAPT gene expression. Mol. Neurodegener. 2015, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, C.; Díaz-Nido, J.; Avila, J. Phosphorylation of microtubule-associated protein 2 (MAP2) and its relevance for the regulation of the neuronal cytoskeleton function. Prog. Neurobiol. 2000, 61, 133–168. [Google Scholar] [CrossRef]
- Kanai, Y.; Hirokawa, N. Sorting mechanisms of Tau and MAP2 in neurons: Suppressed axonal transit of MAP2 and locally regulated microtubule binding. Neuron 1995, 14, 421–432. [Google Scholar] [CrossRef]
- Chen, J.; Kanai, Y.; Cowan, N.J.; Hirokawa, N. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature 1992, 360, 674–677. [Google Scholar] [CrossRef]
- Rosenberg, K.J.; Ross, J.L.; Feinstein, H.E.; Feinstein, S.C.; Israelachvili, J. Complementary dimerization of microtubule-associated tau protein: Implications for microtubule bundling and tau-mediated pathogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 7445–7450. [Google Scholar] [CrossRef]
- Chung, P.J.; Choi, M.C.; Miller, H.P.; Feinstein, H.E.; Raviv, U.; Li, Y.; Wilson, L.; Feinstein, S.C.; Safinya, C.R. Direct force measurements reveal that protein Tau confers short-range attractions and isoform-dependent steric stabilization to microtubules. Proc. Natl. Acad. Sci. USA 2015, 112, E6416–E6425. [Google Scholar] [CrossRef]
- Cabrales Fontela, Y.; Kadavath, H.; Biernat, J.; Riedel, D.; Mandelkow, E.; Zweckstetter, M. Multivalent cross-linking of actin filaments and microtubules through the microtubule-associated protein Tau. Nat. Commun. 2017, 8, 1981. [Google Scholar] [CrossRef]
- Elie, A.; Prezel, E.; Guérin, C.; Denarier, E.; Ramirez-Rios, S.; Serre, L.; Andrieux, A.; Fourest-Lieuvin, A.; Blanchoin, L.; Arnal, I. Tau co-organizes dynamic microtubule and actin networks. Sci. Rep. 2015, 5, 9964. [Google Scholar] [CrossRef] [PubMed]
- Goode, B.L.; Chau, M.; Denis, P.E.; Feinstein, S.C. Structural and functional differences between 3-repeat and 4-repeat tau isoforms: Implications for normal tau function and the onset of neurodegenerative disease. J. Biol. Chem. 2000, 275, 38182–38189. [Google Scholar] [CrossRef] [PubMed]
- Harada, A.; Oguchi, K.; Okabe, S.; Kuno, J.; Terada, S.; Ohshima, T.; Sato-Yoshitake, R.; Takei, Y.; Noda, T.; Hirokawa, N. Altered microtubule organization in small-calibre axons of mice lacking tau protein. Nature 1994, 369, 488–491. [Google Scholar] [CrossRef]
- Teng, J.; Takei, Y.; Harada, A.; Nakata, T.; Chen, J.; Hirokawa, N. Synergistic effects of MAP2 and MAP1B knockout in neuronal migration, dendritic outgrowth, and microtubule organization. J. Cell Biol. 2001, 155, 65–76. [Google Scholar] [CrossRef]
- Takei, Y.; Teng, J.; Harada, A.; Hirokawa, N. Defects in Axonal Elongation and Neuronal Migration in Mice with Disrupted tau and map1b Genes. J. Cell Biol. 2000, 150, 989–1000. [Google Scholar] [CrossRef]
- Mukaetova-Ladinska, E.B.; Xuereb, J.H.; Garcia-Sierra, F.; Hurt, J.; Gertz, H.J.; Hills, R.; Brayne, C.; Huppert, F.A.; Paykel, E.S.; McGee, M.A.; et al. Lewy body variant of Alzheimer’s disease: Selective neocortical loss of t-SNARE proteins and loss of MAP2 and α-Synuclein in medial temporal lobe. Sci. World J. 2009, 9, 1463–1475. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, M.R.; Ilyin, S.; Plata-Salaman, C.R. Abnormal patterns of microtubule-associated protein-2 (MAP-2) immunolabeling in neuronal nuclei and Lewy bodies in Parkinson’s disease substantia nigra brain tissues. Neurosci. Lett. 2001, 306, 137–140. [Google Scholar] [CrossRef]
- Cabrera, J.R.; Lucas, J.J. MAP2 Splicing is Altered in Huntington’s Disease. Brain Pathol. 2016, 27, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, M.; Baulieu, E.E. 3β-Methoxy-pregnenolone (MAP4343) as an innovative therapeutic approach for depressive disorders. Proc. Natl. Acad. Sci. USA 2012, 109, 1713–1718. [Google Scholar] [CrossRef]
- Goedert, M. Tau filaments in neurodegenerative diseases. FEBS Lett. 2018, 592, 2383–2391. [Google Scholar] [CrossRef] [PubMed]
- Congdon, E.E.; Sigurdsson, E.M. Tau-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2018, 14, 399–415. [Google Scholar] [CrossRef]
- Novak, P.; Cehlar, O.; Skrabana, R.; Novak, M. Tau Conformation as a Target for Disease-Modifying Therapy: The Role of Truncation. J. Alzheimers Dis. 2018, 64, S535–S546. [Google Scholar] [CrossRef]
- Jadhav, S.; Avila, J.; Schöll, M.; Kovacs, G.G.; Kövari, E.; Skrabana, R.; Evans, L.D.; Kontsekova, E.; Malawska, B.; de Silva, R.; et al. A walk through tau therapeutic strategies. Acta Neuropathol. Commun. 2019, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.W.P.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H.W. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017, 547, 185–190. [Google Scholar] [CrossRef] [PubMed]
- DeTure, M.A.; Di Noto, L.; Purich, D.L. In vitro assembly of Alzheimer-like filaments: How a small cluster of charged residues in Tau and MAP2 controls filament morphology. J. Biol. Chem. 2002, 277, 34755–34759. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Soeda, Y.; Shinzaki, Y.; In, Y.; Tomoo, K.; Ihara, Y.; Miyasaka, T. Identification of key amino acids responsible for the distinct aggregation properties of microtubule-associated protein 2 and tau. J. Neurochem. 2015, 135, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mandelkow, E. Tau in physiology and pathology. Nat. Rev. Nerocsi. 2016, 17, 5–21. [Google Scholar] [CrossRef]
- Sánchez, C.; Pérez, M.; Avila, J. GSK3β-mediated phosphorylation of the microtubule-associated protein 2C (MAP2C) prevents microtubule bundling. Eur. J. Cell Biol. 2000, 79, 252–260. [Google Scholar] [CrossRef]
- Fischer, D.; Mukrasch, M.D.; Biernat, J.; Bibow, S.; Blackledge, M.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M. Conformational Changes Specific for Pseudophosphorylation at Serine 262 Selectively Impair Binding of Tau to Microtubules. Biochemistry 2009, 48, 10047–10055. [Google Scholar] [CrossRef] [PubMed]
- Schwalbe, M.; Biernat, J.; Bibow, S.; Ozenne, V.; Jensen, M.R.; Kadavath, H.; Blackledge, M.; Mandelkow, E.; Zweckstetter, M. Phosphorylation of human tau protein by microtubule affinity-regulating kinase 2. Biochemistry 2013, 52, 9068–9079. [Google Scholar] [CrossRef] [PubMed]
- Schwalbe, M.; Kadavath, H.; Biernat, J.; Ozenne, V.; Blackledge, M.; Mandelkow, E.; Zweckstetter, M. Structural Impact of Tau Phosphorylation at Threonine 231. Structure 2015, 23, 1448–1458. [Google Scholar] [CrossRef] [PubMed]
- Tholey, A.; Lindemann, A.; Kinzel, V.; Reed, J. Direct effects of phosphorylation on the preferred backbone conformation of peptides: A nuclear magnetic resonance study. Biophys. J. 1999, 76, 76–87. [Google Scholar] [CrossRef]
- Newberry, R.W.; Raines, R.T. The n→π* Interaction. Acc. Chem. Res. 2017, 50, 1838–1846. [Google Scholar] [CrossRef] [PubMed]
- Bielska, A.A.; Zondlo, N.J. Hyperphosphorylation of Tau Induces Local Polyproline II Helix. Biochemistry 2006, 45, 5527–5537. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Latypova, X.; Wilson, C.M.; Magnaudeix, A.; Perrin, M.L.; Yardin, C.; Terro, F. Tau protein kinases: Involvement in Alzheimer’s disease. Ageing Res. Rev. 2013, 12, 289–309. [Google Scholar] [CrossRef] [PubMed]
- Lebouvier, T.; Scales, T.M.E.; Williamson, R.; Noble, W.; Duyckaerts, C.; Hanger, D.P.; Reynolds, C.H.; Anderton, B.H.; Derkinderen, P. The Microtubule-Associated Protein Tau is Also Phosphorylated on Tyrosine. J. Alzheimers Dis. 2009, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, M.A.; Acker, C.M.; Davies, P. Tau phosphorylated at tyrosine 394 is found in Alzheimer’s disease tangles and can be a product of the abl-related kinase, Arg. J. Alzheimers Dis. 2010, 19, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Mukrasch, M.D.; Bibow, S.; Korukottu, J.; Jeganathan, S.; Biernat, J.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M. Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol. 2009, 7, e34. [Google Scholar] [CrossRef] [PubMed]
- Schwalbe, M.; Ozenne, V.; Bibow, S.; Jaremko, M.; Jaremko, L.; Gajda, M.; Jensen, M.R.; Biernat, J.; Becker, S.; Mandelkow, E.; et al. Predictive Atomic Resolution Descriptions of Intrinsically Disordered hTau40 and α-Synuclein in Solution from NMR and Small Angle Scattering. Structure 2014, 22, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Jansen, S.; Melková, K.; Trošanová, Z.; Hanáková, K.; Zachrdla, M.; Nováček, J.; Župa, E.; Zdráhal, Z.; Hritz, J.; Žídek, L. Quantitative mapping of microtubule-associated protein 2c (MAP2c) phosphorylation and regulatory protein 14-3-3ζ-binding sites reveals key differences between MAP2c and its homolog Tau. J. Biol. Chem. 2017, 292, 6715–6727. [Google Scholar] [CrossRef] [PubMed]
- Kadavath, H.; Jaremko, M.; Jaremko, Ł.; Biernat, J.; Mandelkow, E.; Zweckstetter, M. Folding of the Tau Protein on Microtubules. Angew. Chem. Int. Ed. 2015, 54, 10347–10351. [Google Scholar] [CrossRef] [PubMed]
- Kadavath, H.; Hofele, R.V.; Biernat, J.; Kumar, S.; Tepper, K.; Urlaub, H.; Mandelkow, E.; Zweckstetter, M. Tau stabilizes microtubules by binding at the interface between tubulin heterodimers. Proc. Natl. Acad. Sci. USA 2015, 112, 7501–7506. [Google Scholar] [CrossRef]
- Kellogg, E.H.; Hejab, N.M.A.; Poepsel, S.; Downing, K.H.; DiMaio, F.; Nogales, E. Near-atomic model of microtubule-tau interactions. Science 2018, 360, 1242–1246. [Google Scholar] [CrossRef] [PubMed]
- Bibow, S.; Mukrasch, M.D.; Chinnathambi, S.; Biernat, J.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M. The dynamic structure of filamentous Tau. Angew. Chem. Int. Ed. 2011, 50, 11520–11524. [Google Scholar] [CrossRef] [PubMed]
- Sündermann, F.; Fernandez, M.P.; Morgan, R.O. An evolutionary roadmap to the microtubule-associated protein MAP Tau. BMC Genom. 2016, 17, 264. [Google Scholar] [CrossRef] [PubMed]
- Smet, C.; Leroy, A.; Sillen, A.; Wieruszeski, J.M.; Landrieu, I.; Lippens, G. Accepting its Random Coil Nature Allows a Partial NMR Assignment of the Neuronal Tau Protein. ChemBioChem 2004, 5, 1639–1646. [Google Scholar] [CrossRef]
- Lippens, G.; Wieruszeski, J.M.; Leroy, A.; Smet, C.; Sillen, A.; Buée, L.; Landrieu, I. Proline-Directed Random-Coil Chemical Shift Values as a Tool for the NMR Assignment of the Tau Phosphorylation Sites. ChemBioChem 2004, 5, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Mukrasch, M.D.; Biernat, J.; von Bergen, M.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M. Sites of tau important for aggregation populate β-structure and bind to microtubules and polyanions. J. Biol. Chem. 2005, 280, 24978–24986. [Google Scholar] [CrossRef]
- Mukrasch, M.D.; von Bergen, M.; Biernat, J.; Fischer, D.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M. The “jaws” of the tau-microtubule interaction. J. Biol. Chem. 2007, 282, 12230–12239. [Google Scholar] [CrossRef]
- Verdegem, D.; Dijkstra, K.; Hanoulle, X.; Lippens, G. Graphical interpretation of Boolean operators for protein NMR assignments. J. Biomol. NMR 2008, 42, 11–21. [Google Scholar] [CrossRef]
- Sibille, N.; Hanoulle, X.; Fanny, B.; Dries, V.; Isabelle, L.; Jean-Michel, W.; Guy, L. Selective backbone labelling of ILV methyl labelled proteins. J. Biomol. NMR 2009, 43, 219–227. [Google Scholar] [CrossRef]
- Lopez, J.; Ahuja, P.; Gerard, M.; Wieruszeski, J.M.; Lippens, G. A new strategy for sequential assignment of intrinsically unstructured proteins based on 15N single isotope labelling. J. Magn. Reson. 2013, 236, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, R.L.; Dürr, U.H.N.; Bibow, S.; Biernat, J.; Mandelkow, E.; Zweckstetter, M. Automatic Assignment of the Intrinsically Disordered Protein Tau with 441-Residues. J. Am. Chem. Soc. 2010, 132, 11906–11907. [Google Scholar] [CrossRef] [PubMed]
- Harbison, N.W.; Bhattacharya, S.; Eliezer, D. Assigning Backbone NMR Resonances for Full Length Tau Isoforms: Efficient Compromise between Manual Assignments and Reduced Dimensionality. PLoS ONE 2012, 7, e34679. [Google Scholar] [CrossRef] [PubMed]
- Nováček, J.; Janda, L.; Dopitová, R.; Žídek, L.; Sklenář, V. Efficient protocol for backbone and side-chain assignments of large, intrinsically disordered proteins: Transient secondary structure analysis of 49.2 kDa microtubule associated protein 2c. J. Biomol. NMR 2013, 56, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Nodet, G.; Salmon, L.; Ozenne, V.; Meier, S.; Jensen, M.R.; Blackledge, M. Quantitative description of backbone conformational sampling of unfolded proteins at amino acid resolution from NMR residual dipolar couplings. J. Am. Chem. Soc. 2009, 131, 17908–17918. [Google Scholar] [CrossRef]
- Melková, K.; Zapletal, V.; Jansen, S.; Nomilner, E.; Zachrdla, M.; Hritz, J.; Nováček, J.; Zweckstetter, M.; Jensen, M.R.; Blackledge, M.; et al. Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics. J. Biol. Chem. 2018, 293, 13297–13309. [Google Scholar] [CrossRef] [PubMed]
- Kovacech, B.; Skrabana, R.; Novak, M. Transition of Tau Protein from Disordered to Misordered in Alzheimer’s Disease. Neurodegener. Dis. 2010, 7, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Kontsekova, E.; Zilka, N.; Kovacech, B.; Skrabana, R.; Novak, M. Identification of structural determinants on tau protein essential for its pathological function: Novel therapeutic target for tau immunotherapy in Alzheimer’s disease. Alzheimers Res. Ther. 2014, 6, 45. [Google Scholar] [CrossRef] [PubMed]
- de Brevern, A.G. Extension of the classical classification of β-turns. Sci. Rep. 2016, 6, 33191. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Jeganathan, S.; von Bergen, M.; Brutlach, H.; Steinhoff, H.J.; Mandelkow, E. Global hairpin folding of tau in solution. Biochemistry 2006, 45, 2283–2293. [Google Scholar] [CrossRef] [PubMed]
- LaPointe, N.E.; Morfini, G.; Pigino, G.; Gaisina, I.N.; Kozikowski, A.P.; Binder, L.I.; Brady, S.T. The amino terminus of tau inhibits kinesin-dependent axonal transport: Implications for filament toxicity. J. Neurosci. Res. 2009, 87, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Kanaan, N.M.; Morfini, G.A.; LaPointe, N.E.; Pigino, G.F.; Patterson, K.R.; Song, Y.; Andreadis, A.; Fu, Y.; Brady, S.T.; Binder, L.I. Pathogenic Forms of Tau Inhibit Kinesin-Dependent Axonal Transport through a Mechanism Involving Activation of Axonal Phosphotransferases. J. Neurosci. 2011, 31, 9858–9868. [Google Scholar] [CrossRef]
- Liao, H.; Li, Y.; Brautigan, D.L.; Gundersen, G.G. Protein phosphatase 1 is targeted to microtubules by the microtubule- associated protein tau. J. Biol. Chem. 1998, 273, 21901–21908. [Google Scholar] [CrossRef]
- Dente, L.; Vetriani, C.; Zucconi, A.; Pelicci, G.; Lanfrancone, L.; Pelicci, P.; Cesareni, G. Modified phage peptide libraries as a tool to study specificity of phosphorylation and recognition of tyrosine containing peptides. J. Mol. Biol. 1997, 269, 694–703. [Google Scholar] [CrossRef]
- Lee, G.; Thangavel, R.; Sharma, V.M.; Litersky, J.M.; Bhaskar, K.; Fang, S.M.; Do, L.H.; Andreadis, A.; Van Hoesen, G.; Ksiezak-Reding, H. Phosphorylation of Tau by Fyn: Implications for Alzheimers Disease. J. Neurosci. 2004, 24, 2304–2312. [Google Scholar] [CrossRef]
- Stern, J.L.; Lessard, D.V.; Hoeprich, G.J.; Morfini, G.A.; Berger, C.L.; Drubin, D.G. Phosphoregulation of Tau modulates inhibition of kinesin-1 motility. Mol. Biol. Cell 2017, 28, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Schroer, T.A. Dynactin. Annu. Rev. Cell Dev. Biol. 2004, 20, 759–779. [Google Scholar] [CrossRef]
- Carter, A.P.; Diamant, A.G.; Urnavicius, L. How dynein and dynactin transport cargos: A structural perspective. Curr. Opin. Struct. Biol. 2016, 37, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Magnani, E.; Fan, J.; Gasparini, L.; Golding, M.; Williams, M.; Schiavo, G.; Goedert, M.; Amos, L.A.; Spillantini, M.G. Interaction of tau protein with the dynactin complex. EMBO J. 2007, 26, 4546–4554. [Google Scholar] [CrossRef]
- Brandt, R.; Léger, J.; Lee, G. Interaction of tau with the neural plasma membrane mediated by taus amino-terminal projection domain. J. Cell Biol. 1995, 131, 1327–1340. [Google Scholar] [CrossRef]
- Usardi, A.; Pooler, A.M.; Seereeram, A.; Reynolds, C.H.; Derkinderen, P.; Anderton, B.; Hanger, D.P.; Noble, W.; Williamson, R. Tyrosine phosphorylation of tau regulates its interactions with Fyn SH2 domains, but not SH3 domains, altering the cellular localization of tau. FEBS J. 2011, 278, 2927–2937. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, P.; Lee, G.; Sjoberg, M.; MacCioni, R.B. Tau phosphorylation by cdk5 and Fyn in response to amyloid peptide Aβ25–35: Involvement of lipid rafts. J. Alzheimers Dis. 2009, 16, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Baulieu, E.E. Neurosteroids: Of the Nervous System, By the Nervous System, For the Nervous System. Recent Progr. Horm. Res. 1997, 52, 1–32. [Google Scholar] [PubMed]
- Fontaine-Lenoir, V.; Chambraud, B.; Fellous, A.; David, S.; Duchossoy, Y.; Baulieu, E.E.; Robel, P. Microtubule-associated protein 2 (MAP2) is a neurosteroid receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 4711–4716. [Google Scholar] [CrossRef] [PubMed]
- Laurine, E.; Lafitte, D.; Grégoire, C.; Sérée, E.; Loret, E.; Douillard, S.; Michel, B.; Briand, C.; Verdier, J.M. Specific binding of dehydroepiandrosterone to the N terminus of the microtubule-associated protein MAP2. J. Biol. Chem. 2003, 278, 29979–29986. [Google Scholar] [CrossRef] [PubMed]
- Mizota, K.; Ueda, H. N-terminus of MAP2C as a neurosteroid-binding site. NeuroReport 2008, 19, 1529–1533. [Google Scholar] [CrossRef] [PubMed]
- Götz, F.; Roske, Y.; Schulz, M.S.; Autenrieth, K.; Bertinetti, D.; Faelber, K.; Zühlke, K.; Kreuchwig, A.; Kennedy, E.J.; Krause, G.; et al. AKAP18:PKA-RIIα structure reveals crucial anchor points for recognition of regulatory subunits of PKA. Biochem. J. 2016, 473, 1881–1894. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Leon, S.P.; Bresnick, A.; Backer, J.M.; Shafit-Zagardo, B. Fyn phosphorylates human MAP-2c on tyrosine 67. J. Biol. Chem. 2005, 280, 1962–1970. [Google Scholar] [CrossRef] [PubMed]
- Majumder, P.; Roy, K.; Singh, B.K.; Jana, N.R.; Mukhopadhyay, D. Cellular levels of Grb2 and cytoskeleton stability are correlated in a neurodegenerative scenario. Dis. Models Mech. 2017, 10, 655–669. [Google Scholar] [CrossRef]
- Illenberger, S.; Zheng-Fischhöfer, Q.; Preuss, U.; Stamer, K.; Baumann, K.; Trinczek, B.; Biernat, J.; Godemann, R.; Mandelkow, E.M.; Mandelkow, E. The endogenous and cell cycle-dependent phosphorylation of tau protein in living cells: Implications for Alzheimer’s disease. Mol. Biol. Cell 1998, 9, 1495–1512. [Google Scholar] [CrossRef] [PubMed]
- Hanger, D.P.; Byers, H.L.; Wray, S.; Leung, K.Y.; Saxton, M.J.; Seereeram, A.; Reynolds, C.H.; Ward, M.A.; Anderton, B.H. Novel phosphorylation sites in Tau from Alzheimer brain support a role for casein kinase 1 in disease pathogenesis. J. Biol. Chem. 2007, 282, 23645–23654. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Prabakaran, S.; Cantrelle, F.X.; Chambraud, B.; Gunawardena, J.; Lippens, X.G.; Landrieu, I. Characterization of neuronal tau protein as a target of extracellular signal-regulated kinase. J. Biol. Chem. 2016, 291, 7742–7753. [Google Scholar] [CrossRef] [PubMed]
- Feijoo, C.; Campbell, D.G.; Jakes, R.; Goedert, M.; Cuenda, A. Evidence that phosphorylation of the microtubule-associated protein Tau by SAPK4/p38δ at Thr50 promotes microtubule assembly. J. Cell Sci. 2005, 118, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Wray, S.; Saxton, M.; Anderton, B.; Hanger, D. Direct analysis of tau from PSP brain identifies new phosphorylation sites and a major fragment of N-terminally cleaved tau containing four microtubule-binding repeats. J. Neurochem. 2008, 105, 2343–2352. [Google Scholar] [CrossRef]
- Derisbourg, M.; Leghay, C.; Chiappetta, G.; Fernandez-Gomez, F.J.; Laurent, C.; Demeyer, D.; Carrier, S.; Buée-Scherrer, V.; Blum, D.; Vinh, J.; et al. Role of the Tau N-terminal region in microtubule stabilization revealed by new endogenous truncated forms. Sci. Rep. 2015, 5, 9659. [Google Scholar] [CrossRef] [PubMed]
- Zilka, N.; Kovacech, B.; Barath, P.; Kontsekova, E.; Novák, M. The self-perpetuating tau truncation circle. Biochem. Soc. Trans. 2012, 40, 681–686. [Google Scholar] [CrossRef]
- Skrabana, R.; Kovacech, B.; Filipcik, P.; Zilka, N.; Jadhav, S.; Smolek, T.; Kontsekova, E.; Novak, M.; Deli, M. Neuronal Expression of Truncated Tau Efficiently Promotes Neurodegeneration in Animal Models: Pitfalls of Toxic Oligomer Analysis. J. Alzheimers Dis. 2017, 58, 1017–1025. [Google Scholar] [CrossRef]
- Berling, B.; Wille, H.; Roll, B.; Mandelkow, E.M.; Garner, C.; Mandelkow, E. Phosphorylation of microtubule-associated proteins MAP2a,b and MAP2c at Ser136 by proline-directed kinases in vivo and in vitro. Eur. J. Cell Biol. 1994, 64, 120–130. [Google Scholar]
- Philpot, B.D.; Lim, J.H.; Halpain, S.; Brunjes, P.C. Experience-Dependent Modifications in MAP2 Phosphorylation in Rat Olfactory Bulb. J. Neurosci. 1997, 17, 9596–9604. [Google Scholar] [CrossRef] [PubMed]
- Woolf, N.; Zinnerman, M.; Johnson, G. Hippocampal microtubule-associated protein-2 alterations with contextual memory. Brain Res. 1999, 821, 241–249. [Google Scholar] [CrossRef]
- Tie, L.; Zhang, J.Z.; Lin, Y.H.; Su, T.H.; Li, Y.H.; Wu, H.L.; Zhang, Y.Y.; Yu, H.M.; Li, X.J. Epinephrine increases phosphorylation of MAP-2c in rat pheochromocytoma cells (PC12 Cells) via a protein kinase C- and mitogen activated protein kinase-dependent mechanism. J. Proteome. Res. 2008, 7, 1704–1711. [Google Scholar] [CrossRef]
- Alexa, A.; Schmidt, G.; Tompa, P.; Ogueta, S.; Vázquez, J.; Kulcsár, P.; Kovács, J.; Dombrádi, V.; Friedrich, P. The phosphorylation state of threonine-220, a uniquely phosphatase-sensitive protein kinase A site in microtubule-associated protein MAP2c, regulates microtubule binding and stability. Biochemistry 1992, 41, 12427–12435. [Google Scholar] [CrossRef]
- Joo, Y.; Schumacher, B.; Landrieu, I.; Bartel, M.; Smet-Nocca, C.; Jang, A.; Choi, H.S.; Jeon, N.L.; Chang, K.A.; Kim, H.S.; et al. Involvement of 14-3-3 in tubulin instability and impaired axon development is mediated by Tau. FASEB J. 2015, 29, 4133–4144. [Google Scholar] [CrossRef] [PubMed]
- Valencia, R.; Walko, G.; Janda, L.; Novaček, J.; Mihailovska, E.; Reipert, S.; Andrä-Marobela, K.; Wiche, G. Intermediate filament-associated cytolinker plectin 1c destabilizes microtubules in keratinocytes. Mol. Biol. Cell 2013, 24, 768–784. [Google Scholar] [CrossRef] [PubMed]
- He, H.J.; Wang, X.S.; Pan, R.; Wang, D.L.; Liu, M.N.; He, R.Q. The proline-rich domain of tau plays a role in interactions with actin. BMC Cell Biol. 2009, 10, 81. [Google Scholar] [CrossRef]
- Gohar, M.; Yang, W.; Strong, W.; Volkening, K.; Leystra-Lantz, C.; Strong, M.J. Tau phosphorylation at threonine-175 leads to fibril formation and enhanced cell death: Implications for amyotrophic lateral sclerosis with cognitive impairment. J. Neurochem. 2009, 108, 634–643. [Google Scholar] [CrossRef]
- Moszczynski, A.J.; Strong, W.; Xu, K.; McKee, A.; Brown, A.; Strong, M.J. Pathologic Thr175 tau phosphorylation in CTE and CTE with ALS. Neurology 2018, 90, e380–e387. [Google Scholar] [CrossRef]
- Moszczynski, A.J.; Gohar, M.; Volkening, K.; Leystra-Lantz, C.; Strong, W.; Strong, M.J. Thr175-phosphorylated tau induces pathologic fibril formation via GSK3β-mediated phosphorylation of Thr231 in vitro. Neurobiol. Aging 2015, 36, 1590–1599. [Google Scholar] [CrossRef]
- Gandhi, N.S.; Landrieu, I.; Byrne, C.; Kucic, P.; Cantrelle, F.X.; Wieruszeski, J.M.; L, M.R.; Jacquot, Y.; Lippens, G. A Phosphorylation-Induced Turn Defines the Alzheimer’s Disease AT8 Antibody Epitope on the Tau Protein. Angew. Chem. Int. Ed. 2015, 54, 6819–6823. [Google Scholar] [CrossRef] [PubMed]
- Ittner, A.; Chua, S.W.; Bertz, J.; Volkerling, A.; van der Hoven, J.; Gladbach, A.; Przybyla, M.; Bi, M.; van Hummel, A.; Stevens, C.H.; et al. Site-specific phosphorylation of tau inhibits amyloid-β toxicity in Alzheimer’s mice. Science 2016, 354, 904–908. [Google Scholar] [CrossRef]
- Despres, C.; Byrne, C.; Qi, H.; Cantrelle, F.X.; Huvent, I.; Chambraud, B.; Baulieu, E.E.; Jacquot, Y.; Landrieu, I.; Lippens, G.; et al. Identification of the Tau phosphorylation pattern that drives its aggregation. Proc. Natl. Acad. Sci. USA 2017, 114, 9080–9085. [Google Scholar] [CrossRef] [PubMed]
- Malia, T.J.; Teplyakov, A.; Ernst, R.; Wu, S.J.; Lacy, E.R.; Liu, X.; Vandermeeren, M.; Mercken, M.; Luo, J.; Sweet, R.W.; et al. Epitope mapping and structural basis for the recognition of phosphorylated tau by the anti-tau antibody AT8. Proteins Struct. Funct. Bioinf. 2016, 84, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, M.; Hashiguchi, T. Chapter Four—Kinase-Kinase Interaction and Modulation of Tau Phosphorylation. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Academic Press: Cambridge, MA, USA, 2013; Volume 300, pp. 121–160. [Google Scholar]
- Yang, P.H.; Zhu, J.X.; Huang, Y.D.; Zhang, X.Y.; Lei, P.; Bush, A.; Xiang, Q.; Su, Z.; Zhang, Q.H. Human Basic Fibroblast Growth Factor Inhibits Tau Phosphorylation via the PI3K/Akt-GSK3β Signaling Pathway in a 6-Hydroxydopamine-Induced Model of Parkinson’s Disease. Neurodegener. Dis. 2016, 16, 357–369. [Google Scholar] [CrossRef]
- Amniai, L.; Barbier, P.; Sillen, A.; Wieruszeski, J.M.; Peyrot, V.; Lippens, G.; Landrieu, I. Alzheimer disease specific phosphoepitopes of Tau interfere with assembly of tubulin but not binding to microtubules. FASEB J. 2009, 23, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Komulainen, E.; Zdrojewska, J.; Freemantle, E.; Mohammad, H.; Kulesskaya, N.; Deshpande, P.; Marchisella, F.; Mysore, R.; Hollos, P.; Michelsen, K.A.; et al. JNK1 controls dendritic field size in L2/3 and L5 of the motor cortex, constrains soma size, and influences fine motor coordination. Front. Cell. Neurosci. 2014, 8, 272. [Google Scholar] [CrossRef]
- Reynolds, C.H.; Garwood, C.J.; Wray, S.; Price, C.; Kellie, S.; Perera, T.; Zvelebil, M.; Yang, A.; Sheppard, P.W.; Varndell, I.M.; et al. Phosphorylation regulates tau interactions with Src homology 3 domains of phosphatidylinositol 3-kinase, phospholipase Cγ1, Grb2, and Src family kinases. J. Biol. Chem. 2008, 283, 18177–18186. [Google Scholar] [CrossRef]
- Yoshida, H.; Goedert, M. Sequential phosphorylation of tau protein by cAMP-dependent protein kinase and SAPK4/p38delta or JNK2 in the presence of heparin generates the AT100 epitope. J. Neurochem. 2006, 99, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Landrieu, I.; Lacosse, L.; Leroy, A.; Wieruszeski, J.M.; Trivelli, X.; Sillen, A.; Sibille, N.; Schwalbe, H.; Saxena, K.; Langer, T.; et al. NMR analysis of a Tau phosphorylation pattern. J. Am. Chem. Soc. 2006, 128, 3575–3583. [Google Scholar] [CrossRef]
- Sadik, G.; Tanaka, T.; Kato, K.; Yamamori, H.; Nessa, B.N.; Morihara, T.; Takeda, M. Phosphorylation of tau at Ser214 mediates its interaction with 14-3-3 protein: Implications for the mechanism of tau aggregation. J. Neurochem. 2009, 108, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Sluchanko, N.N.; Seit-Nebi, A.S.; Gusev, N.B. Effect of phosphorylation on interaction of human tau protein with 14-3-3zeta. Biochem. Biophys. Res. Commun. 2009, 379, 990–994. [Google Scholar] [CrossRef] [PubMed]
- Sluchanko, N.N.; Seit-Nebi, A.S.; Gusev, N.B. Phosphorylation of more than one site is required for tight interaction of human tau protein with 14-3-3zeta. FEBS Lett. 2009, 583, 2739–4272. [Google Scholar] [CrossRef] [PubMed]
- Sluchanko, N.N.; Gusev, N.B. 14-3-3 proteins and regulation of cytoskeleton. Biochemistry 2010, 75, 1528–1546. [Google Scholar] [CrossRef]
- Johnson, C.; Crowther, S.; Stafford, M.; Campbell, D.; Toth, R.; MacKintosh, C. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem. J. 2010, 427, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Leon, S.P.; Lee, G.; Davies, P.; Shafit-Zagardo, B. Binding of Fyn to MAP-2c through an SH3 binding domain. J. Biol. Chem. 2001, 276, 39950–39958. [Google Scholar] [CrossRef]
- Andrei, S.A.; Meijer, F.A.; Neves, J.F.; Brunsveld, L.; Landrieu, I.; Ottmann, C.; Milroy, L.G. Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes. ACS Chem. Neurocsi. 2018, 9, 2639–2654. [Google Scholar] [CrossRef]
- Gigant, B.; Landrieu, I.; Fauquan, C.; Barbie, P.; Huvent, I.; Wieruszeski, J.M.; Knossow, M.; Lippens, G. Mechanism of Tau-Promoted Microtubule Assembly As Probed by NMR Spectroscopy. J. Am. Chem. Soc. 2014, 136, 12615–12623. [Google Scholar] [CrossRef] [PubMed]
- von Bergen, M.; Friedhoff, P.; Biernat, J.; Heberle, J.; Mandelkow, E.M.; Mandelkow, E. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc. Natl. Acad. Sci. USA 2000, 97, 5129–5134. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.Y.R.; Song, Y.; Barad, B.A.; Cheng, Y.; Fraser, J.S.; DiMaio, F. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife 2016, 5, e17219. [Google Scholar] [CrossRef]
- Al-Bassam, J.; Ozer, R.S.; Safer, D.; Halpain, S.; Milligan, R.A. MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J. Cell Biol. 2002, 157, 1187–1196. [Google Scholar] [CrossRef]
- Kar, S.; Fan, J.; Smith, M.J.; Goedert, M.; Amos, L.A. Repeat motifs of tau bind to the insides of microtubules in the absence of taxol. EMBO J. 2003, 22, 70–77. [Google Scholar] [CrossRef]
- Drewes, G.; Trinczek, B.; Illenberger, S.; Biernat, J.; Schmitt-Ulms, G.; Meyer, H.E.; Mandelkow, E.M.; Mandelkow, E. Microtubule-associated Protein/Microtubule Affinity-regulating Kinase (p110mark): A novel protein kinase that regulates tau-microtubule interactions and dynamic instability by phosphorylation at the Alzheimer-specific site serine 262. J. Biol. Chem. 1995, 270, 7679–7688. [Google Scholar] [CrossRef] [PubMed]
- Illenberger, S.; Drewes, G.; Trinczek, B.; Biernat, J.; Meyer, H.E.; Olmsted, J.B.; Mandelkow, E.M.; Mandelkow, E. Phosphorylation of microtubule-associated proteins MAP2 and MAP4 by the protein kinase p110mark. Phosphorylation sites and regulation of microtubule dynamics. J. Biol. Chem. 1996, 271, 10834–10843. [Google Scholar] [CrossRef]
- Brandt, R.; Lee, G.; Teplow, D.B.; Shalloway, D.; Abdel-Ghany, M. Differential Effect of Phosphorylation and Substrate Modulation on Tau’s Ability to Promote Microtubule Growth and Nucleation. J. Biol. Chem. 1994, 269, 11776–11782. [Google Scholar]
- Itoh, T.J.; Hisanaga, S.; Hosoi, T.; Kishimoto, T.; Hotani, H. Phosphorylation states of microtubule-associated protein 2 (MAP2) determine the regulatory role of MAP2 in microtubule dynamics. Biochemistry 1997, 36, 12574–12582. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.; Biernat, J.; von Bergen, M.; Mandelkow, E.; Mandelkow, E.M. Phosphorylation that Detaches Tau Protein from Microtubules (Ser262, Ser214) Also Protects It against Aggregation into Alzheimer Paired Helical Filaments. Biochemistry 1999, 38, 3549–3558. [Google Scholar] [CrossRef] [PubMed]
- Sattilaro, R.F. Interaction of microtubule-associated protein 2 with actin filaments. Biochemistry 1986, 25, 2003–2009. [Google Scholar] [CrossRef] [PubMed]
- Ozer, R.S.; Halpain, S. Phosphorylation-dependent localization of microtubule-associated protein MAP2c to the actin cytoskeleton. Mol. Biol. Cell 2000, 11, 3573–3587. [Google Scholar] [CrossRef]
- Al-Hilaly, Y.K.; Pollack, S.J.; Vadukul, D.M.; Citossi, F.; Rickard, J.E.; Simpson, M.; Storey, J.M.; Harrington, C.R.; Wischik, C.M.; Serpell, L.C. Alzheimer’s Disease-like Paired Helical Filament Assembly from Truncated Tau Protein Is Independent of Disulfide Crosslinking. J. Mol. Biol. 2017, 429, 3650–3665. [Google Scholar] [CrossRef]
- Xie, C.; Miyasaka, T.; Yoshimura, S.; Hatsuta, H.; Yoshina, S.; Kage-Nakadai, E.; Mitani, S.; Murayama, S.; Ihara, Y. The homologous carboxyl-terminal domains of microtubule-associated protein 2 and TAU induce neuronal dysfunction and have differential fates in the evolution of neurofibrillary tangles. PLoS ONE 2014, 9, e89796. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Ramos, A.; Díaz-Hernández, M.; Rubio, A.; Miras-Portugal, M.T.; Avila, J. Extracellular tau promotes intracellular calcium increase through M1 and M3 muscarinic receptors in neuronal cells. Mol. Cell. Neurosci. 2008, 37, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Paudel, H.K. Glycogen Synthase Kinase 3β Phosphorylates Alzheimer’s Disease-Specific Ser396 of Microtubule-Associated Protein Tau by a Sequential Mechanism. Biochemistry 2006, 45, 3125–3133. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Hernández, M.; Gómez-Ramos, A.; Rubio, A.; Gómez-Villafuertes, R.; Naranjo, J.R.; Teresa Miras-Portugal, M.; Avila, J. Tissue-nonspecific alkaline phosphatase promotes the neurotoxicity effect of extracellular tau. J. Biol. Chem. 2010, 285, 32539–35248. [Google Scholar] [CrossRef] [PubMed]
- Berry, R.W.; Abraha, A.; Lagalwar, S.; LaPointe, N.; Gamblin, T.C.; Cryns, V.L.; Binder, L.I. Inhibition of Tau Polymerization by Its Carboxy-Terminal Caspase Cleavage Fragment. Biochemistry 2003, 42, 8325–8331. [Google Scholar] [CrossRef] [PubMed]
- Fifre, A.; Sponne, I.; Koziel, V.; Kriem, B.; Potin, F.T.Y.; Bihain, B.E.; Olivier, J.L.; Oster, T.; Pillot, T. Microtubule-associated Protein MAP1A, MAP1B, and MAP2 Proteolysis during Soluble Amyloid β-Peptide-induced Neuronal Apoptosis: Synergistic Involvement of Calpain and Caspase-3. J. Biol. Chem. 2006, 281, 229–240. [Google Scholar] [CrossRef]
- Walker, S.; Ullman, O.; Stultz, C.M. Using intramolecular disulfide bonds in tau protein to deduce structural features of aggregation-resistant conformations. J. Biol. Chem. 2012, 287, 9591–9600. [Google Scholar] [CrossRef]
- Crowe, A.; James, M.J.; Virginia, M.Y.; Smith, A.B.; Trojanowski, J.Q.; Ballatore, C.; Brunden, K.R. Aminothienopyridazines and methylene blue affect Tau fibrillization via cysteine oxidation. J. Biol. Chem. 2013, 288, 11024–11037. [Google Scholar] [CrossRef]
- De Ancos, J.G.; Correas, I.; Avila, J. Differences in microtubule binding and self-association abilities of bovine brain tau isoforms. J. Biol. Chem. 1993, 268, 7976–7982. [Google Scholar]
- Paudel, H.K. Phosphorylation by neuronal cdc2-like protein kinase promotes dimerization of tau protein in vitro. J. Biol. Chem. 1997, 272, 28328–28334. [Google Scholar] [CrossRef] [PubMed]
- Wille, H.; Mandelkow, E.M.; Mandelkow, E. The Juvenile Microtubule-associated Protein MAP2c Is a Rod-like Molecule That Forms Antiparallel Dimer. J. Biol. Chem. 1992, 267, 10737–10742. [Google Scholar]
- Goode, B.L.; Denis, P.E.; Panda, D.; Radeke, M.J.; Miller, H.P.; Wilson, L.; Feinstein, S.C. Functional interactions between the proline-rich and repeat regions of tau enhance microtubule binding and assembly. Mol. Biol. Cell 1997, 8, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gong, H.S.; Zhang, J.; Xie, W.L.; Tian, C.; Chen, C.; Shi, Q.; Wang, S.B.; Xu, Y.; Zhang, B.Y.; et al. Remarkable reduction of MAP2 in the brains of scrapie-infected rodents and human prion disease possibly correlated with the increase of calpain. PLoS ONE 2012, 7, e30163. [Google Scholar] [CrossRef] [PubMed]
- Ackmann, M.; Wiech, H.; Mandelkow, E. Nonsaturable binding indicates clustering of tau on the microtubule surface in a paired helical filament-like conformation. J. Biol. Chem. 2000, 275, 30335–30343. [Google Scholar] [CrossRef] [PubMed]
- Meixner, A.; Haverkamp, S.; Wässle, H.; Führer, S.; Thalhammer, J.; Kropf, N.; Bittner, R.E.; Lassmann, H.; Wiche, G.; Propst, F. MAP1B is required for axon guidance and Is involved in the development of the central and peripheral nervous system. J. Cell Biol. 2000, 151, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Iqbal, K.; Grundke-Iqbal, I.; Rossie, S.; Gong, C.X. Dephosphorylation of tau by protein phosphatase 5: Impairment in Alzheimer’s disease. J. Biol. Chem. 2005, 280, 1790–1796. [Google Scholar] [CrossRef]
- Tompa, P.; Schad, E.; Tantos, A.; Kalmar, L. Intrinsically disordered proteins: Emerging interaction specialists. Curr. Opin. Struct. Biol. 2015, 35, 49–59. [Google Scholar] [CrossRef] [PubMed]
| Kinase | Abbreviation | Proline-Directed | Activator |
|---|---|---|---|
| cAMP-dependent protein kinase | PKA | no | – |
| MT-affinity-regulating kinases | MARK1–4 | no | – |
| extracellular signal-regulated kinase 2 | ERK2 | yes | MEK |
| glycogen-synthase kinase 3 | GSK3 | yes | – |
| cyclin dependent kinase 5 | CDK5 | yes | p35, p39 |
| c-Jun N-terminal kinase 1 | JNK1 | yes | – |
| stress-activated protein kinase 4 | SAPK4/p38 | yes | – |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melková, K.; Zapletal, V.; Narasimhan, S.; Jansen, S.; Hritz, J.; Škrabana, R.; Zweckstetter, M.; Ringkjøbing Jensen, M.; Blackledge, M.; Žídek, L. Structure and Functions of Microtubule Associated Proteins Tau and MAP2c: Similarities and Differences. Biomolecules 2019, 9, 105. https://doi.org/10.3390/biom9030105
Melková K, Zapletal V, Narasimhan S, Jansen S, Hritz J, Škrabana R, Zweckstetter M, Ringkjøbing Jensen M, Blackledge M, Žídek L. Structure and Functions of Microtubule Associated Proteins Tau and MAP2c: Similarities and Differences. Biomolecules. 2019; 9(3):105. https://doi.org/10.3390/biom9030105
Chicago/Turabian StyleMelková, Kateřina, Vojtěch Zapletal, Subhash Narasimhan, Séverine Jansen, Jozef Hritz, Rostislav Škrabana, Markus Zweckstetter, Malene Ringkjøbing Jensen, Martin Blackledge, and Lukáš Žídek. 2019. "Structure and Functions of Microtubule Associated Proteins Tau and MAP2c: Similarities and Differences" Biomolecules 9, no. 3: 105. https://doi.org/10.3390/biom9030105
APA StyleMelková, K., Zapletal, V., Narasimhan, S., Jansen, S., Hritz, J., Škrabana, R., Zweckstetter, M., Ringkjøbing Jensen, M., Blackledge, M., & Žídek, L. (2019). Structure and Functions of Microtubule Associated Proteins Tau and MAP2c: Similarities and Differences. Biomolecules, 9(3), 105. https://doi.org/10.3390/biom9030105