Proteomic Response of Pseudomonas putida KT2440 to Dual Carbon-Phosphorus Limitation during mcl-PHAs Synthesis
Abstract
1. Introduction
2. Results and Discussion
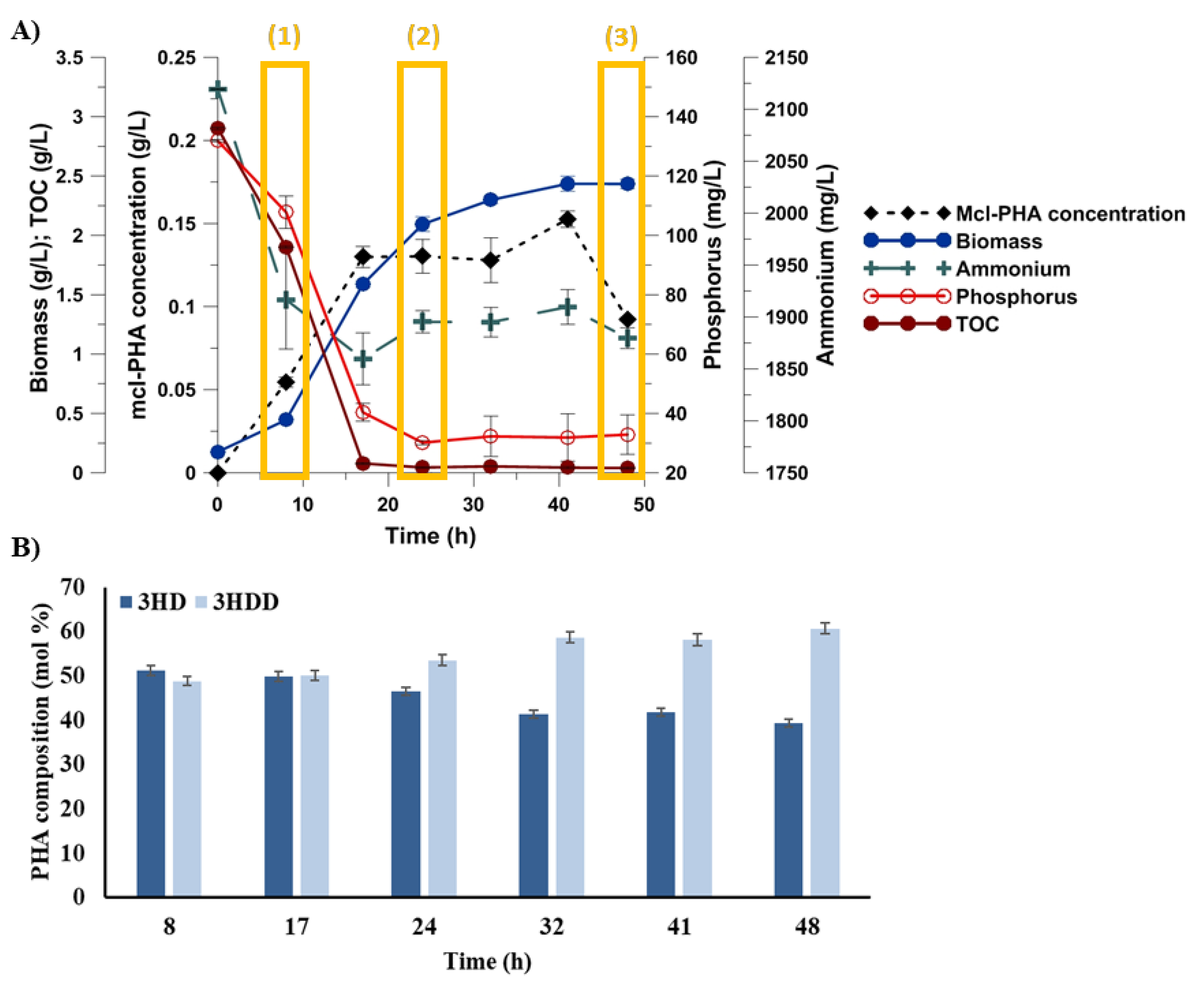
2.1. The Influence of Phosphorus Availability on Growth, and mcl-PHAs Synthesis and Composition
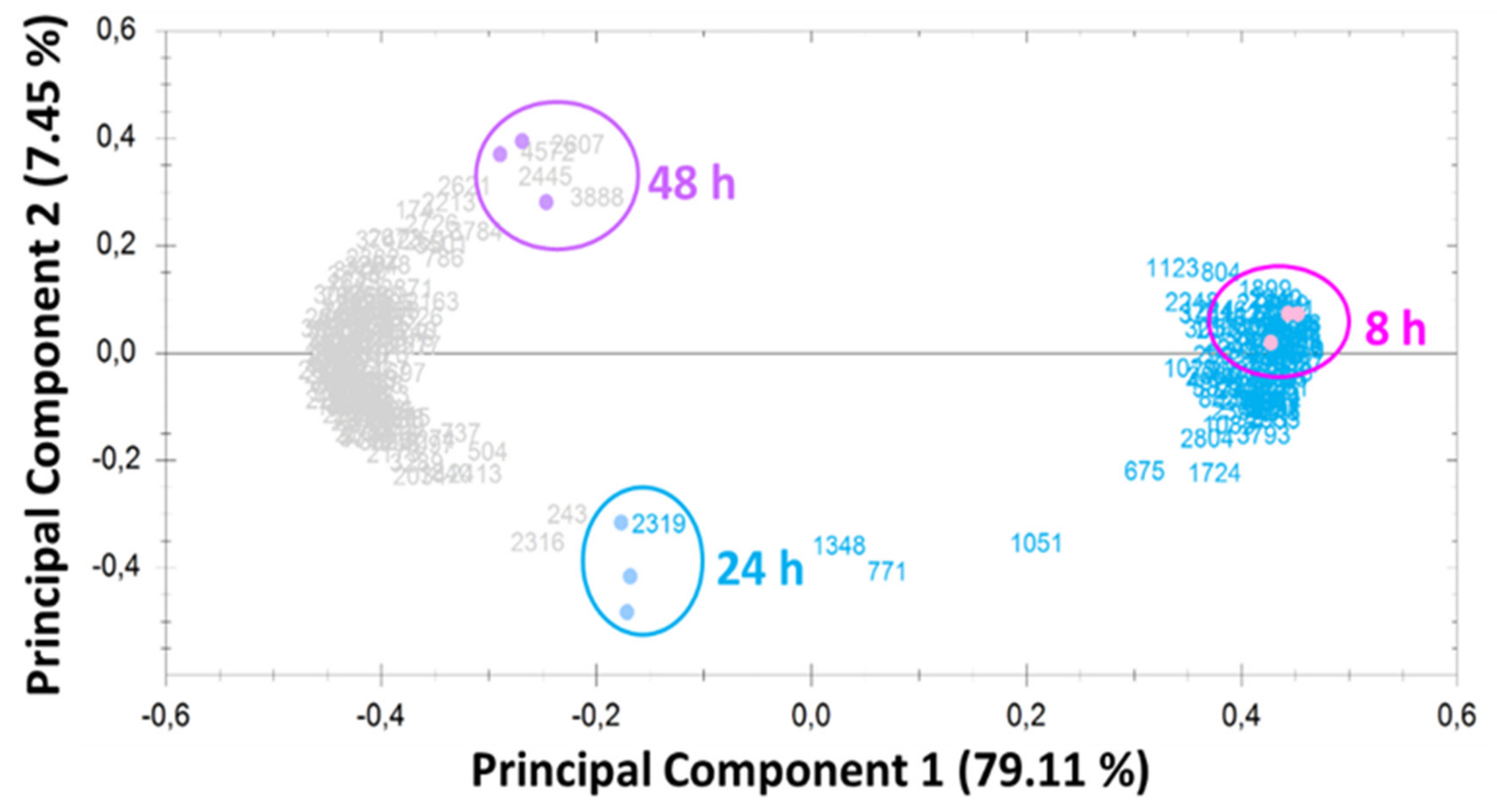
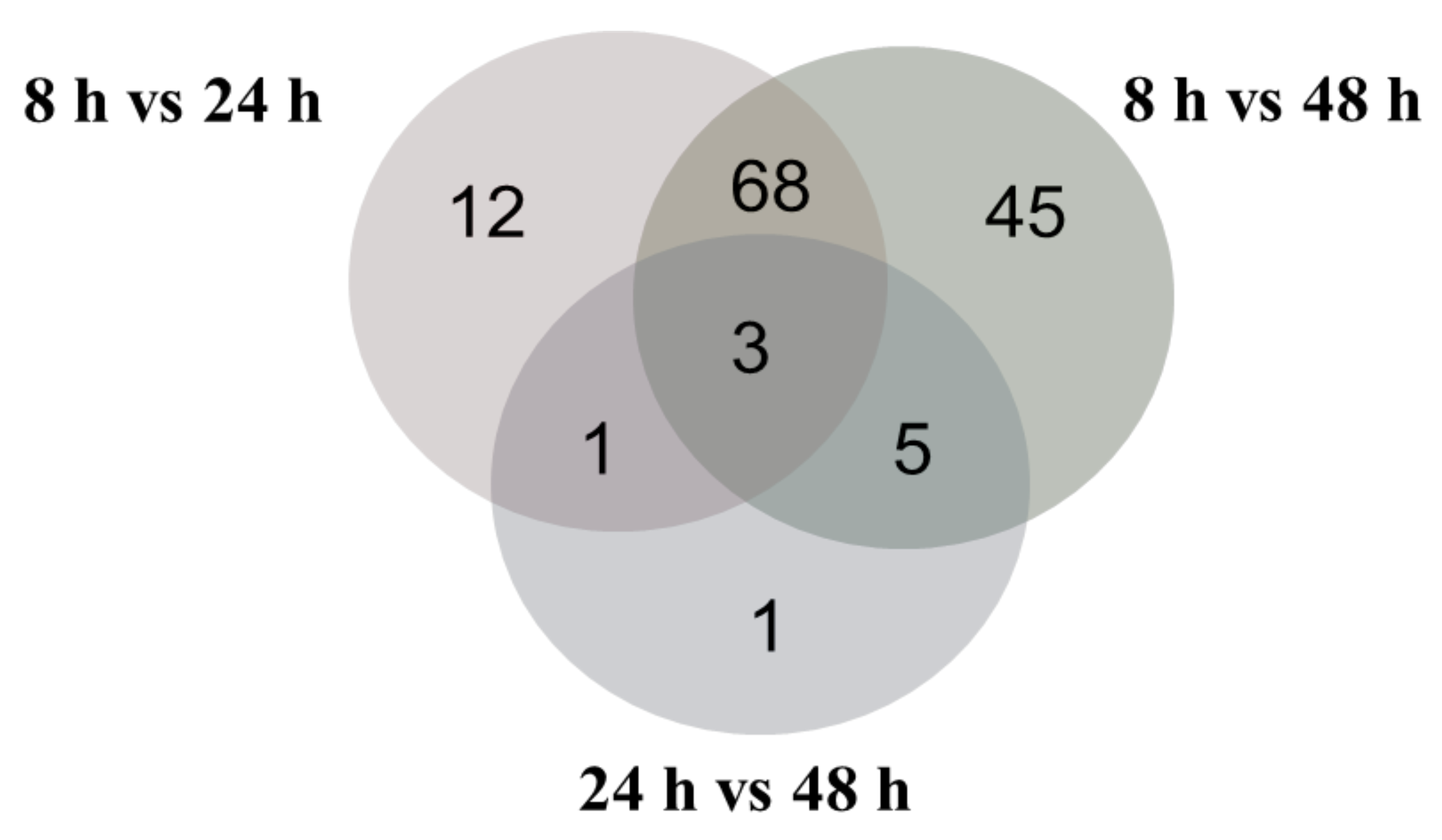
2.2. Overview of Proteomic Analysis
2.3. Proteome Changes during mcl-PHAs Synthesis and Different Growth Phases
2.3.1. Carbon and Energy Metabolism
2.3.2. PHA Biosynthesis
2.3.3. Amino Acid Metabolism and Biosynthesis
2.3.4. Stress Response
2.3.5. Replication, Transcription, and Translation
2.3.6. Transport
2.3.7. Other Proteins
3. Material and Methods
3.1. Cultivation and Analytical Procedures
3.2. Protein Extraction and Two-Dimensional Gel Electrophoresis (2-DE)
3.3. Image Analysis
3.4. Spots Identification Using Matrix-Assisted Laser Desorption/ Ionization Time-of-Flight Tandem Mass Spectrometry
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tan, G.Y.A.; Chen, C.L.; Li, L.; Ge, L.; Wang, L.; Razaad, I.M.N.; Li, Y.; Zhao, L.; Mo, Y.; Wang, J.Y. Start a research on biopolymer polyhydroxyalkanoate (PHA): A Review. Polymers 2014, 6, 706–754. [Google Scholar] [CrossRef]
- Butt, F.; Muhammad, N.; Hamid, A.; Moniruzzaman, M.; Sharif, F. Recent progress in the utilization of biosynthesized polyhydroxyalkanoates for biomedical applications—Review. Int. J. Biol. Macromol. 2018, 120, 1294–1305. [Google Scholar] [CrossRef]
- Belda, E.; van Heck, R.G.; José Lopez-Sanchez, M.; Cruveiller, S.; Barbe, V.; Fraser, C.; Klenk, H.P.; Petersen, J.; Morgat, A.; Nikel, P.I.; et al. The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ. Microbiol. 2016, 18, 3403–3424. [Google Scholar] [CrossRef]
- Nikel, P.I.; Martínez-García, E.; de Lorenzo, V. Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol. 2014, 12, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, A.; Arias-Barrau, E.; Arcos, M.; Naharro, G.; Olivera, E.R.; Luengo, J.M. Genetic and ultra-structural analysis of different mutants of Pseudomonas putida affected in the poly-3-hydroxy-n-alkanoate gene cluster. Environ. Microbiol. 2007, 9, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Prieto, A.; Escapa, I.F.; Martínez, V.; Dinjaski, N.; Herencias, C.; de la Peña, F.; Tarazona, N.; Revelles, O. A holistic view of polyhydroxyalkanoate metabolism in Pseudomonas putida. Environ. Microbiol. 2016, 18, 341–357. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Tappel, R.C.; Zhu, C.; Nomura, C.T. Development of a New Strategy for Production of Medium-Chain-Length Polyhydroxyalkanoates by Recombinant Escherichia coli via Inexpensive Non-Fatty Acid Feedstocks. Appl. Environ. Microbiol. 2012, 78, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Mozejko-Ciesielska, J.; Pokoj, T.; Ciesielski, S. Transcriptome remodeling of Pseudomonas putida KT2440 during mcl-PHAs synthesis: Effect of different carbon sources and response to nitrogen stress. J. Ind. Microbiol. Biotechnol. 2018, 45, 433–446. [Google Scholar] [CrossRef]
- Nikodinovic-Runic, J.; Flanagan, M.; Hume, A.R.; Cagney, G.; O’Connor, K.E. Analysis of the Pseudomonas putida CA-3 proteome during growth on styrene under nitrogen-limiting and non-limiting conditions. Microbiol. 2009, 155, 3348–3361. [Google Scholar] [CrossRef][Green Version]
- Poblete-Castro, I.; Escapa, I.F.; Jäger, C.; Puchalka, J.; Lam, C.M.C.; Schomburg, D.; Prieto, M.A.; dos Santos, V.M. The metabolic response of P. putida KT2442 producing high levels of polyhydroxyalkanoate under single- and multiple-nutrient-limited growth: Highlights from a multi-level omics approach. Microb. Cell Fact. 2012, 11, 34. [Google Scholar] [CrossRef]
- Fu, J.; Sharma, P.; Spicer, V.; Krokhin, O.V.; Zhang, X.; Fristensky, B.; Cicek, N.; Sparling, R.; Levin, D.B. Quantitative Omics Analyses of Medium Chain Length Polyhydroxyalkanaote Metabolism in Pseudomonas putida LS46 Cultured with Waste Glycerol and Waste Fatty Acids. PLoS ONE 2015, 10, e0142322. [Google Scholar] [CrossRef] [PubMed]
- Możejko-Ciesielska, J.; Mostek, A. Time-Course Proteomic Analysis of Pseudomonas putida KT2440 during Mcl-Polyhydroxyalkanoate Synthesis under Nitrogen Deficiency. Polymers 2019, 26, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, S.; Zhu, Z.; Shen, H.; Lin, X.; Jin, X.; Jiao, X.; Zhao, Z.K. Systems analysis of phosphate-limitation-induced lipid accumulation by the oleaginous yeast Rhodosporidium toruloides. Biotechnol. Biofuels 2018, 11, 148. [Google Scholar] [CrossRef]
- Eberl, L.; Givskov, M.; Sternberg, C.; Møller, S.; Christiansen, G.; Molin, S. Physiological responses of Pseudomonas putida KT2442 to phosphate starvation. Microbiology 1996, 142, 155–163. [Google Scholar] [CrossRef]
- Bains, M.; Fernández, L.; Hancock, R.E. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2012, 78, 6762–6768. [Google Scholar] [CrossRef]
- Lee, S.H.; Oh, D.H.; Ahn, W.S.; Lee, Y.; Choi, J.; Lee, S.Y. Production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) by high-cell-density cultivation of Aeromonas hydrophila. Biotechnol. Bioeng. 2000, 67, 240–244. [Google Scholar] [CrossRef]
- Diniz, S.C.; Taciro, M.K.; Gomez, J.G.; da Cruz Pradella, J.G. High-cell-density cultivation of Pseudomonas putida IPT 046 and medium-chain-length polyhydroxyalkanoate production from sugarcane carbohydrates. Appl. Biochem. Biotechnol. 2004, 119, 51–70. [Google Scholar] [CrossRef]
- Możejko-Ciesielska, J.; Pokój, T. Exploring nutrient limitation for polyhydroxyalkanoates synthesis by newly isolated strains of Aeromonas sp. using biodiesel-derived glycerol as a substrate. PeerJ 2018, 6, e5838. [Google Scholar] [CrossRef]
- Możejko-Ciesielska, J.; Mostek, A. A 2D-DIGE-based proteomic analysis brings new insights into cellular responses of Pseudomonas putida KT2440 during polyhydroxyalkanoates synthesis. Microb. Cell Fact. 2019, 18, 93. [Google Scholar] [CrossRef]
- Wang, Q.; Nomura, C.T. Monitoring differences in gene expression levels and polyhydroxyalkanoate (PHA) production in Pseudomonas putida KT2440 grown on different carbon sources. J. Biosci. Bioeng. 2010, 110, 653–659. [Google Scholar] [CrossRef]
- Dinjaski, N.; Prieto, M.A. Swapping of phasin modules to optimize the in vivo immobilization of proteins to medium-chain-length polyhydroxyalkanoate granules in Pseudomonas putida. Biomacromolecules 2013, 14, 3285–3293. [Google Scholar] [CrossRef] [PubMed]
- Maestro, B.; Galan, B.; Alfonso, C.; Rivas, G.; Prieto, M.A.; Sanz, J.M. A new family of intrinsically disordered proteins: Structural characterization of the major phasin PhaF from Pseudomonas putida KT2440. PLoS ONE 2013, 8, e56904. [Google Scholar] [CrossRef] [PubMed]
- Mato, A.; Tarazona, N.A.; Hidalgo, A.; Cruz, A.; Jiménez, M.; Pérez-Gil, J.; Prieto, M.A. Interfacial Activity of Phasin PhaF from Pseudomonas putida KT2440 at Hydrophobic-Hydrophilic Biointerfaces. Langmuir 2019, 35, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Galan, B.; Dinjaski, N.; Maestro, B.; de Eugenio, L.I.; Escapa, I.F.; Sanz, J.M. Nucleoid-associated PhaF phasin drives intracellular location and segregation of polyhydroxyalkanoate granules in Pseudomonas putida KT2442. Mol. Microbiol. 2011, 79, 402–418. [Google Scholar] [CrossRef] [PubMed]
- Prieto, M.A.; Bühler, B.; Jung, K.; Witholt, B.; Kessler, B. PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GPo1 involved in the regulatory expression system for pha genes. J. Bacteriol. 1999, 181, 858–868. [Google Scholar] [PubMed]
- Chowdhury, E.K.; Nagata, S.; Misono, H. 3-Hydroxyisobutyrate dehydrogenase from Pseudomonas putida E23: Purification and characterization. Biosci. Biotechnol. Biochem. 1996, 60, 2043–2047. [Google Scholar] [CrossRef]
- Joshi, V.; Joung, J.G.; Fei, Z.; Jander, G. Interdependence of threonine, methionine and isoleucine metabolism in plants: Accumulation and transcriptional regulation under abiotic stress. Amino Acids 2010, 39, 933–947. [Google Scholar] [CrossRef]
- Schertl, P.; Danne, L.; Braun, H.P. 3-Hydroxyisobutyrate Dehydrogenase Is Involved in Both, Valine and Isoleucine Degradation in Arabidopsis thaliana. Plant Physiol. 2017, 175, 51–61. [Google Scholar] [CrossRef]
- Fukui, K.A.; Hongo, K.; Mizobata, T.; Kawata, Y. Modulating the Effects of the Bacterial Chaperonin GroEL on Fibrillogenic Polypeptides through Modification of Domain Hinge Architecture. J. Biol. Chem. 2016, 291, 25217–25226. [Google Scholar] [CrossRef]
- Li, S.S.; Hu, X.; Zhao, H.; Li, Y.X.; Zhang, L.; Gong, L.J.; Guo, J.; Zhao, H.B. Quantitative analysis of cellular proteome alterations of Pseudomonas putida to naphthalene-induced stress. Biotechnol. Lett. 2015, 37, 1645–1654. [Google Scholar] [CrossRef]
- Kurbatov, L.; Albrecht, D.; Herrmann, H.; Petruschka, L. Analysis of the proteome of Pseudomonas putida KT2440 grown on different sources of carbon and energy. Environ. Microbiol. 2006, 8, 466–478. [Google Scholar] [CrossRef] [PubMed]
- Kluck, C.J.; Patzelt, H.; Genevaux, P.; Brehmer, D.; Rist, W.; Schneider-Mergener, J.; Bukau, B.; Mayer, M.P. Structure-function analysis of HscC, the Escherichia coli member of a novel subfamily of specialized Hsp70 chaperones. J. Biol. Chem. 2002, 277, 41060–41069. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, R.; Pastore, A. The role of chaperones in iron-sulfur cluster biogenesis. FEBS Lett. 2018, 592, 4011–4019. [Google Scholar] [CrossRef] [PubMed]
- Genevaux, P.; Georgopoulos, C.; Kelley, W.L. The Hsp70 chaperone machines of Escherichia coli: A paradigm for the repartition of chaperone functions. Mol. Microbiol. 2007, 66, 840–857. [Google Scholar] [CrossRef] [PubMed]
- Régnier, P.; Arraiano, C.M. Degradation of mRNA in bacteria: Emergence of ubiquitous features. Bioessays 2000, 22, 235–244. [Google Scholar] [CrossRef]
- Yehudai-Resheff, S.; Hirsh, M.; Schuster, G. Polynucleotide Phosphorylase Functions as Both an Exonuclease and a Poly (A) Polymerase in Spinach Chloroplasts. Mol. Cell. Biol. 2001, 21, 5408–5416. [Google Scholar] [CrossRef]
- Zou, Z.; Qin, H.; Brenner, A.E.; Raghavan, R.; Millar, J.A.; Gu, Q.; Xie, Z.; Kreth, J.; Merritt, J. LytTR Regulatory Systems: A potential new class of prokaryotic sensory system. PLoS Genet. 2018, 14, e1007709. [Google Scholar] [CrossRef]
- Talavera, A.; Hendrix, J.; Versées, W.; Jurėnas, D.; Van Nerom, K.; Vandenberk, N.; Singh, R.K.; Konijnenberg, A.; De Gieter, S.; Castro-Roa, D.; et al. Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors. Sci. Adv. 2018, 4, eaap9714. [Google Scholar] [CrossRef]
- Li, K.; Jiang, T.; Yu, B.; Wang, L.; Gao, C.; Ma, C.; Xu, P.; Ma, Y. Transcription Elongation Factor GreA Has Functional Chaperone Activity. PLoS ONE. 2012, 7, e47521. [Google Scholar] [CrossRef][Green Version]
- Guha, S.; Udupa, S.; Ahmed, W.; Nagaraja, V. Rewired downregulation of DNA Gyrase impacts cell division, expression of topology modulators, and transcription in Mycobacterium smegmatis. J. Mol. Biol. 2018, 430, 4986–5001. [Google Scholar] [CrossRef]
- Fang, H.; Huangfu, L.; Chen, R.; Li, P.; Xu, S.; Zhang, E.; Cao, W.; Liu, L.; Yao, Y.; Liang, G.; et al. Ancestor of land plants acquired the DNA-3-methyladenine glycosylase (MAG) gene from bacteria through horizontal gene transfer. Sci. Rep. 2017, 7, 9324. [Google Scholar] [CrossRef] [PubMed]
- Wilkens, S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Eitinger, T.; Rodionov, D.A.; Grote, M.; Schneider, E. Canonical and ECF-typeATP-binding cassette importers in prokaryotes: Diversity inmodular organization and cellular functions. FEMS Microbiol. Rev. 2011, 35, 3–67. [Google Scholar] [CrossRef] [PubMed]
- Clavel, T.; Germon, P.; Vianney, A.; Portalier, R.; Lazzaroni, J.C. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol. Microbiol. 1998, 29, 359–367. [Google Scholar] [CrossRef]
- Clements, J.M.; Coignard, F.; Johnson, I.; Chandler, S.; Palan, S.; Waller, A.; Wijkmans, J.; Hunter, M.G. Antibacterial activities and characterization of novel inhibitors of LpxC. Antimicrob. Agents Chemother. 2002, 46, 1793–1799. [Google Scholar] [CrossRef]
- Smith, J.L. The physiological role of ferritin-like compounds in bacteria. Crit. Rev. Microbiol. 2004, 30, 173–185. [Google Scholar] [CrossRef]
- Chekabab, S.M.; Harel, J.; Dozois, C.M. Interplay between genetic regulation of phosphate homeostasis and bacterial virulence. Virulence 2014, 5, 786–793. [Google Scholar] [CrossRef]
- Mozejko, J.; Wilke, A.; Przybyłek, G.; Ciesielski, S. Mcl-PHAs produced by Pseudomonas sp. Gl01 using fed-batch cultivation with waste rapeseed oil as carbon source. J. Microbiol. Biotechnol. 2012, 22, 371–377. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Możejko-Ciesielska, J.; Serafim, L.S. Proteomic Response of Pseudomonas putida KT2440 to Dual Carbon-Phosphorus Limitation during mcl-PHAs Synthesis. Biomolecules 2019, 9, 796. https://doi.org/10.3390/biom9120796
Możejko-Ciesielska J, Serafim LS. Proteomic Response of Pseudomonas putida KT2440 to Dual Carbon-Phosphorus Limitation during mcl-PHAs Synthesis. Biomolecules. 2019; 9(12):796. https://doi.org/10.3390/biom9120796
Chicago/Turabian StyleMożejko-Ciesielska, Justyna, and Luísa S. Serafim. 2019. "Proteomic Response of Pseudomonas putida KT2440 to Dual Carbon-Phosphorus Limitation during mcl-PHAs Synthesis" Biomolecules 9, no. 12: 796. https://doi.org/10.3390/biom9120796
APA StyleMożejko-Ciesielska, J., & Serafim, L. S. (2019). Proteomic Response of Pseudomonas putida KT2440 to Dual Carbon-Phosphorus Limitation during mcl-PHAs Synthesis. Biomolecules, 9(12), 796. https://doi.org/10.3390/biom9120796





