Integrative Analyses of Genes Associated with Subcutaneous Insulin Resistance
Abstract
1. Introduction
2. Materials and Methods
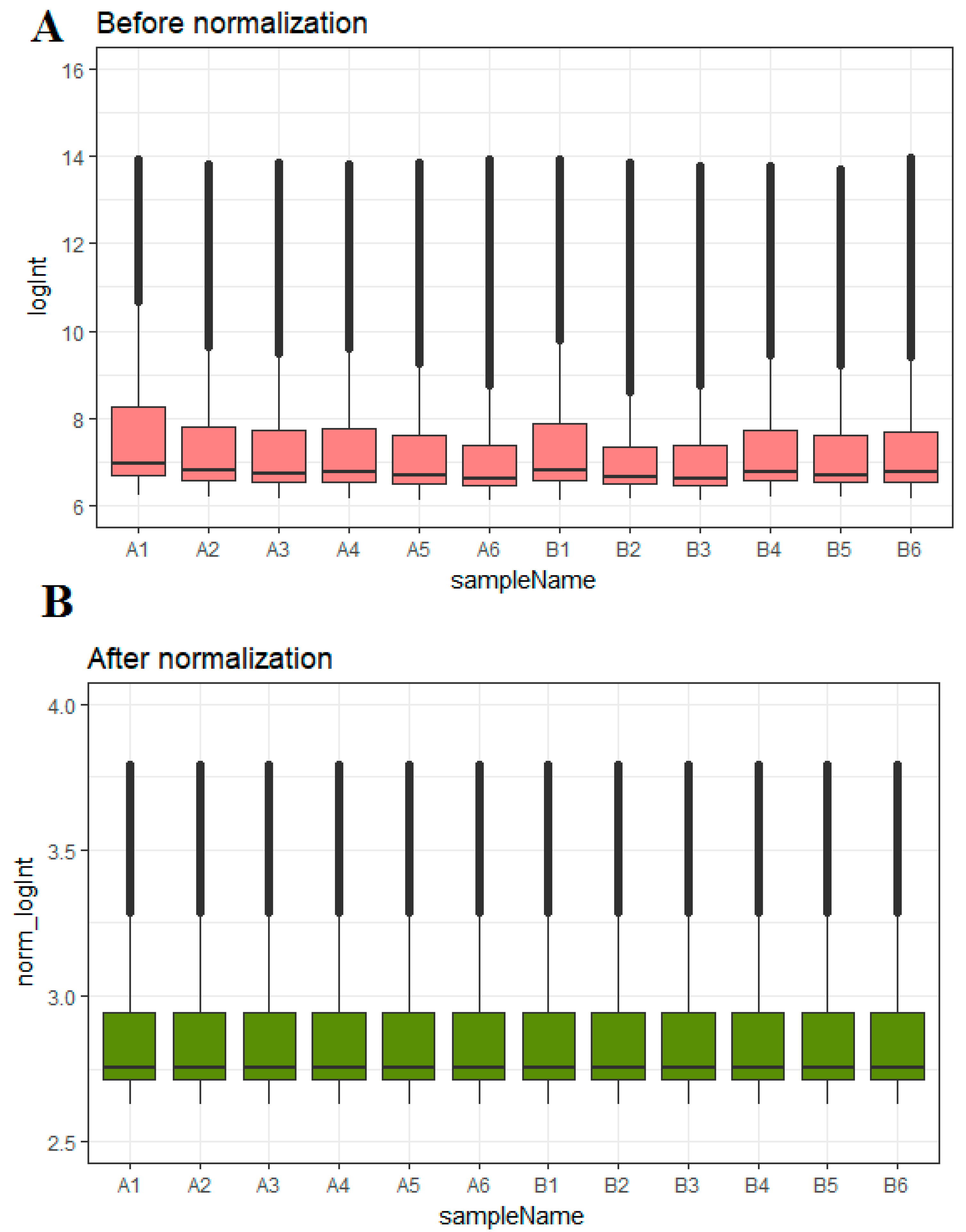
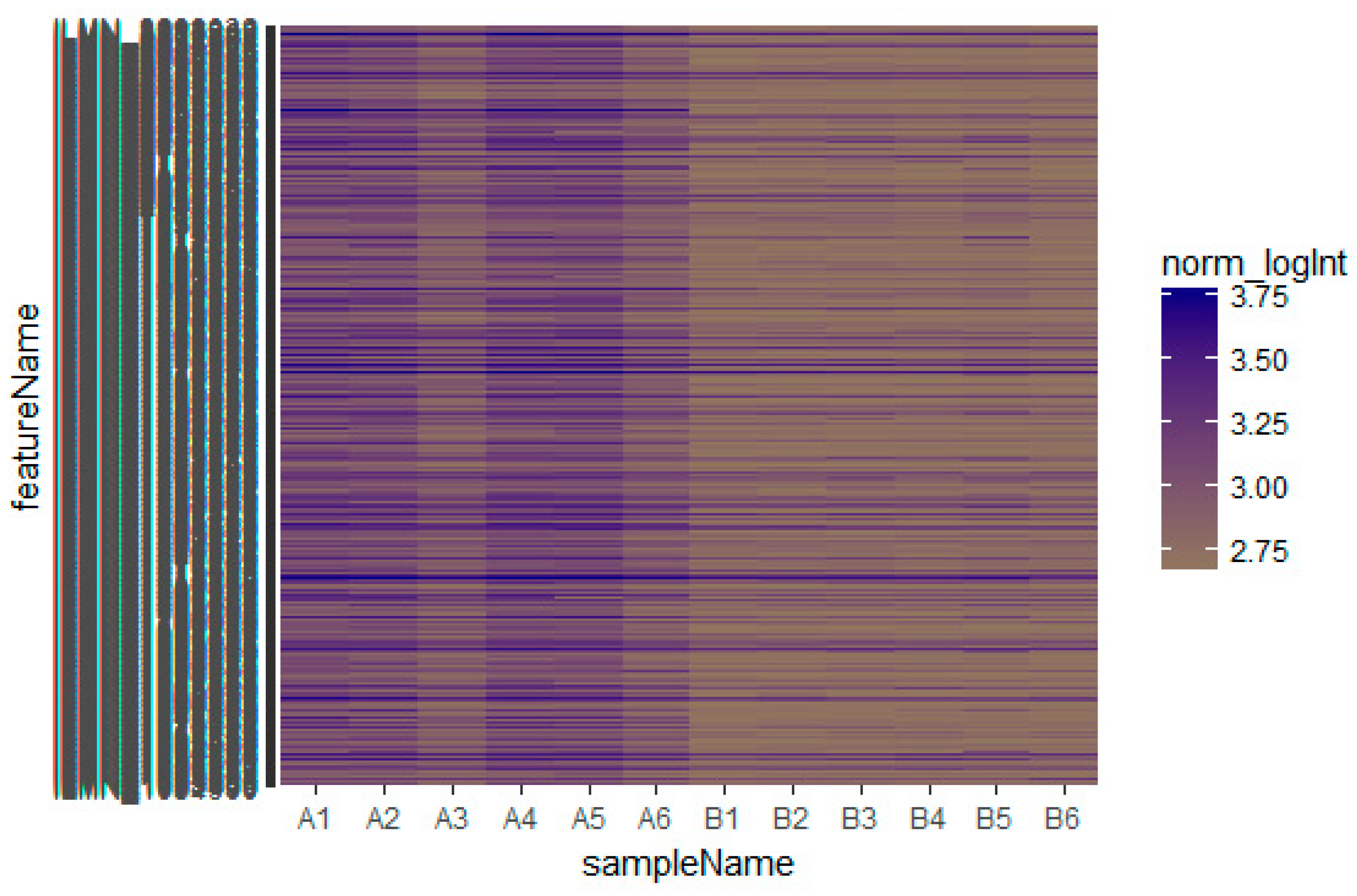
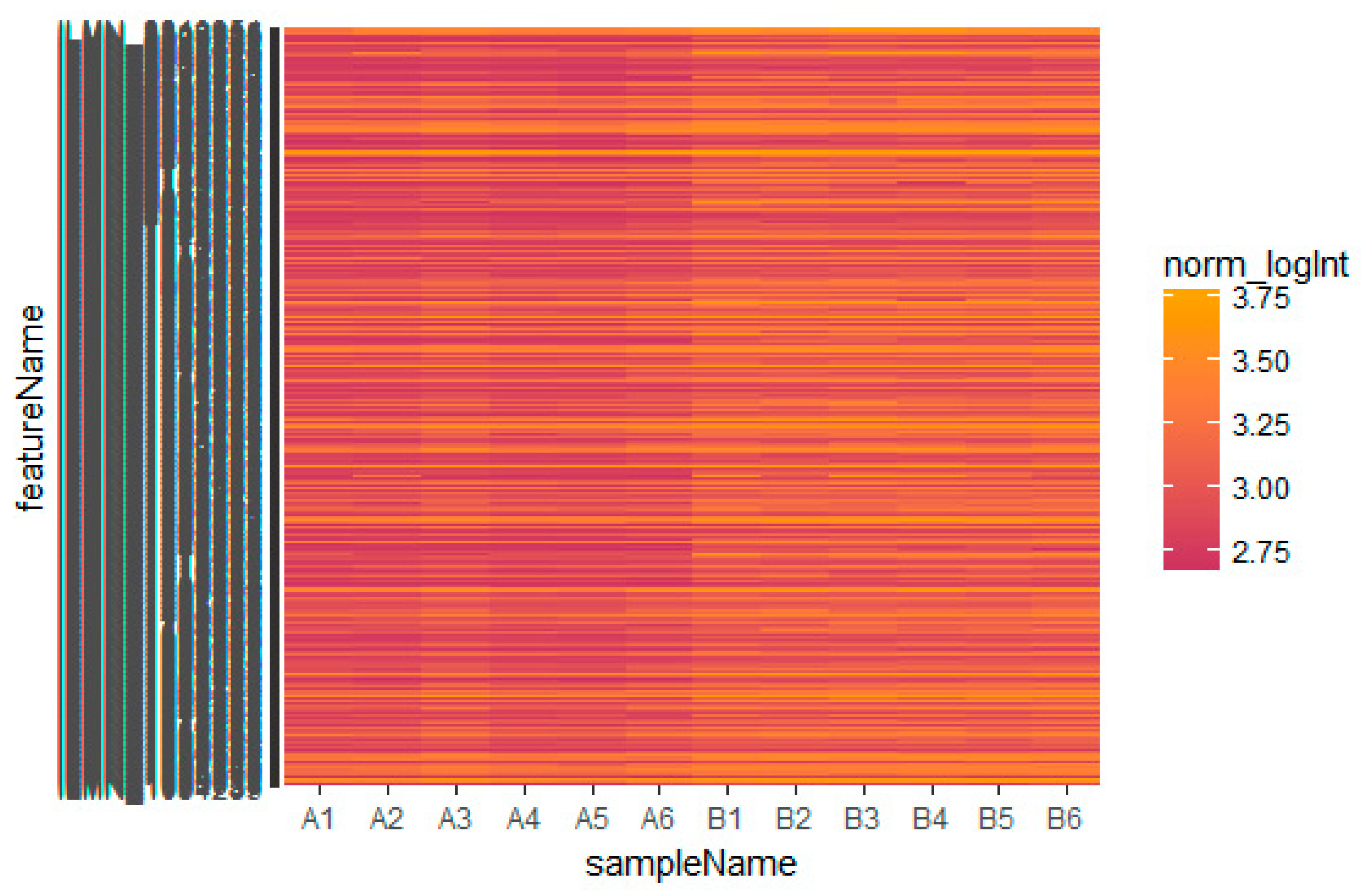
2.1. Microarray Data
2.2. Identification of Differentially Expressed Geness
2.3. Pathway Enrichment Analyses of Differentially Expressed Genes
2.4. Gene OntologyEnrichment Analyses of Differentially Expressed Genes
2.5. Construction of a Protein–Protein Interaction Network and Topological Analysis
2.6. Module Analysis
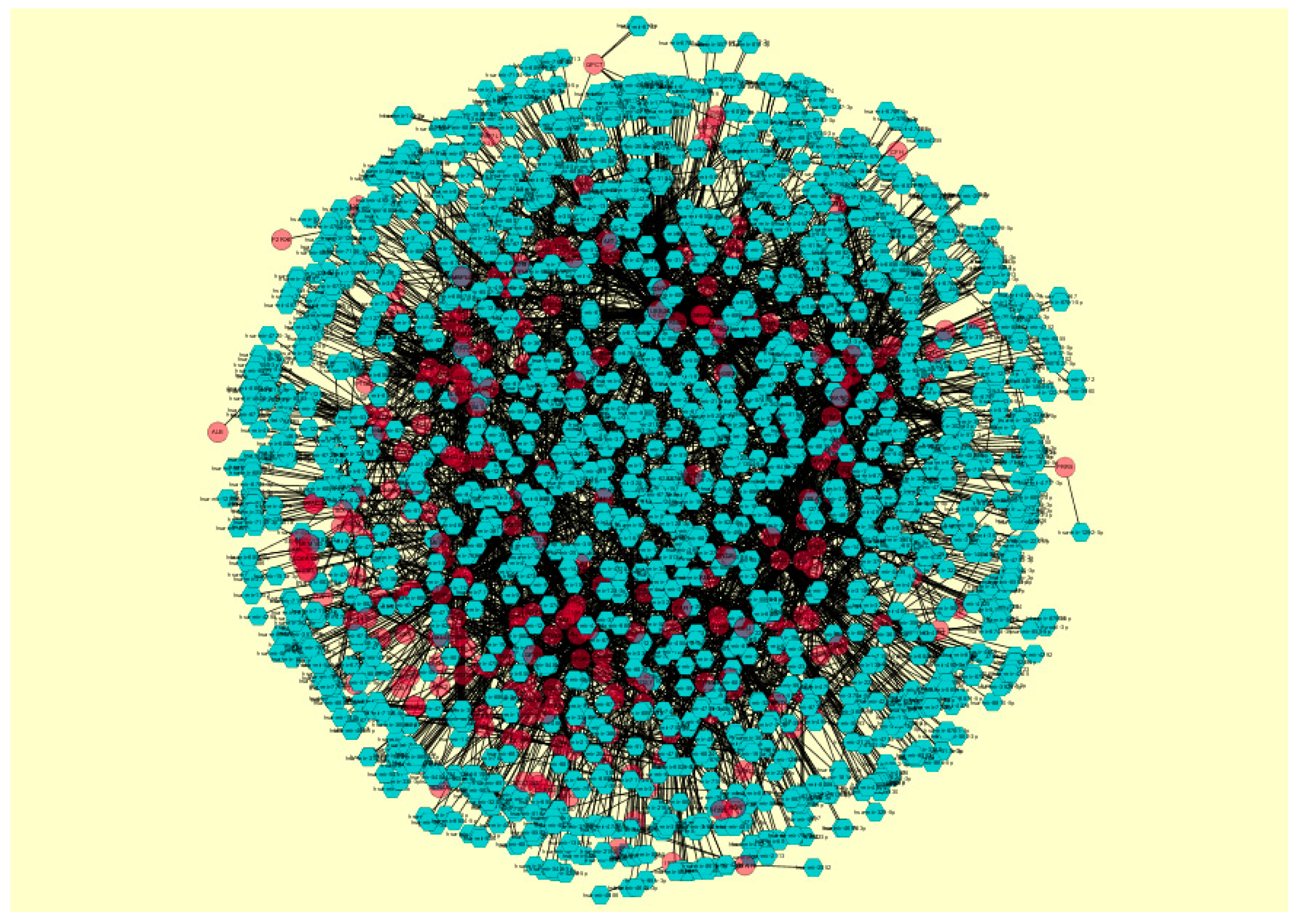
2.7. Construction of the Target Gene–MicroRNA Network
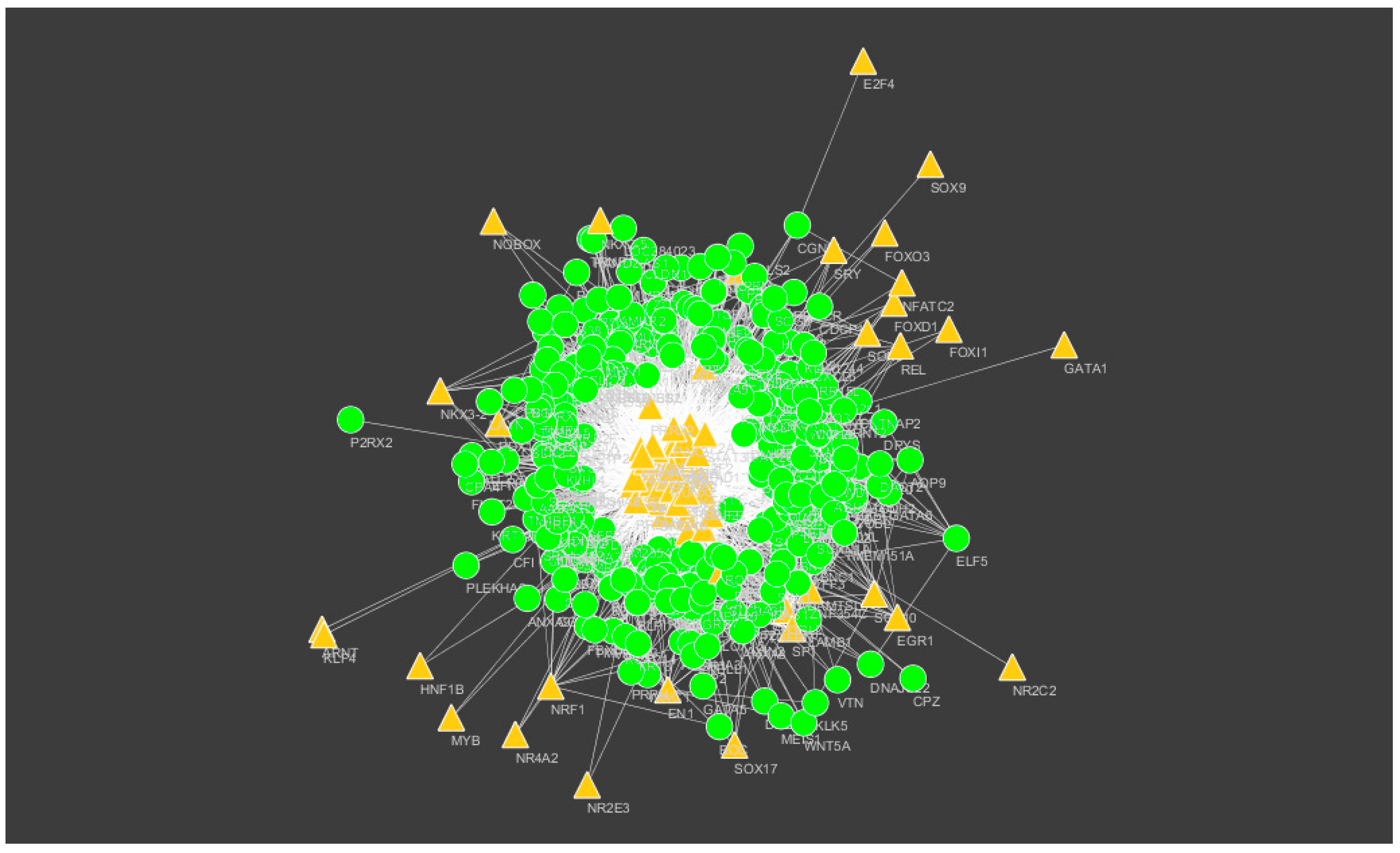
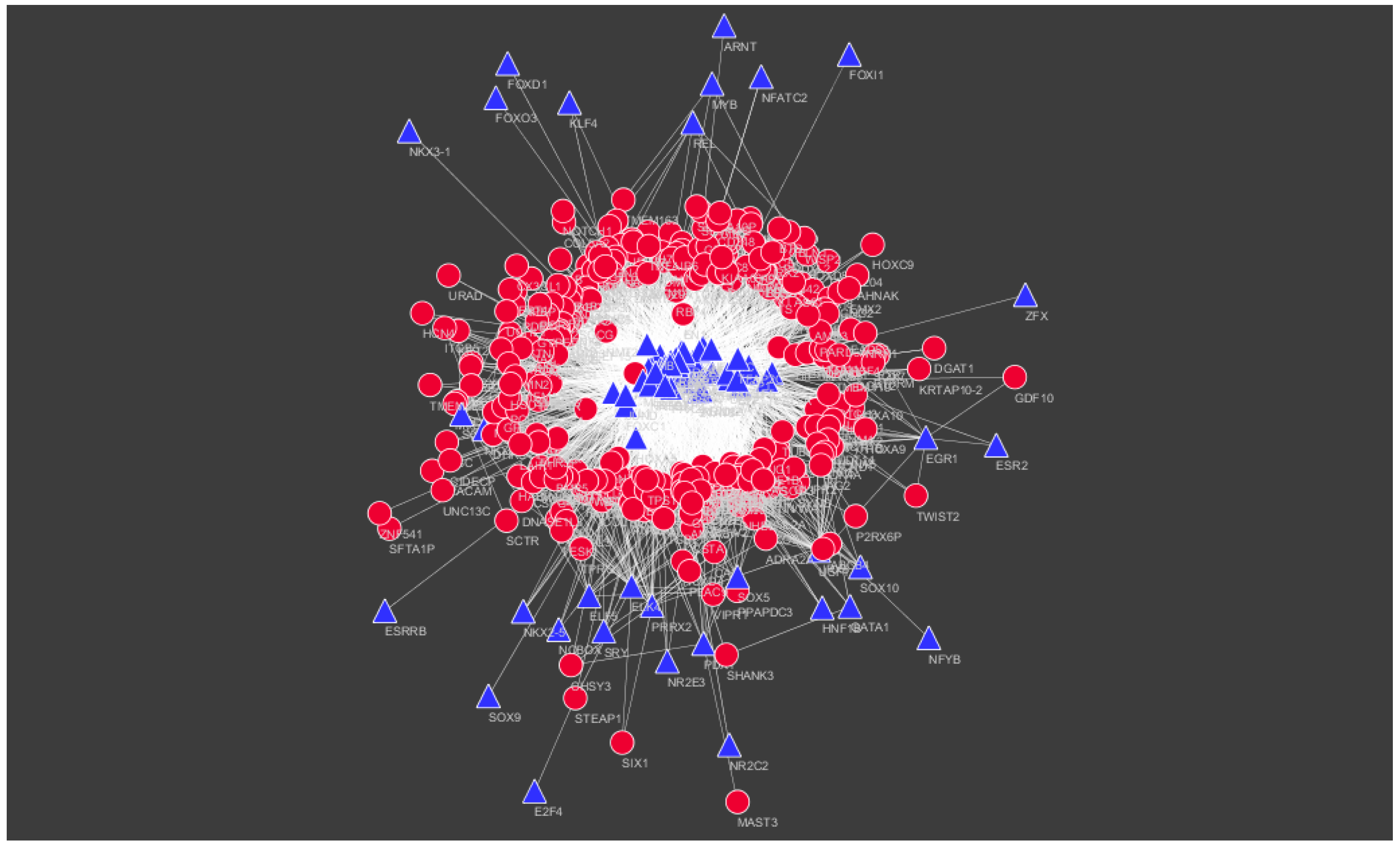
2.8. Construction of the Target Gene–Tanscription FactorNetwork
3. Results
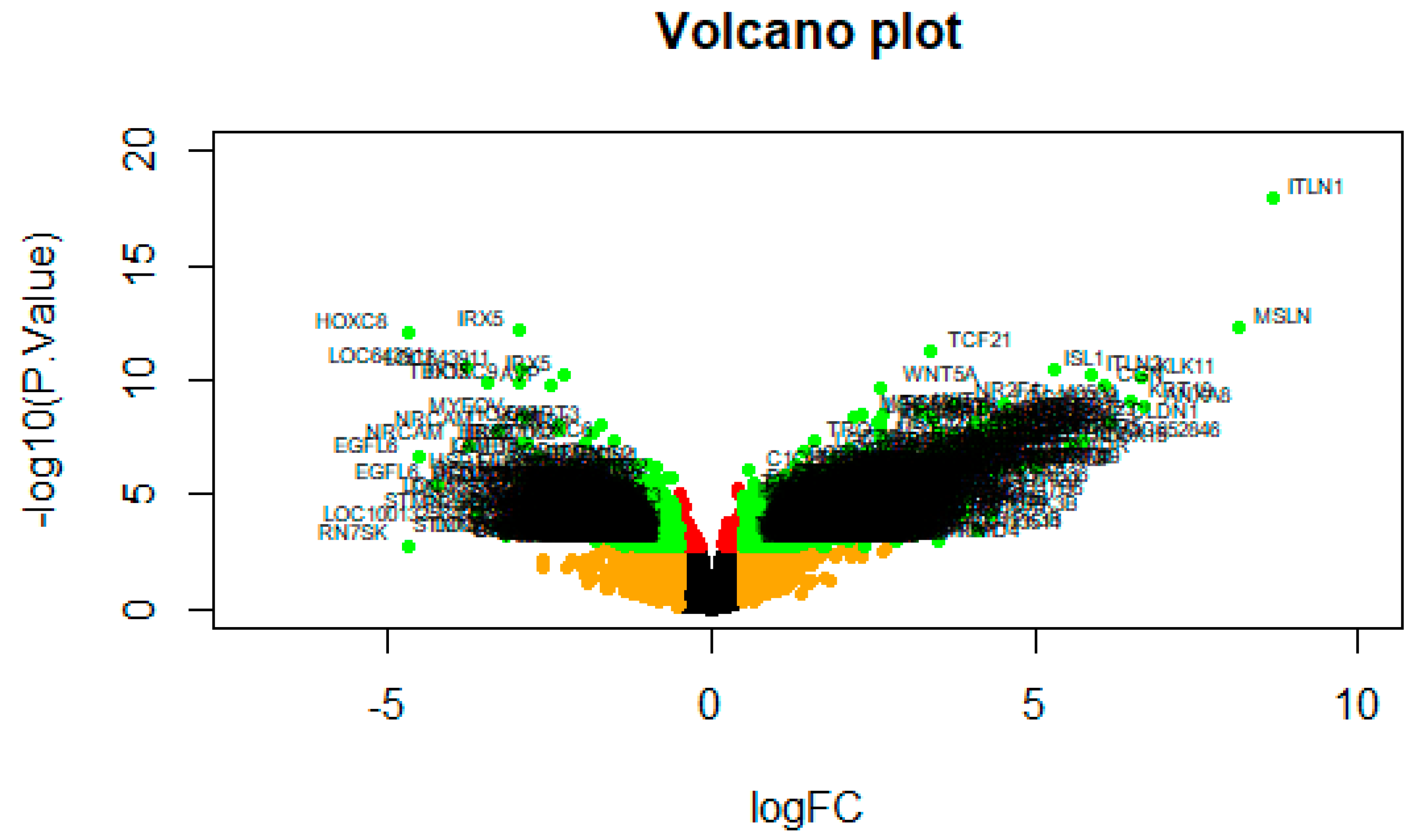
3.1. Identification of Differentially Expressed Genes in Insulin Resistance
3.2. Pathway Enrichment Analysis of Differentially Expressed Genes
3.3. Gene OntologyEnrichment Analyses of Differentially Expressed Genes
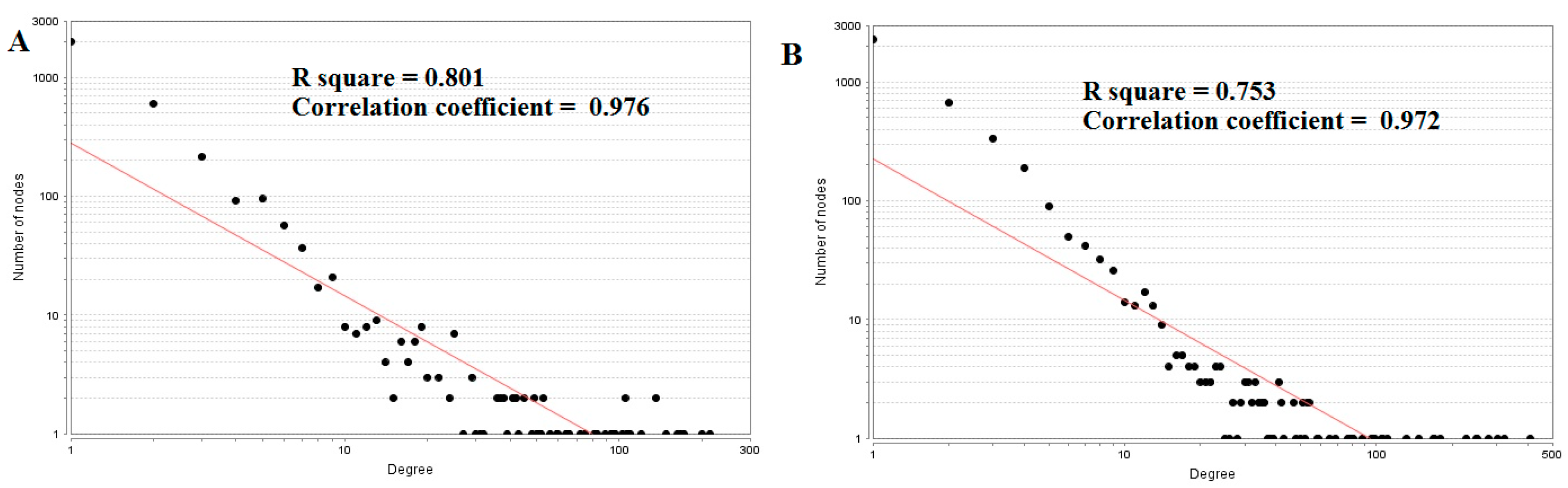
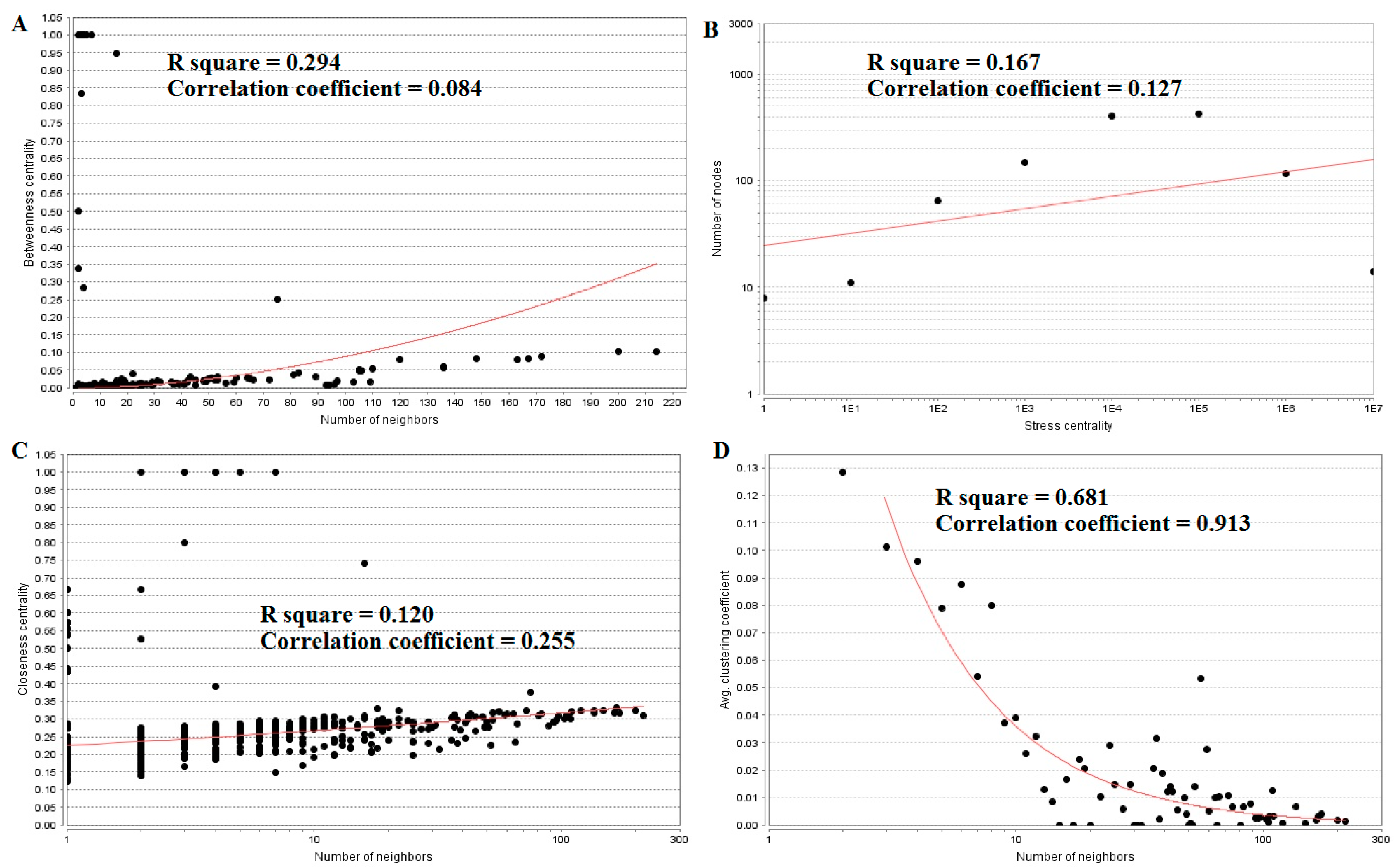
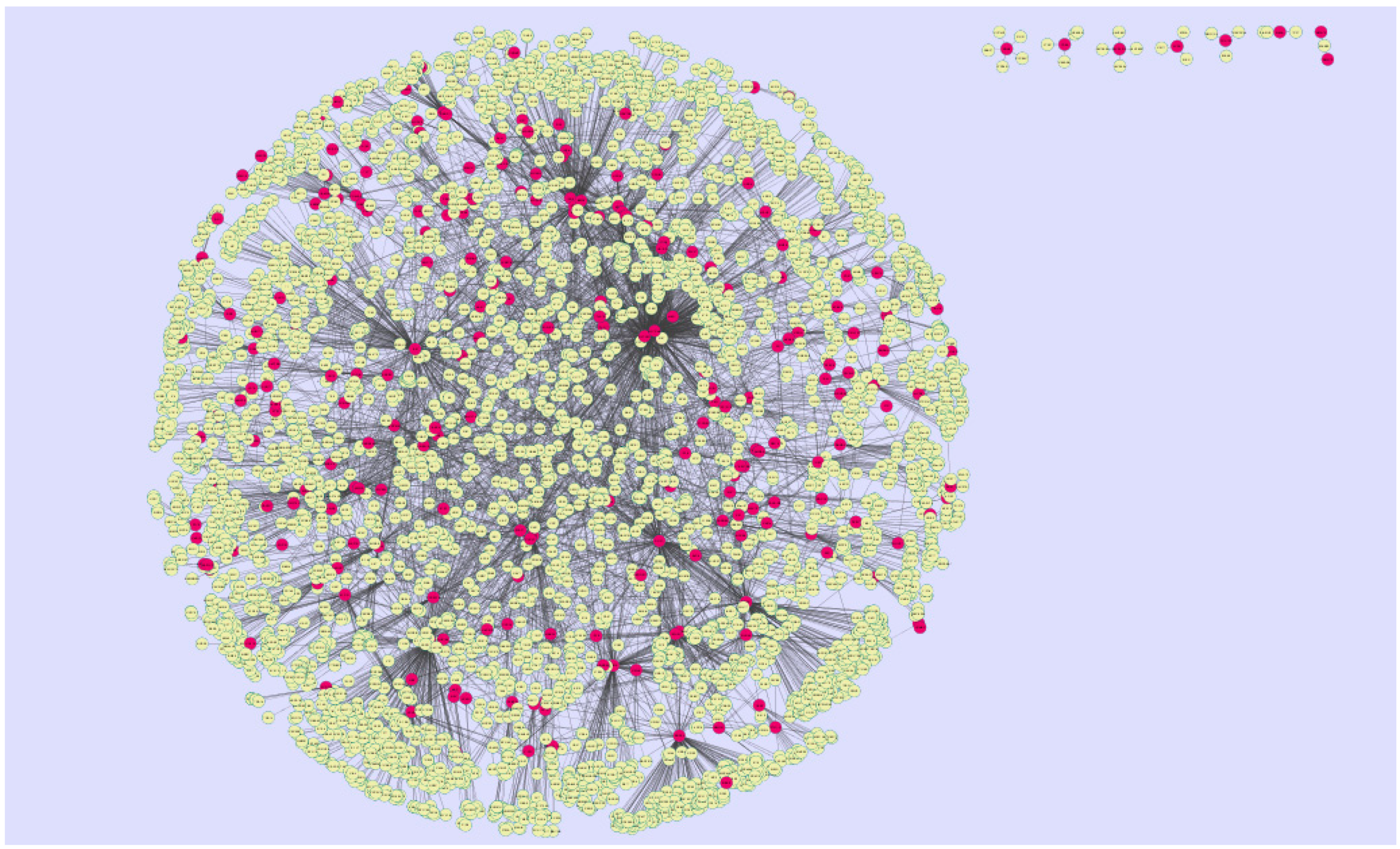
3.4. Construction of the Protein–Protein InteractionNetwork and Topological Analysis
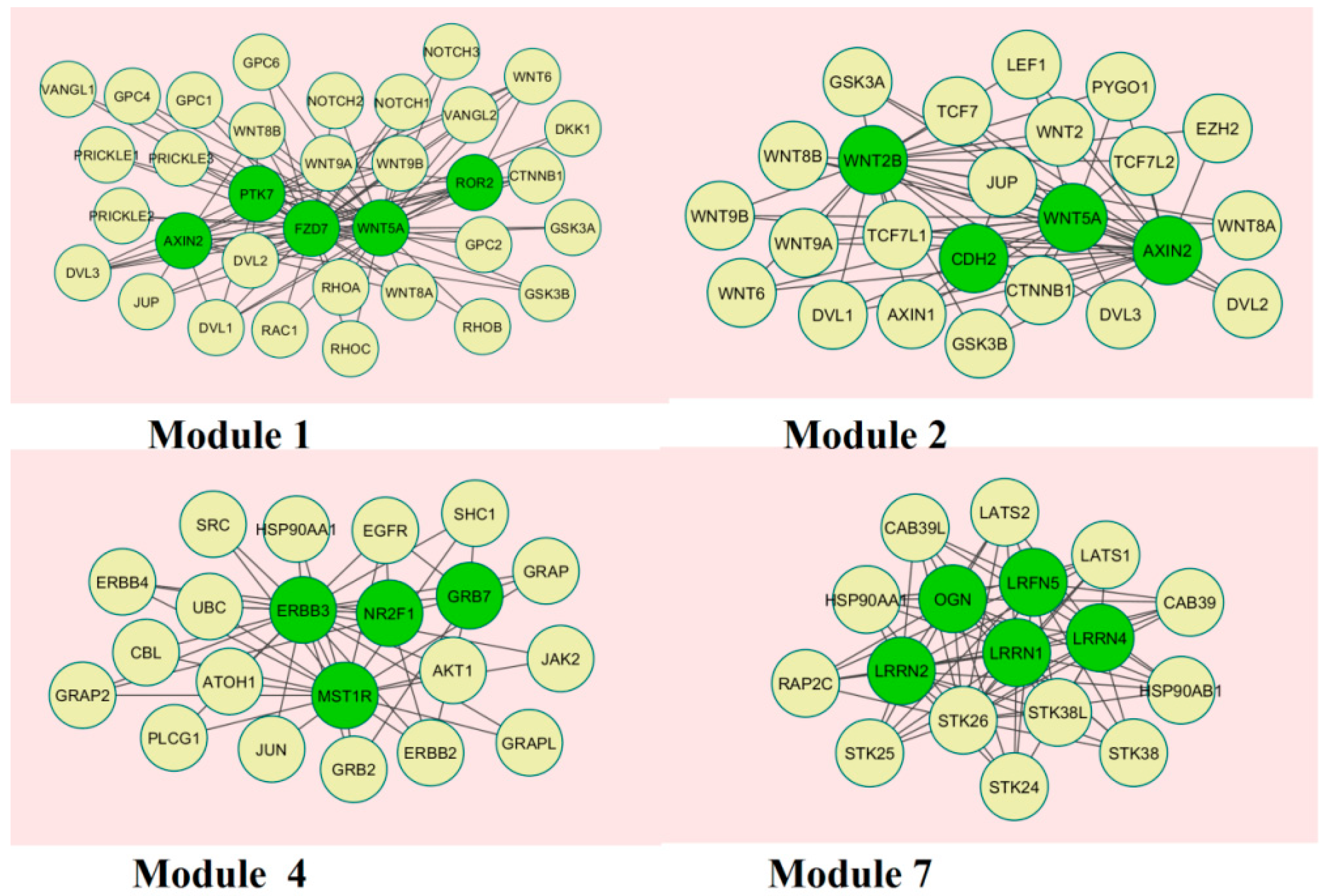
3.5. Module Analysis
3.6. Construction of the Target Gene–MicroRNA Network
3.7. Construction of the Target Gene–Transcription FactorNetwork
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethical Approval
Informed Consent
References
- Hossain, P.; Kawar, B.; El Nahas, M. Obesity and diabetes in the developing world—A growing challenge. N. Engl. J. Med. 2007, 356, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Myles, S.; Hradetzky, E.; Engelken, J.; Lao, O.; Nürnberg, P.; Trent, R.J.; Wang, X.; Kayser, M.; Stoneking, M. Identification of a candidate genetic variant for the high prevalence of type II diabetes in Polynesians. Eur. J. Hum. Genet. 2007, 15, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Yaribeygi, H.; Farrokhi, F.R.; Butler, A.E.; Sahebkar, A. Insulin resistance: Review of the underlying molecular mechanisms. J. Cell. Physiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- DeFronzo, R.A.; Ferrannini, E. Insulin resistance. A multifaceted syndrome responsible for NIDDM, obesity, hypertension, dyslipidemia, and atherosclerotic cardiovascular disease. Diabetes Care 1991, 14, 173–194. [Google Scholar] [CrossRef]
- Boden, G. Role of fatty acids in the pathogenesis of insulin resistance and NIDDM. Diabetes 1997, 46, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Koves, T.R.; Ussher, J.R.; Noland, R.C.; Slentz, D.; Mosedale, M.; Ilkayeva, O.; Bain, J.; Stevens, R.; Dyck, J.R.; Newgard, C.B.; et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008, 7, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S.; Shargill, N.S.; Spiegelman, B.M. Adipose expression of tumor necrosis factor-alpha: Direct role in obesity-linked insulin resistance. Science 1993, 259, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Rehman, K.; Akash, M.S.H.; Liaqat, A.; Kamal, S.; Qadir, M.I.; Rasul, A. Role of Interleukin-6 in Development of Insulin Resistance and Type 2 Diabetes Mellitus. Crit. Rev. Eukaryot. Gene Expr. 2017, 27, 229–236. [Google Scholar] [CrossRef]
- Rull, A.; Camps, J.; Alonso-Villaverde, C.; Joven, J. Insulin resistance, inflammation, and obesity: Role of monocyte chemoattractant protein-1 (or CCL2) in the regulation of metabolism. Mediat. Inflamm. 2010, 2010. [Google Scholar] [CrossRef]
- Dai, M.; Wu, L.; Tu, L.; Xu, X.; Wang, D.W. The immune-metabolic regulatory roles of epoxyeicosatrienoic acids on macrophages phenotypic plasticity in obesity-related insulin resistance. Prostaglandins Other Lipid Mediat. 2018, 139, 36–40. [Google Scholar] [CrossRef]
- Evans, J.L.; Goldfine, I.D.; Maddux, B.A.; Grodsky, G.M. Are oxidative stress-activated signaling pathways mediators of insulin resistance and beta-cell dysfunction? Diabetes 2003, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Moller, D.E.; Bjørbaek, C.; Vidal-Puig, A. Candidate genes for insulin resistance. Diabetes Care 1996, 19, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Graham, T.E.; Mody, N.; Preitner, F.; Peroni, O.D.; Zabolotny, J.M.; Kotani, K.; Quadro, L.; Kahn, B.B. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 2005, 436, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Pizzuti, A.; Frittitta, L.; Argiolas, A.; Baratta, R.; Goldfine, I.D.; Bozzali, M.; Ercolino, T.; Scarlato, G.; Iacoviello, L.; Vigneri, R.; et al. A polymorphism (K121Q) of the human glycoprotein PC-1 gene coding region is strongly associated with insulin resistance. Diabetes 1999, 48, 1881–1884. [Google Scholar] [CrossRef]
- Collins, S.C.; Luan, J.; Thompson, A.J.; Daly, A.; Semple, R.K.; O’Rahilly, S.; Wareham, N.J.; Barroso, I. Adiponectin receptor genes: Mutation screening in syndromes of insulin resistance and association studies for type 2 diabetes and metabolic traits in UK populations. Diabetologia 2007, 50, 555–562. [Google Scholar] [CrossRef]
- Cederberg, A.; Grønning, L.M.; Ahrén, B.; Taskén, K.; Carlsson, P.; Enerbäck, S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell 2001, 106, 563–573. [Google Scholar] [CrossRef]
- Cusi, K.; Maezono, K.; Osman, A.; Pendergrass, M.; Patti, M.E.; Pratipanawatr, T.; DeFronzo, R.A.; Kahn, C.R.; Mandarino, L.J. Insulin resistance differentially affects the PI 3-kinase- and MAP kinase-mediated signaling in human muscle. J. Clin. Investig. 2000, 105, 311–320. [Google Scholar] [CrossRef]
- Arkan, M.C.; Hevener, A.L.; Greten, F.R.; Maeda, S.; Li, Z.W.; Long, J.M.; Wynshaw-Boris, A.; Poli, G.; Olefsky, J.; Karin, M. IKK-beta links inflammation to obesity-induced insulin resistance. Nat. Med. 2005, 11, 191–198. [Google Scholar] [CrossRef]
- Shah, O.J.; Wang, Z.; Hunter, T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 2004, 14, 1650–1656. [Google Scholar] [CrossRef]
- Solinas, G.; Becattini, B. JNK at the crossroad of obesity, insulin resistance, and cell stress response. Mol. Metab. 2016, 6, 174–184. [Google Scholar] [CrossRef]
- Barrett, T.; Troup, D.B.; Wilhite, S.E.; Ledoux, P.; Rudnev, D.; Evangelista, C.; Kim, I.F.; Soboleva, A.; Tomashevsky, M.; Marshall, K.A.; et al. NCBI GEO: Archive for high-throughput functional genomic data. Nucleic Acids Res. 2009, 37, D885–D890. [Google Scholar] [CrossRef] [PubMed]
- Wentworth, J.M.; Naselli, G.; Ngui, K.; Smyth, G.K.; Liu, R.; O’Brien, P.E.; Bruce, C.; Weir, J.; Cinel, M.; Meikle, P.J.; et al. GM3 ganglioside and phosphatidylethanolamine-containing lipids are adipose tissue markers of insulin resistance in obese women. Int. J. Obes. (Lond.) 2016, 40, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Slenter, D.N.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Mélius, J.; Cirillo, E.; Coort, S.L.; Digles, D.; et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018, 46, D661–D667. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.F.; Jia, P.; Zhao, Z.; Kuo, P.H. Enriched pathways for major depressive disorder identified from a genome-wide association study. Int. J. Neuropsychopharmacol. 2012, 15, 1401–1411. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Wagg, J.; Green, M.L.; Kaiser, D.; Krummenacker, M.; Karp, P.D. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005, 6, R2. [Google Scholar] [CrossRef]
- Mi, H.; Thomas, P. PANTHER pathway: An ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol. 2009, 563, 123–140. [Google Scholar] [CrossRef]
- Schaefer, C.F.; Anthony, K.; Krupa, S.; Buchoff, J.; Day, M.; Hannay, T.; Buetow, K.H. PID: The Pathway Interaction Database. Nucleic Acids Res. 2009, 37, D674–D679. [Google Scholar] [CrossRef]
- Bader, G.D.; Cary, M.P.; Sander, C. Pathguide: A pathway resource list. Nucleic Acids Res. 2006, 34, D504–D506. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Przulj, N. Biological network comparison using graphlet degree distribution. Bioinformatics 2007, 23, e177–e183. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Blumer, A.; Lee, K. An algorithm for modularity analysis of directed and weighted biological networks based on edge-betweenness centrality. Bioinformatics 2006, 22, 3106–3108. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Assenov, Y.; Domingues, F.S.; Albrecht, M. Topological analysis and interactive visualization of biological networks and protein structures. Nat. Protoc. 2012, 7, 670–685. [Google Scholar] [CrossRef]
- Hahn, M.W.; Kern, A.D. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol. Biol. Evol. 2005, 22, 803–806. [Google Scholar] [CrossRef]
- Barabási, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef]
- Zaki, N.; Efimov, D.; Berengueres, J. Protein complex detection using interaction reliability assessment and weighted clustering coefficient. BMC Bioinform. 2013, 14, 163. [Google Scholar] [CrossRef]
- Soifer, H.S.; Rossi, J.J.; Saetrom, P. MicroRNAs in disease and potential therapeutic applications. Mol. Ther. 2007, 15, 2070–2079. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Gill, E.E.; Hancock, R.E. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015, 10, 823–844. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015, 43, D153–D159. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.H.; Shrestha, S.; Yang, C.D.; Chang, N.W.; Lin, Y.L.; Liao, K.W.; Huang, W.C.; Sun, T.H.; Tu, S.J.; Lee, W.H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, A.S. Series introduction: The transcription factor NF-kappaB and human disease. J. Clin. Investig. 2001, 107, 3–6. [Google Scholar] [CrossRef]
- Khan, A.; Fornes, O.; Stigliani, A.; Gheorghe, M.; Castro-Mondragon, J.A.; Lee, R.; Bessy, A.; Chèneby, J.; Kulkarni, S.R.; Tan, G.; et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018, 46, D1284. [Google Scholar] [CrossRef]
- Buettner, R.; Schölmerich, J.; Bollheimer, L.C. High-fat diets: Modeling the metabolic disorders of human obesity in rodents. Obesity 2007, 15, 798–808. [Google Scholar] [CrossRef]
- Tabassum, R.; Mahajan, A.; Dwivedi, O.P.; Chauhan, G.; Spurgeon, C.J.; Kumar, M.V.; Ghosh, S.; Madhu, S.V.; Mathur, S.K.; Chandak, G.R.; et al. Common variants of SLAMF1 and ITLN1 on 1q21 are associated with type 2 diabetes in Indian population. J. Hum. Genet. 2012, 57, 184–190. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, W.P.; Guo, T.; Yang, J.C.; Chen, P.; Ma, K.T.; Guan, Y.F.; Zhou, C.Y. The LIM-homeodomain protein ISL1 activates insulin gene promoter directly through synergy with BETA2. J. Mol. Biol. 2009, 392, 566–577. [Google Scholar] [CrossRef]
- Riggs, A.C.; Tanizawa, Y.; Aoki, M.; Wasson, J.; Ferrer, J.; Rabin, D.U.; Vaxillaire, M.; Froguel, P.; Permutt, M.A. Characterization of the LIM/homeodomain gene islet-1 and single nucleotide screening in NIDDM. Diabetes 1995, 44, 689–694. [Google Scholar] [CrossRef]
- Catalán, V.; Gómez-Ambrosi, J.; Rodríguez, A.; Pérez-Hernández, A.I.; Gurbindo, J.; Ramírez, B.; Méndez-Giménez, L.; Rotellar, F.; Valentí, V.; Moncada, R.; et al. Activation of noncanonical Wnt signaling through WNT5A in visceral adipose tissue of obese subjects is related to inflammation. J. Clin. Endocrinol. Metab. 2014, 99, E1407–E1417. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.C.; Wang, C.P.; Hsu, C.C.; Chiu, C.A.; Yu, T.H.; Hung, W.C.; Lu, L.F.; Chung, F.M.; Tsai, I.T.; Lin, H.C.; et al. Circulating secreted frizzled-related protein 5 (Sfrp5) and wingless-type MMTV integration site family member 5a (Wnt5a) levels in patients with type 2 diabetes mellitus. Diabetes Metab. Res. Rev. 2013, 29, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Zieger, K.; Weiner, J.; Kunath, A.; Gericke, M.; Krause, K.; Kern, M.; Stumvoll, M.; Klöting, N.; Blüher, M.; Heiker, J.T. Ablation of kallikrein 7 (KLK7) in adipose tissue ameliorates metabolic consequences of high fat diet-induced obesity by counteracting adipose tissue inflammation in vivo. Cell. Mol. Life Sci. 2018, 75, 727–742. [Google Scholar] [CrossRef] [PubMed]
- Tong, Q. Synaptotagmin 4: A new antiobesity target? Neuron 2011, 69, 401–403. [Google Scholar] [CrossRef] [PubMed]
- Hunt, L.E.; Noyvert, B.; Bhaw-Rosun, L.; Sesay, A.K.; Paternoster, L.; Nohr, E.A.; Davey Smith, G.; Tommerup, N.; Sørensen, T.I.; Elgar, G. Complete re-sequencing of a 2Mb topological domain encompassing the FTO/IRXB genes identifies a novel obesity-associated region upstream of IRX5. Genome Med. 2015, 7, 126. [Google Scholar] [CrossRef]
- Lee, K.Y.; Sharma, R.; Gase, G.; Ussar, S.; Li, Y.; Welch, L.; Berryman, D.E.; Kispert, A.; Bluher, M.; Kahn, C.R. Tbx15 Defines a Glycolytic Subpopulation and White Adipocyte Heterogeneity. Diabetes 2017, 66, 2822–2829. [Google Scholar] [CrossRef]
- Brune, J.E.; Kern, M.; Kunath, A.; Flehmig, G.; Schön, M.R.; Lohmann, T.; Dressler, M.; Dietrich, A.; Fasshauer, M.; Kovacs, P.; et al. Fat depot-specific expression of HOXC9 and HOXC10 may contribute to adverse fat distribution and related metabolic traits. Obesity 2016, 24, 51–59. [Google Scholar] [CrossRef]
- Coiro, V.; Chiodera, P. Effect of obesity and weight loss on the arginine vasopressin response to insulin-induced hypoglycaemia. Clin. Endocrinol. 1987, 27, 253–258. [Google Scholar] [CrossRef]
- Thun, G.A.; Imboden, M.; Berger, W.; Rochat, T.; Probst-Hensch, N.M. The association of a variant in the cell cycle control gene CCND1 and obesity on the development of asthma in the Swiss SAPALDIA study. J. Asthma 2013, 50, 147–154. [Google Scholar] [CrossRef]
- Hernández-Mijares, A.; Jarabo-Bueno, M.M.; López-Ruiz, A.; Solá-Izquierdo, E.; Morillas-Ariño, C.; Martínez-Triguero, M.L. Levels of C3 in patients with severe, morbid and extreme obesity: Its relationship to insulin resistance and different cardiovascular risk factors. Int. J. Obes. 2007, 31, 927–932. [Google Scholar] [CrossRef]
- Alessi, M.C.; Nicaud, V.; Scroyen, I.; Lange, C.; Saut, N.; Fumeron, F.; Marre, M.; Lantieri, O.; Fontaine-Bisson, B.; Juhan-Vague, I. Association of vitronectin and plasminogen activator inhibitor-1 levels with the risk of metabolic syndrome and type 2 diabetes mellitus. Results from the D.E.S.I.R. prospective cohort. Thromb. Haemost. 2011, 106, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Gabrielsson, B.G.; Johansson, J.M.; Lönn, M.; Jernås, M.; Olbers, T.; Peltonen, M.; Larsson, I.; Lönn, L.; Sjöström, L.; Carlsson, B.; et al. High expression of complement components in omental adipose tissue in obese men. Obes. Res. 2003, 11, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Kim-Muller, J.Y.; Fan, J.; Kim, Y.J.; Lee, S.A.; Ishida, E.; Blaner, W.S.; Accili, D. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic β cells in diabetic mice. Nat. Commun. 2016, 7, 12631. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.E.; Kim, J.M.; Yi, H.S.; Joung, K.H.; Lee, J.H.; Kim, H.J.; Ku, B.J. Serum R-Spondin 1 Is a New Surrogate Marker for Obesity and Insulin Resistance. Diabetes Metab. J. 2018. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Demeterco, C.; Geron, I.; Abrahamsson, A.; Levine, F.; Itkin-Ansari, P. Islet specific Wnt activation in human type II diabetes. Exp. Diabetes Res. 2008, 2008, 728763. [Google Scholar] [CrossRef]
- Hemi, R.; Yochananov, Y.; Barhod, E.; Kasher-Meron, M.; Karasik, A.; Tirosh, A.; Kanety, H. p38 mitogen-activated protein kinase-dependent transactivation of ErbB receptor family: A novel common mechanism for stress-induced IRS-1 serine phosphorylation and insulin resistance. Diabetes 2011, 60, 1134–1145. [Google Scholar] [CrossRef] [PubMed]
- Kumashiro, N.; Beddow, S.A.; Vatner, D.F.; Majumdar, S.K.; Cantley, J.L.; Guebre-Egziabher, F.; Fat, I.; Guigni, B.; Jurczak, M.J.; Birkenfeld, A.L.; et al. Targeting pyruvate carboxylase reduces gluconeogenesis and adiposity and improves insulin resistance. Diabetes 2013, 62, 2183–2194. [Google Scholar] [CrossRef]
- Griffin, J.W.D.; Liu, Y.; Bradshaw, P.C.; Wang, K. In Silico Preliminary Association of Ammonia Metabolism Genes GLS, CPS1, and GLUL with Risk of Alzheimer’s Disease, Major Depressive Disorder, and Type 2 Diabetes. J. Mol. Neurosci. 2018, 64, 385–396. [Google Scholar] [CrossRef]
- Yu, J.; Xiao, F.; Guo, Y.; Deng, J.; Liu, B.; Zhang, Q.; Li, K.; Wang, C.; Chen, S.; Guo, F. Hepatic Phosphoserine Aminotransferase 1 Regulates Insulin Sensitivity in Mice via Tribbles Homolog 3. Diabetes 2015, 64, 1591–1602. [Google Scholar] [CrossRef]
- Li, H.; Lee, J.; He, C.; Zou, M.H.; Xie, Z. Suppression of the mTORC1/STAT3/Notch1 pathway by activated AMPK prevents hepatic insulin resistance induced by excess amino acids. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E197–E209. [Google Scholar] [CrossRef]
- Pajvani, U.B.; Shawber, C.J.; Samuel, V.T.; Birkenfeld, A.L.; Shulman, G.I.; Kitajewski, J.; Accili, D. Inhibition of Notch signaling ameliorates insulin resistance in a FoxO1-dependent manner. Nat. Med. 2011, 17, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Strakovsky, R.S.; Pan, Y.X. A decrease in DKK1, a WNT inhibitor, contributes to placental lipid accumulation in an obesity-prone rat model. Biol. Reprod. 2012, 86, 81. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Nair, A.K.; Muller, Y.L.; Piaggi, P.; Bian, L.; Del Rosario, M.; Knowler, W.C.; Kobes, S.; Hanson, R.L.; Bogardus, C.; et al. Whole exome sequencing identifies variation in CYB5A and RNF10 associated with adiposity and type 2 diabetes. Obesity 2014, 22, 984–988. [Google Scholar] [CrossRef] [PubMed]
- Bajzová, M.; Kováciková, M.; Vítková, M.; Klimcáková, E.; Polák, J.; Kovácová, Z.; Viguerie, N.; Vedral, T.; Mikulásek, L.; Srámková, P.; et al. Retinol-binding protein 4 expression in visceral and subcutaneous fat in human obesity. Physiol. Res. 2008, 57, 927–934. [Google Scholar] [PubMed]
- Cho, Y.M.; Youn, B.S.; Lee, H.; Lee, N.; Min, S.S.; Kwak, S.H.; Lee, H.K.; Park, K.S. Plasma retinol-binding protein-4 concentrations are elevated in human subjects with impaired glucose tolerance and type 2 diabetes. Diabetes Care 2006, 29, 2457–2461. [Google Scholar] [CrossRef]
- Kovacs, P.; Geyer, M.; Berndt, J.; Klöting, N.; Graham, T.E.; Böttcher, Y.; Enigk, B.; Tönjes, A.; Schleinitz, D.; Schön, M.R.; et al. Effects of genetic variation in the human retinol binding protein-4 gene (RBP4) on insulin resistance and fat depot-specific mRNA expression. Diabetes 2007, 56, 3095–3100. [Google Scholar] [CrossRef] [PubMed]
- Svensson, P.A.; Gabrielsson, B.G.; Jernås, M.; Gummesson, A.; Sjöholm, K. Regulation of human aldoketoreductase 1C3 (AKR1C3) gene expression in the adipose tissue. Cell. Mol. Biol. Lett. 2008, 13, 599–613. [Google Scholar] [CrossRef]
- Cho, M.K. Decreased Expression of Type 5 17β-Hydroxysteroid Dehydrogenase (AKR1C3) Protein Identified in Human Diabetic Skin Tissue. Ann. Dermatol. 2013, 25, 423–427. [Google Scholar] [CrossRef]
- O’Reilly, M.; Gathercole, L.; Capper, F.; Arlt, W.; Tomlinson, J. Effect of insulin on AKR1C3 expression in female adipose tissue: In-vivo and in-vitro study of adipose androgen generation in polycystic ovary syndrome. Lancet 2015, 385, S0140–S6736. [Google Scholar] [CrossRef]
- Fujino, T.; Asaba, H.; Kang, M.J.; Ikeda, Y.; Sone, H.; Takada, S.; Kim, D.H.; Ioka, R.X.; Ono, M.; Tomoyori, H.; et al. Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc. Natl. Acad. Sci. USA 2003, 100, 229–234. [Google Scholar] [CrossRef]
- Zenibayashi, M.; Miyake, K.; Horikawa, Y.; Hirota, Y.; Teranishi, T.; Kouyama, K.; Sakaguchi, K.; Takeda, J.; Kasuga, M. Lack of association of LRP5 and LRP6 polymorphisms with type 2 diabetes mellitus in the Japanese population. Endocr. J. 2008, 55, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.F.; Xiong, D.H.; Shen, H.; Zhao, L.J.; Xiao, P.; Guo, Y.; Wang, W.; Yang, T.L.; Recker, R.R.; Deng, H.W. Polymorphisms of the low-density lipoprotein receptor-related protein 5 (LRP5) gene are associated with obesity phenotypes in a large family-based association study. J. Med. Genet. 2006, 43, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.F.; Ma, G.; Feng, N.H.; Yu, D.; Wu, Y.; Li, C. Twist2 and CD24 expression alters renal microenvironment in obesity associated kidney cancer. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, A.T.; Mejhert, N.; Jernås, M.; Carlsson, L.M.; Dahlman, I.; Laurencikiene, J.; Arner, P.; Rydén, M. Twist1 in human white adipose tissue and obesity. J. Clin. Endocrinol. Metab. 2011, 96, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Maj, M.; Gartner, W.; Ilhan, A.; Neziri, D.; Attems, J.; Wagner, L. Expression of TAU in insulin-secreting cells and its interaction with the calcium-binding protein secretagogin. J. Endocrinol. 2010, 205, 25–36. [Google Scholar] [CrossRef]
- Daimon, M.; Oizumi, T.; Karasawa, S.; Kaino, W.; Takase, K.; Tada, K.; Jimbu, Y.; Wada, K.; Kameda, W.; Susa, S.; et al. Association of the clusterin gene polymorphisms with type 2 diabetes mellitus. Metabolism 2011, 60, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.J.; Ju, T.J.; Heo, J.Y.; Kim, Y.W.; Kim, J.Y.; Won, K.C.; Kim, J.R.; Bae, Y.K.; Park, I.S.; Min, B.H.; et al. Deficiency of clusterin exacerbates high-fat diet-induced insulin resistance in male mice. Endocrinology 2014, 155, 2089–2101. [Google Scholar] [CrossRef]
- Iwasaki, T.; Yoneda, M.; Nakajima, A.; Terauchi, Y. Serum butyrylcholinesterase is strongly associated with adiposity, the serum lipid profile and insulin resistance. Intern. Med. 2007, 46, 1633–1639. [Google Scholar] [CrossRef]
- Lin, H.Y.; Yu, I.C.; Wang, R.S.; Chen, Y.T.; Liu, N.C.; Altuwaijri, S.; Hsu, C.L.; Ma, W.L.; Jokinen, J.; Sparks, J.D.; et al. Increased hepatic steatosis and insulin resistance in mice lacking hepatic androgen receptor. Hepatology 2008, 47, 1924–1935. [Google Scholar] [CrossRef]
- Chen, Q.; Yang, H.; Feng, Y.; Zhang, P.; Wu, W.; Li, S.; Thompson, B.; Wang, X.; Peng, T.; Wang, F.; et al. SOS1 gene polymorphisms are associated with gestational diabetes mellitus in a Chinese population: Results from a nested case-control study in Taiyuan, China. Diab. Vasc. Dis. Res. 2018, 15, 158–161. [Google Scholar] [CrossRef]
- Spálová, J.; Zamrazilová, H.; Vcelák, J.; Vanková, M.; Lukásová, P.; Hill, M.; Hlavatá, K.; Srámková, P.; Fried, M.; Aldhoon, B.; et al. Neuromedin beta: P73T polymorphism in overweight and obese subjects. Physiol. Res. 2008, 57 (Suppl. 1), S39–S48. [Google Scholar]
- Luo, Y.; Qu, H.; Wang, H.; Wu, J.; Duan, Y.; Liu, D.; Deng, H. Plasma Periostin Levels Are Increased in Chinese Subjects with Obesity and Type 2 Diabetes and Are Positively Correlated with Glucose and Lipid Parameters. Mediat. Inflamm. 2016, 2016, 6423637. [Google Scholar] [CrossRef]
- De Franco, E.; Shaw-Smith, C.; Flanagan, S.E.; Shepherd, M.H.; Hattersley, A.T.; Ellard, S. GATA6 mutations cause a broad phenotypic spectrum of diabetes from pancreatic agenesis to adult-onset diabetes without exocrine insufficiency. Diabetes 2013, 62, 993–997. [Google Scholar] [CrossRef] [PubMed]
- Ziolkowski, A.F.; Popp, S.K.; Freeman, C.; Parish, C.R.; Simeonovic, C.J. Heparan sulfate and heparanase play key roles in mouse β cell survival and autoimmune diabetes. J. Clin. Investig. 2012, 122, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Talmud, P.J.; Cooper, J.A.; Gaunt, T.; Holmes, M.V.; Shah, S.; Palmen, J.; Drenos, F.; Shah, T.; Kumari, M.; Kivimaki, M.; et al. Variants of ADRA2A are associated with fasting glucose, blood pressure, body mass index and type 2 diabetes risk: Meta-analysisof four prospective studies. Diabetologia 2011, 54, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- Shunmugam, V.; Say, Y.H. Evaluation of Association of ADRA2A rs553668 and ACE I/D Gene Polymorphisms with Obesity Traits in the Setapak Population, Malaysia. Iran. Red. Crescent Med. J. 2016, 18, e22452. [Google Scholar] [CrossRef] [PubMed]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pujar, M.K.; Vastrad, B.; Vastrad, C. Integrative Analyses of Genes Associated with Subcutaneous Insulin Resistance. Biomolecules 2019, 9, 37. https://doi.org/10.3390/biom9020037
Pujar MK, Vastrad B, Vastrad C. Integrative Analyses of Genes Associated with Subcutaneous Insulin Resistance. Biomolecules. 2019; 9(2):37. https://doi.org/10.3390/biom9020037
Chicago/Turabian StylePujar, Manoj Kumar, Basavaraj Vastrad, and Chanabasayya Vastrad. 2019. "Integrative Analyses of Genes Associated with Subcutaneous Insulin Resistance" Biomolecules 9, no. 2: 37. https://doi.org/10.3390/biom9020037
APA StylePujar, M. K., Vastrad, B., & Vastrad, C. (2019). Integrative Analyses of Genes Associated with Subcutaneous Insulin Resistance. Biomolecules, 9(2), 37. https://doi.org/10.3390/biom9020037




