Phenolic Composition Influences the Health-Promoting Potential of Bee-Pollen
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Bee-Pollen Samples
2.3. Preparation of Bee-Pollen Extracts
2.4. Determination of the Total Phenolic Content
2.5. High-Performance Thin-Layer Chromatography
2.6. UHPLC–LTQ Orbitrap MS
2.7. Statistical Analysis
3. Results and Discussion
3.1. Floral Origin of Bee-Pollen Loads
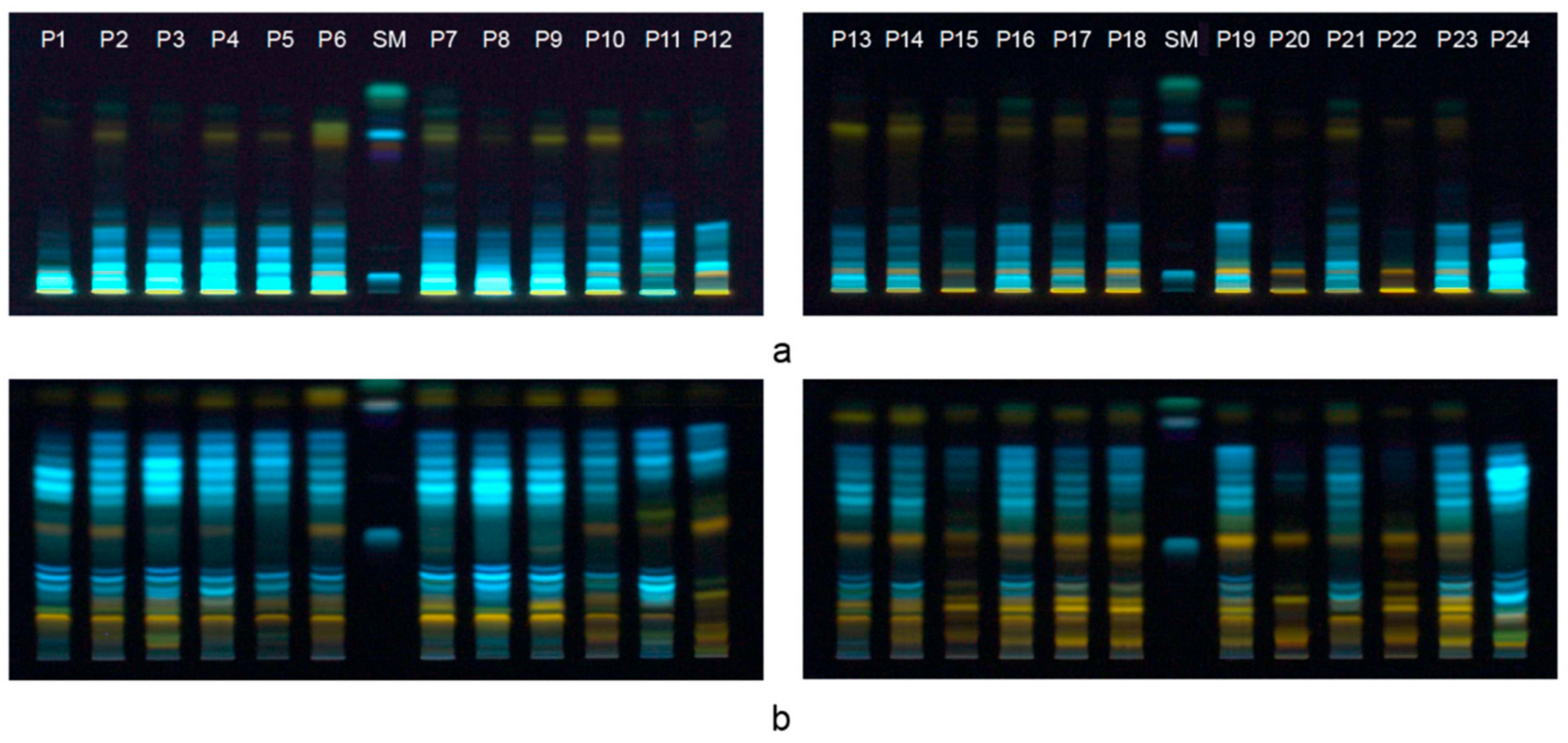
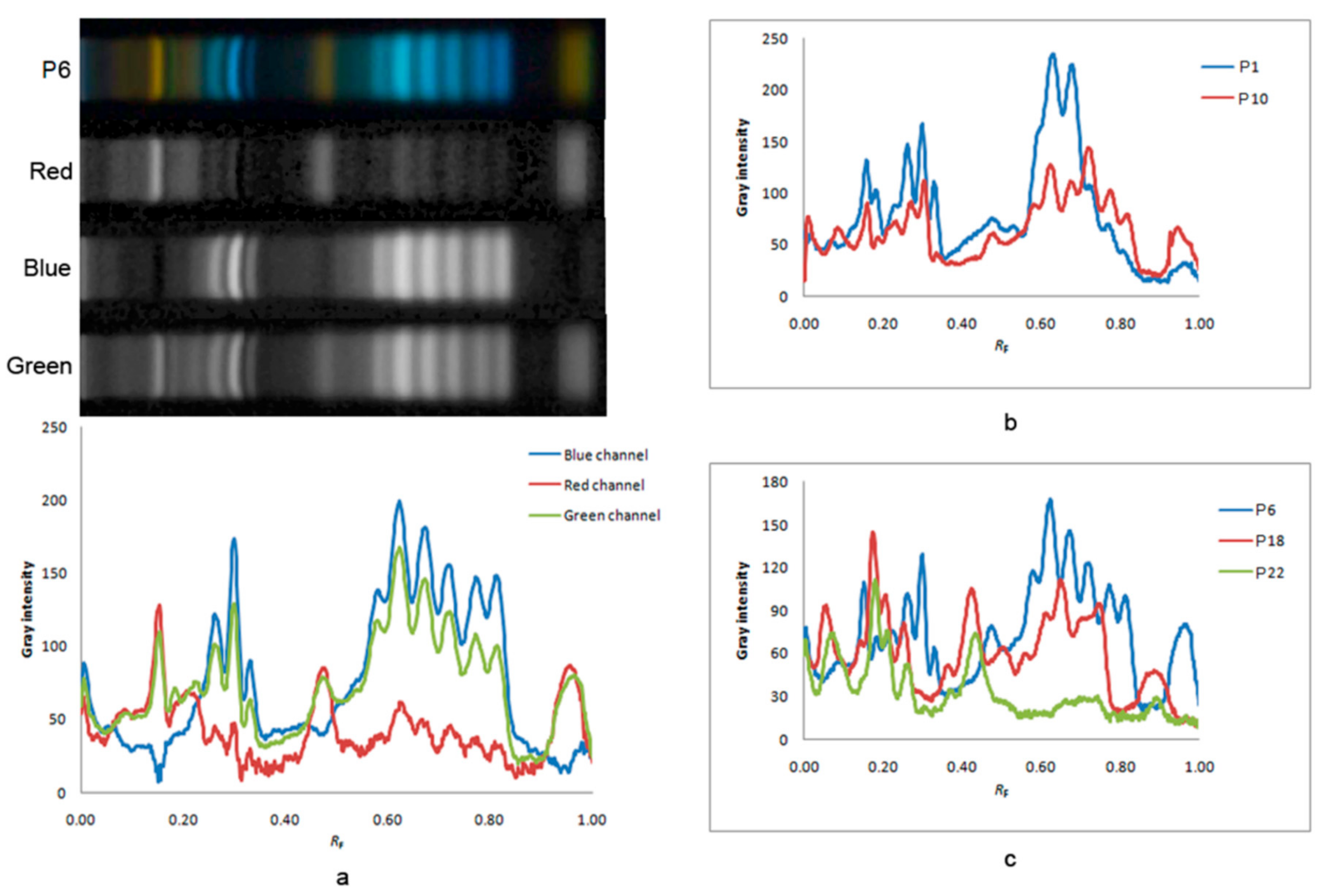
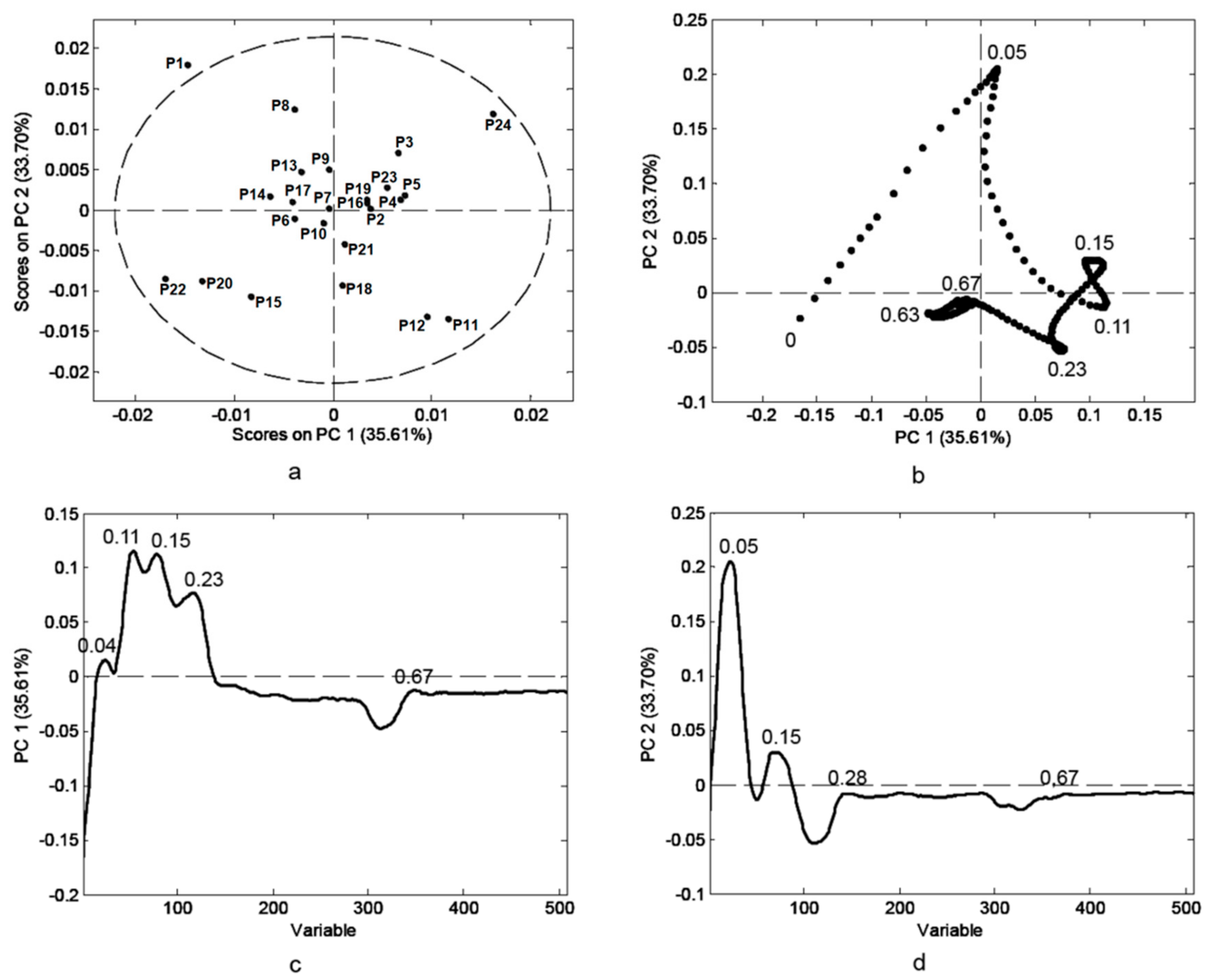
3.2. HPTLC Fingerprint of Phenolics in Bee-Pollen Samples
3.3. Identification of Flavonoid Glycosides by UHPLC–LTQ Orbitrap MS
3.4. The Total Phenolic Content of Bee-Pollen Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shoushtari, M.J.; Majd, A.; Pourpak, Z. Bee Pollen Flavonoids as a Therapeutic Agent in Allergic and Immunological Disorders. Iran. J. Allergy Asthma Immunol. 2017, 16, 171–182. [Google Scholar]
- Manach, C.; Scalbert, A.; Morand, C.; Rémésy, C.; Jiménez, L. Polyphenols: Food sources and bioavailability. Am. J. Clin. Nutr. 2004, 79, 727–747. [Google Scholar] [CrossRef]
- Campos, M.; Markham, K.R.; Mitchell, K.A.; da Cunha, A.P. An Approach to the Characterization of Bee Pollens via their Flavonoid/Phenolic Profiles. Phytochem. Anal. 1997, 8, 181–185. [Google Scholar] [CrossRef]
- Negri, G.; Teixeira, E.W.; Alves, M.L.; Moreti, A.C.; Otsuk, I.P.; Borguini, R.G.; Salatino, A. Hydroxycinnamic Acid Amide Derivatives, Phenolic Compounds and Antioxidant Activities of Extracts of Pollen Samples from Southeast Brazil. J. Agric. Food Chem. 2011, 59, 5516–5522. [Google Scholar] [CrossRef] [PubMed]
- Ferreres, F.; Pereira, D.M.; Valentão, P.; Andrade, P.B. First report of non-coloured flavonoids in Echium plantagineum bee pollen: Differentiation of isomers by liquid chromatography/ion trap mass spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 801–806. [Google Scholar] [CrossRef]
- Kostić, A.; Milinčić, D.; Gašić, U.; Nedić, N.; Stanojević, S.; Tešić, Ž.; Pešić, M. Polyphenolic profile and antioxidant properties of bee-collected pollen from sunflower (Helianthus annuus L.) plant. LWT—Food Sci. Technol. 2019, 112, 108224. [Google Scholar] [CrossRef]
- Rocchetti, G.; Castiglioni, S.; Maldarizzi, G.; Carloni, P.; Lucini, L. UHPLC-ESI-QTOF-MS phenolic profiling and antioxidant capacity of bee pollen from different botanical origin. Int. J. Food Sci. Technol. 2019, 54, 335–346. [Google Scholar] [CrossRef]
- Anjos, O.; Fernandes, R.; Cardoso, S.M.; Delgado, T.; Farinha, N.; Paula, V.; Estevinho, L.M.; Carpes, S.T. Bee pollen as a natural antioxidant source to prevent lipid oxidation in black pudding. LWT—Food Sci. Technol. 2019, 111, 869–875. [Google Scholar] [CrossRef]
- Mudrić, S.Ž.; Gašić, U.M.; Dramićanin, A.M.; Ćirić, I.Ž.; Milojković-Opsenica, D.M.; Popović-Đorđević, J.B.; Momirović, N.M.; Tešić, Ž.L. The polyphenolics and carbohydrates as indicators of botanical and geographical origin of Serbian autochthonous clones of red spice paprika. Food Chem. 2017, 217, 705–715. [Google Scholar] [CrossRef]
- Ferreres, F.; Magalhães, S.C.; Gil-Izquierdo, A.; Valentão, P.; Cabrita, A.R.; Fonseca, A.J.; Andrade, P.B. HPLC-DAD-ESI/MSn profiling of phenolic compounds from Lathyrus cicera L. Seeds. Food Chem. 2017, 214, 678–685. [Google Scholar] [CrossRef]
- Ferreres, F.; Llorach, R.; Gil-Izquierdo, A. Characterization of the interglycosidic linkage in di-, tri-, tetra- and pentaglycosylated flavonoids and differentiation of positional isomers by liquid chromatography/electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 2004, 39, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Cuyckens, F.; Claeys, M. Determination of the glycosylation site in flavonoid mono-O-glycosides by collision-induced dissociation of electrospray-generated deprotonated and sodiated molecules. J. Mass Spectrom. 2005, 40, 364–372. [Google Scholar] [CrossRef] [PubMed]
- De Rijke, E.; Out, P.; Niessen, W.M.A.; Ariese, F.; Gooijer, C.; Brinkman, U.A. Analytical separation and detection methods for flavonoids. J. Chromatogr. A 2006, 1112, 31–63. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, A.L.; Cavaliere, C.; Foglia, P.; Piovesana, S.; Ventura, S. Chromatographic Methods Coupled to Mass Spectrometry Detection for the Determination of Phenolic Acids in Plants and Fruits. J. Liq. Chromatogr. Relat. Technol. 2015, 38, 353–370. [Google Scholar] [CrossRef]
- Almaraz-Abarca, N.; Campos, M.G.; Ávila-Reyes, J.A.; Naranjo-Jiménez, N.; Corral, J.H.; González-Valdez, L.S. Antioxidant activity of polyphenolic extract of monofloral honeybee-collected pollen from mesquite (Prosopis juliflora, Leguminosae). J. Food Compos. Anal. 2007, 20, 119–124. [Google Scholar] [CrossRef]
- Morais, M.; Moreira, L.; Feás, X.; Estevinho, L.M. Honeybee-collected pollen from five Portuguese Natural Parks: Palynological origin, phenolic content, antioxidant properties and antimicrobial activity. Food Chem. Toxicol. 2011, 49, 1096–1101. [Google Scholar] [CrossRef]
- LeBlanc, B.W.; Davis, O.K.; Boue, S.; DeLucca, A.; Deeby, T. Antioxidant activity of Sonoran Desert bee pollen. Food Chem. 2009, 115, 1299–1305. [Google Scholar] [CrossRef]
- Kostić, A.; Barać, M.; Stanojević, S.; Milojković-Opsenica, D.; Tešić, Ž.; Šikoparija, B.; Radišić, P.; Prentović, M.; Pešić, M. Physicochemical composition and techno-functional properties of bee pollen collected in Serbia. LWT—Food Sci. Technol. 2015, 62, 301–309. [Google Scholar] [CrossRef]
- Kostić, A.; Pešić, M.; Mosić, M.; Dojčinović, B.; Natić, M.; Trifković, J. Mineral content of bee pollen from Serbia. Arch. Ind. Hyg. Toxicol. 2015, 66, 251–258. [Google Scholar]
- Kostić, A.; Petrović, T.; Krnjaja, V.; Nedić, N.; Tešić, Ž.; Milojković-Opsenica, D.; Barać, M.; Stanojević, S.; Pešić, M. Mold/aflatoxin contamination of honeybee collected pollen from different Serbian regions. J. Apic. Res. 2017, 56, 13–20. [Google Scholar] [CrossRef]
- Waisi, H.; Kosović, A.; Krstić, Đ.; Milojković-Opsenica, D.; Nikolić, B.; Dragičević, V.; Trifković, J. Polyphenolic Profile of Maize Seedlings Treated with 24-Epibrassinolide. J. Chem. 2015, 2015, 976971. [Google Scholar] [CrossRef]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventos, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of folinciocalteu reagent. Method. Enzymol. 1999, 299, 152–178. [Google Scholar]
- Ristivojević, P.; Andrić, F.; Trifković, J.; Vovk, I.; Stanisavljević, L.; Tešić, Ž.; Milojković-Opsenica, D. Pattern Recognition Methods and Multivariate Image Analysis in HPTLC Fingerprinting of Propolis Extracts. J. Chemom. 2014, 28, 301–310. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Advanced Chemometrics Software for use with MATLAB. Available online: http://eigenvector.com/software/pls-toolbox/ (accessed on 18 November 2019).
- Guffa, B.; Nedić, N.; Dabić Zagorac, D.; Tosti, T.; Gašić, U.; Natić, M.; Fotirić Akšić, M. Characterization of Sugar and Polyphenolic Diversity in Floral Nectar of Different ‘Oblačinska’ Sour Cherry Clones. Chem. Biodivers. 2017, 14, e1700061. [Google Scholar] [CrossRef] [PubMed]
- Barth, O.M.; Freitas, A.S.; Oliveira, E.S.; Silva, R.A.; Maester, F.M.; Andrella, R.R.; Cardozo, G.M. Evaluation of the botanical origin of commercial dry bee pollen load batches using pollen analysis: A proposal for technical standardization. An. Acad. Bras. Cienc. 2010, 82, 893–902. [Google Scholar] [CrossRef]
- Senguttuvan, J.; Subramaniam, P. HPTLC Fingerprints of Various Secondary Metabolites in the Traditional Medicinal Herb Hypochaeris radicata L. J. Bot. 2016, 2016, 5429625. [Google Scholar] [CrossRef]
- Ciesla, Ł.; Staszek, D.; Hajnos, M.; Kowalska, T.; Waksmundzka-Hajnos, M. Development of chromatographic and free radical scavenging activity fingerprints by thin-layer chromatography for selected Salvia species. Phytochem. Anal. 2011, 22, 59–65. [Google Scholar] [CrossRef]
- Ristivojević, P.; Trifković, J.; Vovk, I.; Milojković-Opsenica, D. Comparative study of different approaches for multivariate image analysis in HPTLC fingerprinting of natural products such as plant resin. Talanta 2017, 162, 72–79. [Google Scholar] [CrossRef]
- Castiglioni, S.; Astolfi, P.; Conti, C.; Monaci, E.; Stefano, M.; Carloni, P. Morphological, Physicochemical and FTIR Spectroscopic Properties of Bee Pollen Loads from Different Botanical Origin. Molecules 2019, 24, 3974. [Google Scholar] [CrossRef]
- Costa, M.C.; Morgano, M.A.; Ferreira, M.M.; Milani, R.F. Analysis of bee pollen constituents from different Brazilian regions: Quantification by NIR spectroscopy and PLS regression. LWT—Food Sci. Technol. 2017, 80, 76–83. [Google Scholar] [CrossRef]
- Mihajlović, L.; Radosavljević, J.; Burazer, L.; Smiljanić, K.; Ćirković-Veličković, T. Composition of polyphenol and polyamide compounds in common ragweed (Ambrosia artemisiifolia L.) pollen and sub-pollen particles. Phytochemistry 2015, 109, 125–132. [Google Scholar] [CrossRef] [PubMed]
- March, R.E.; Lewars, E.G.; Stadey, C.J.; Miao, X.S.; Zhao, X.M.; Metcalfe, C.D. A comparison of flavonoid glycosides by electrospray tandem mass spectrometry. Int. J. Mass Spectrom. 2006, 248, 61–85. [Google Scholar] [CrossRef]
- Cuyckens, F.; Rozenberg, R.; de Hoffmann, E.; Claeys, M. Structure characterization of flavonoid O-diglycosides by positive and negative nano-electrospray ionization ion trap mass spectrometry. J. Mass Spectrom. 2001, 36, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lin, Z.; Fang, J.; Liu, M.; Niu, Y.; Chen, S.; Wang, H. An on-line high-performance liquid chromatography–diode-array detector–electrospray ionization–ion-trap–time-of-flight–mass spectrometry–total antioxidant capacity detection system applying two antioxidant methods for activity evaluation of the edible flowers from Prunus mume. J. Chromatogr. A 2015, 1414, 88–102. [Google Scholar] [PubMed]
- Ablajan, K.; Abliz, A.; Shang, X.Y.; He, J.M.; Rui-Ping Zhang, R.P.; Shi, J.G. Structural characterization of flavonol 3,7- di-O glycosides and determination of the glycosylation position by using negative ion electrospray ionisation tandem mass spectrometry. J. Mass Spectrom. 2006, 41, 352–360. [Google Scholar] [CrossRef]
- Gonzales, G.B.; Raes, K.; Coelus, S.; Struijs, K.; Smagghe, G.; Van Campa, J. Ultra(high)-pressure liquid chromatography–electrospray ionization-time-of-flight-ion mobility-high definition mass spectrometry for the rapid identification and structural characterization of flavonoid glycosides from cauliflower waste. J. Chromatogr. A 2014, 1323, 39–48. [Google Scholar] [CrossRef]
- Rösch, D.; Krumbein, A.; Mügge, C.; Kroh, L.W. Structural investigation of flavonol glycosides from sea buchthorn (Hippophae rhamnoides) pomace by NMR spectroscopy and HPLC-ESI-MSn. J. Agric. Food Chem. 2004, 52, 4039–4046. [Google Scholar] [CrossRef]
- Goupy, P.; Vian, M.A.; Chemat, F.; Caris-Veyrat, C. Identification and quantification of flavonols, anthocyanins and lutein diesters in tepals of Crocus sativus by ultra-performance liquid chromatography coupled to diode array and ion trap mass spectrometry detections. Ind. Crops Prod. 2013, 44, 496–510. [Google Scholar] [CrossRef]
- Larbat, R.; Paris, C.; Le Bot, J.; Adamowicz, S. Phenolic characterization and variability in leaves, stems and roots of Micro-Tom and patio tomatoes, in response to nitrogen limitation. Plant Sci. 2014, 224, 62–73. [Google Scholar] [CrossRef]
- Zhou, J.; Qi, Y.; Ritho, J.; Zhang, Y.; Zheng, X.; Wu, L.; Li, Y.; Sun, L. Flavonoid glycosides as floral origin markers to discriminate of unifloral bee pollen by LC-MS/MS. Food Control 2015, 57, 54–61. [Google Scholar] [CrossRef]
- Mărghitaş, L.A.; Stanciu, O.G.; Dezmirean, D.S.; Bobiş, O.; Popescu, O.; Bogdanov, S.; Campos, M.G. In vitro antioxidant capacity of honeybee-collected pollen of selected floral origin harvested from Romania. Food Chem. 2009, 115, 878–883. [Google Scholar] [CrossRef]
- Kalaycıoğlu, Z.; Kaygusuz, H.; Döker, S.; Kolaylı, S.; Erim, B.F. Characterization of Turkish honeybee pollens by principal component analysis based on their individual organic acids, sugars, minerals, and antioxidant activities. LWT—Food Sci. Technol. 2017, 84, 402–408. [Google Scholar] [CrossRef]
- Pascoal, A.; Rodrigues, S.; Teixeira, A.; Feás, X.; Estevinho, L.M. Biological activities of commercial bee pollens: Antimicrobial, antimutagenic, antioxidant and anti-inflammatory. Food Chem. Toxicol. 2014, 63, 233–239. [Google Scholar] [CrossRef] [PubMed]
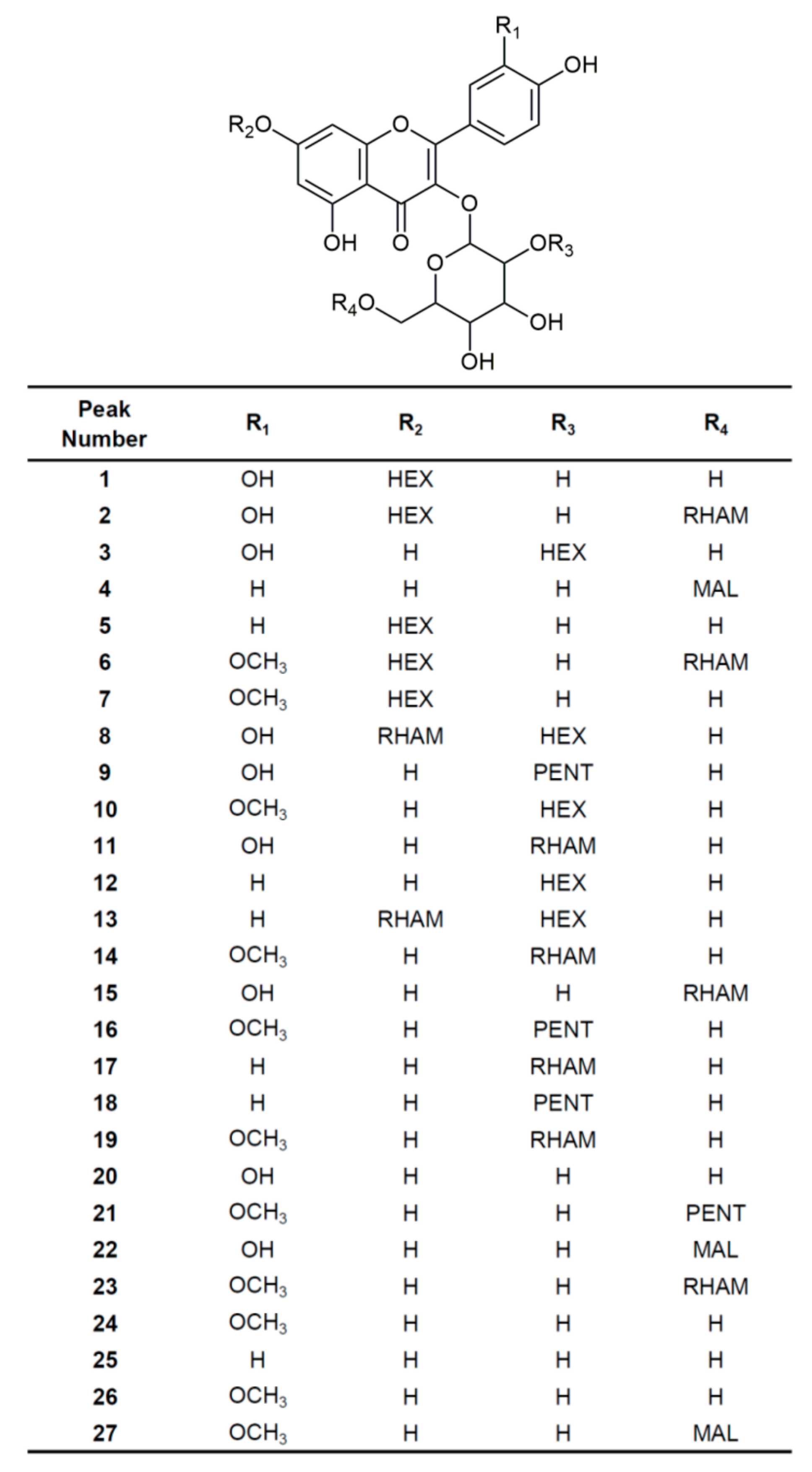
| Peak Number | tR, min | Molecular Formula, [M–H]− | Calculated Mass, [M–H]− | Exact Mass, [M–H]− | Δ, ppm | MS/MS Fragmentation (%) | Identification |
|---|---|---|---|---|---|---|---|
| 1 | 5.30 | C27H29O17− | 625.14102 | 625.14233 | 2.10 | 505(5), 463(100), 462(20), 343(5), 301(30) | Quercetin 3,7-di-O-hexoside |
| 2 | 5.74 | C33H39O21− | 771.19893 | 771.20038 | 1.88 | 609(100), 463(10), 301(5) | Quercetin 3-O-(6″-O-rhamnosyl)hexoside-7-O-hexoside b |
| 3 | 5.85 | C27H29O17− | 625.14102 | 625.14172 | 1.12 | 505(15), 463(15), 445(50), 343 (10), 301(50), 300(100), 271(20), 255(10) | Quercetin 3-O-(2″-O-hexosyl)hexoside |
| 4 | 5.89 | C24H21O14− | 533.09368 | 533.09454 | 1.61 | 489(100), 285(5) | Kaempferol 3-O-(6″-O-malonyl)hexoside |
| 5 | 5.90 | C27H29O16− | 609.14611 | 609.14636 | 0.41 | 581(10), 489(10), 447(70), 446(50), 327(5), 285(100) | Kaempferol 3,7-di-O-hexoside |
| 6 | 5.91 | C34H41O21− | 785.21458 | 785.21509 | 0.65 | 623(100), 477(10), 315(5) | Isorhamnetin 3-O-(6″-O-rhamnosyl)hexoside-7-O-hexosideb |
| 7 | 5.96 | C28H31O17− | 639.15667 | 639.15717 | 0.78 | 477(40), 476(20), 357(5), 315(100), 300(10) | Isorhamnetin 3,7-di-O-hexoside b |
| 8 | 5.98 | C33H39O21− | 771.19893 | 771.20056 | 2.11 | 753(10), 625(100), 609(30), 591(40), 573(20), 445(10), 427(10), 409(10), 301(45), 300(50) | Quercetin 3-O-(2″-O-hexosyl)hexoside-7-O-rhamnosideb |
| 9 | 6.00 | C26H27O16− | 595.13046 | 595.13098 | 0.87 | 475(10), 463(10), 445(15), 301(45), 300(100), 271(20), 255(10) | Quercetin 3-O-(2″-O-pentosyl)hexoside b |
| 10 | 6.02 | C28H31O17− | 639.15667 | 639.15784 | 1.83 | 624(20), 519(15), 491(10), 477(20), 459(65), 444(30), 315(100), 314(50), 300(50), 299(60) | Isorhamnetin 3-O-(2″-O-hexosyl)hexoside |
| 11 | 6.04 | C27H29O16− | 609.14611 | 609.14642 | 0.51 | 489(15), 463(10), 445(20), 429(10), 343(5), 301(20), 300(100), 285(5), 271(15), 255(10) | Quercetin 3-O-(2″-O-rhamnosyl)hexoside |
| 12 | 6.07 | C27H29O16− | 609.14611 | 609.14697 | 1.41 | 489(5), 447(10), 429(100), 327(10), 285(90), 284(80), 255(15) | Kaempferol 3-O-(2″-O-hexosyl)hexoside |
| 13 | 6.19 | C33H39O20− | 755.20402 | 755.20471 | 0.91 | 609(100), 593(90), 575(85), 429(20), 285(90), 284(70), 255(25) | Kaempferol 3-O-(2″-O-hexosyl)hexoside-7-O-rhamnoside b |
| 14 | 6.22 | C28H31O16− | 623.16176 | 623.16211 | 0.56 | 608(10), 503(10), 477(10), 459(30), 444(10), 327(10), 315(15), 314(100), 300(10), 299(65) | Isorhamnetin 3-O-(2″-O-rhamnosyl)hexoside isomer 1 |
| 15 | 6.24 | C27H29O16− | 609.14611 | 609.14679 | 1.12 | 343(5), 301(100), 300(20), 271(10), 255(5) | Quercetin 3-O-(6″-O-rhamnosyl)glucoside (Rutin) a |
| 16 | 6.26 | C27H29O16− | 609.14611 | 609.14716 | 1.72 | 577(15), 489(5), 477(5), 459(50), 357(10), 315(100), 314(90), 300(20), 299(30) | Isorhamnetin 3-O-(2″-O-pentosyl)hexoside b |
| 17 | 6.28 | C27H29O15− | 593.15119 | 593.15161 | 0.71 | 473(5), 447(10), 429(50), 327(5), 285(40), 284(100), 255(20) | Kaempferol 3-O-(2″-O-rhamnosyl)hexoside |
| 18 | 6.30 | C26H27O15− | 579.13554 | 579.13678 | 2.14 | 459(5), 447(20), 429(60), 327(10), 285(75), 284(100), 255(30) | Kaempferol 3-O-(2″-O-pentosyl)hexoside |
| 19 | 6.34 | C28H31O16− | 623.16176 | 623.16248 | 1.16 | 591(5), 503(10), 477(10), 459(25), 357(5), 315(35), 314(100), 300(10), 299(25) | Isorhamnetin 3-O-(2″-O-rhamnosyl)hexoside isomer 2 |
| 20 | 6.39 | C21H19O12− | 463.08820 | 463.08951 | 2.83 | 301(100), 300(20) | Quercetin 3-O-galactoside (Hyperoside) a |
| 21 | 6.43 | C27H29O16− | 609.14611 | 609.14697 | 1.41 | 315(100), 300(20), 271(10), 255(5) | Isorhamnetin 3-O-(6″-O-pentosyl)hexoside b |
| 22 | 6.54 | C24H21O15− | 549.08859 | 549.08923 | 1.17 | 505(100), 301(5) | Quercetin 3-O-(6″-O-malonyl)hexoside |
| 23 | 6.57 | C28H31O16− | 623.16176 | 623.16272 | 1.54 | 315(100), 314(10), 300(20), 271(10), 255(5) | Isorhamnetin 3-O-(6″-O-rhamnosyl)hexoside |
| 24 | 6.65 | C22H21O12− | 477.10385 | 477.10477 | 1.93 | 315(100), 314(20) | Isorhamnetin 3-O-hexoside isomer 1 |
| 25 | 6.70 | C21H19O11− | 447.09329 | 447.09424 | 2.12 | 327(20), 285(80), 284(100), 255(10) | Kaempferol 3-O-glucoside (Astragalin) a |
| 26 | 6.78 | C22H21O12− | 477.10385 | 477.10437 | 1.09 | 449(5), 357(15), 315(30), 314(100) | Isorhamnetin 3-O-hexoside isomer 2 |
| 27 | 6.86 | C25H23O15− | 563.10424 | 563.10486 | 1.10 | 519(100), 315(5) | Isorhamnetin 3-O-(6″-O-malonyl)hexoside |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mosić, M.; Trifković, J.; Vovk, I.; Gašić, U.; Tešić, Ž.; Šikoparija, B.; Milojković-Opsenica, D. Phenolic Composition Influences the Health-Promoting Potential of Bee-Pollen. Biomolecules 2019, 9, 783. https://doi.org/10.3390/biom9120783
Mosić M, Trifković J, Vovk I, Gašić U, Tešić Ž, Šikoparija B, Milojković-Opsenica D. Phenolic Composition Influences the Health-Promoting Potential of Bee-Pollen. Biomolecules. 2019; 9(12):783. https://doi.org/10.3390/biom9120783
Chicago/Turabian StyleMosić, Mirjana, Jelena Trifković, Irena Vovk, Uroš Gašić, Živoslav Tešić, Branko Šikoparija, and Dušanka Milojković-Opsenica. 2019. "Phenolic Composition Influences the Health-Promoting Potential of Bee-Pollen" Biomolecules 9, no. 12: 783. https://doi.org/10.3390/biom9120783
APA StyleMosić, M., Trifković, J., Vovk, I., Gašić, U., Tešić, Ž., Šikoparija, B., & Milojković-Opsenica, D. (2019). Phenolic Composition Influences the Health-Promoting Potential of Bee-Pollen. Biomolecules, 9(12), 783. https://doi.org/10.3390/biom9120783








