Ectopic Expression of Glycine max GmNAC109 Enhances Drought Tolerance and ABA Sensitivity in Arabidopsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of Transgenic Arabidopsis GmNAC109-EX Plants
2.2. Plant Growth Conditions
2.3. Growth Assessment of Transgenic Arabidopsis Plants under Normal Conditions
2.4. Drought Tolerance Assay
2.5. Water Loss Rate Determination
2.6. Assays for Endogenous Hydrogen Peroxide and ROS-Scavenging Enzyme Activities
2.7. Expression Analysis of Antioxidant Enzyme-Encoding Genes
2.8. ABA Sensitivity Assay
2.9. Statistical Analysis
3. Results
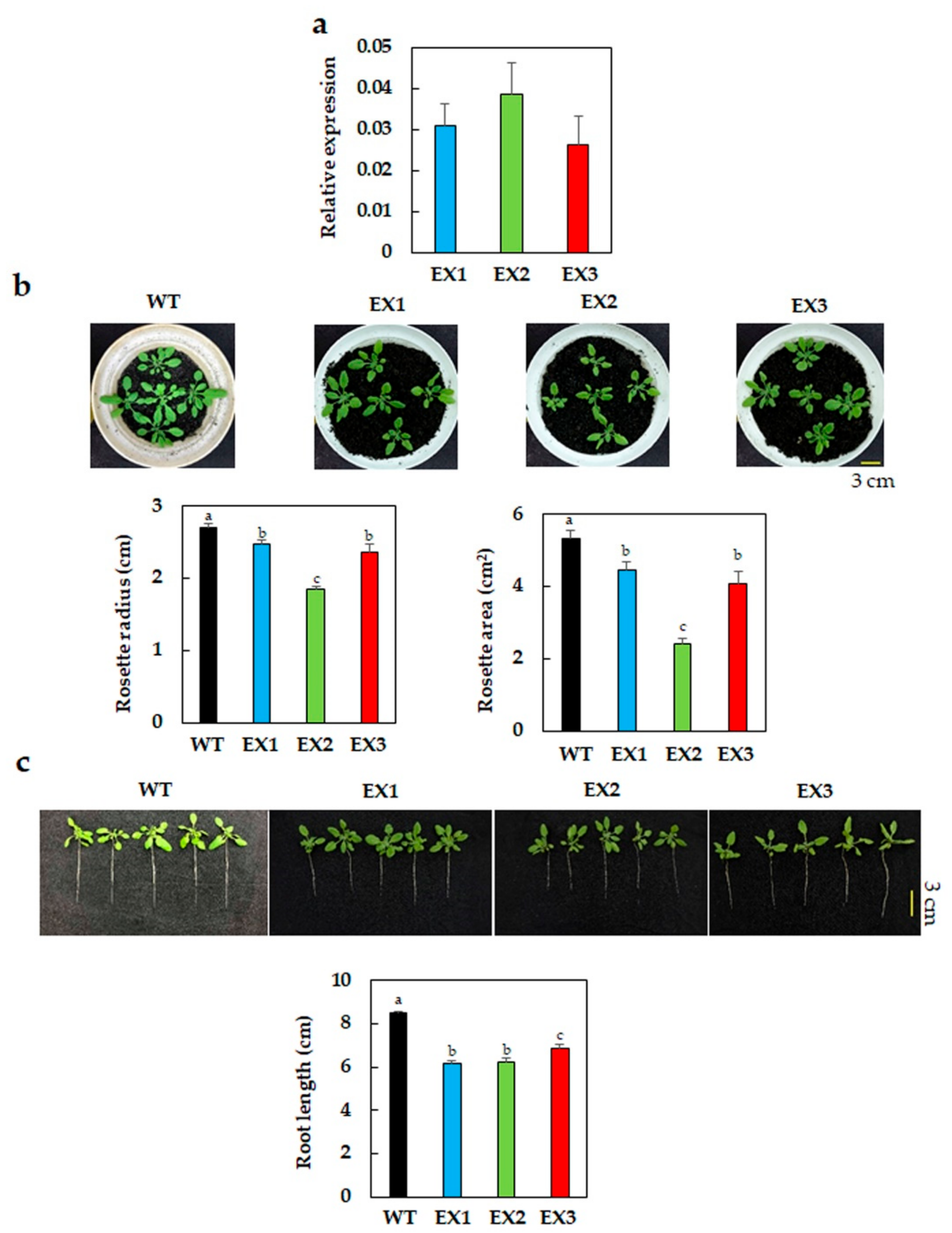
3.1. Reduced Growth Phenotype of GmNAC109-EX Lines under Normal Conditions
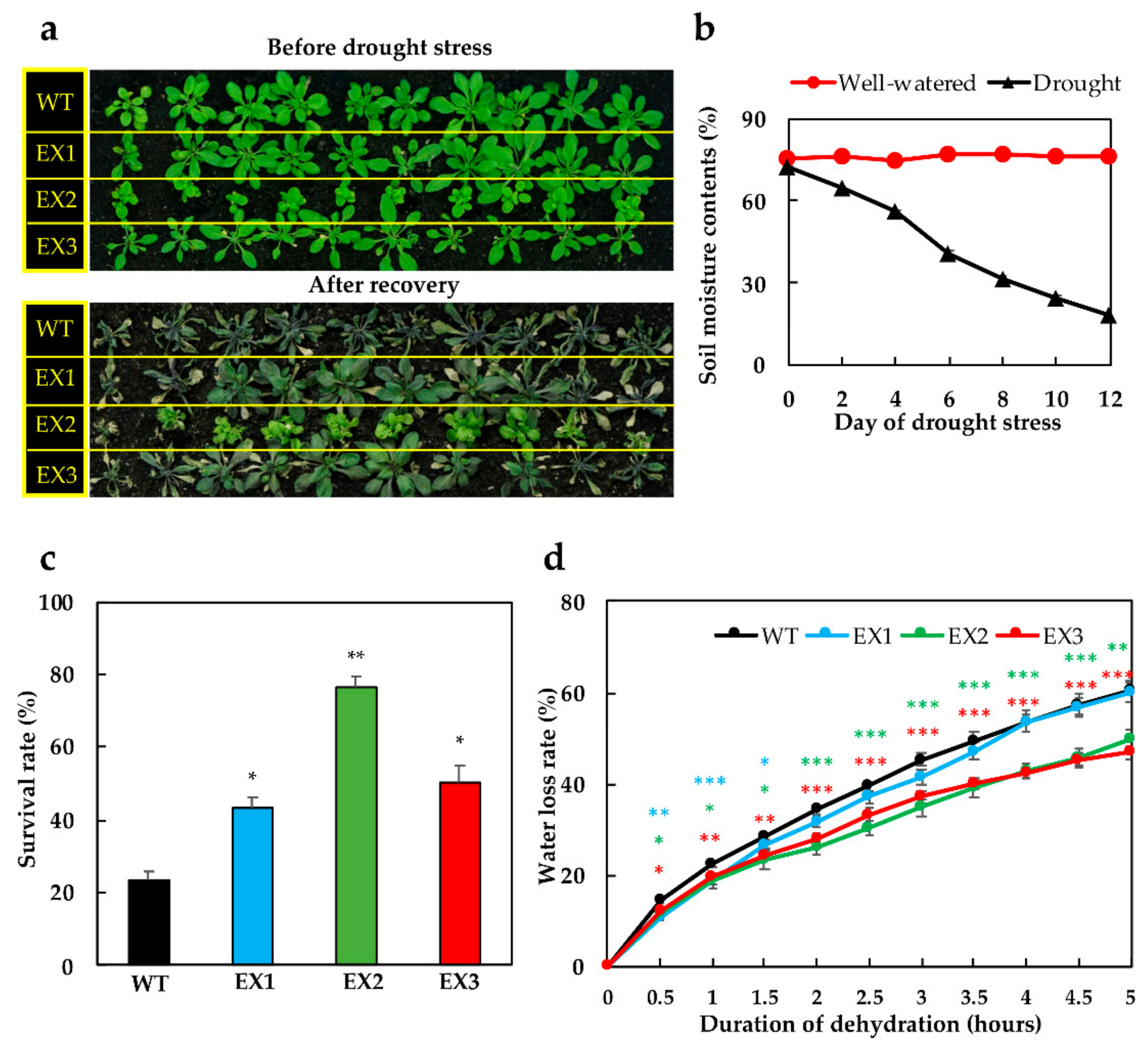
3.2. Improved Tolerance of GmNAC109-EX Plants to Drought
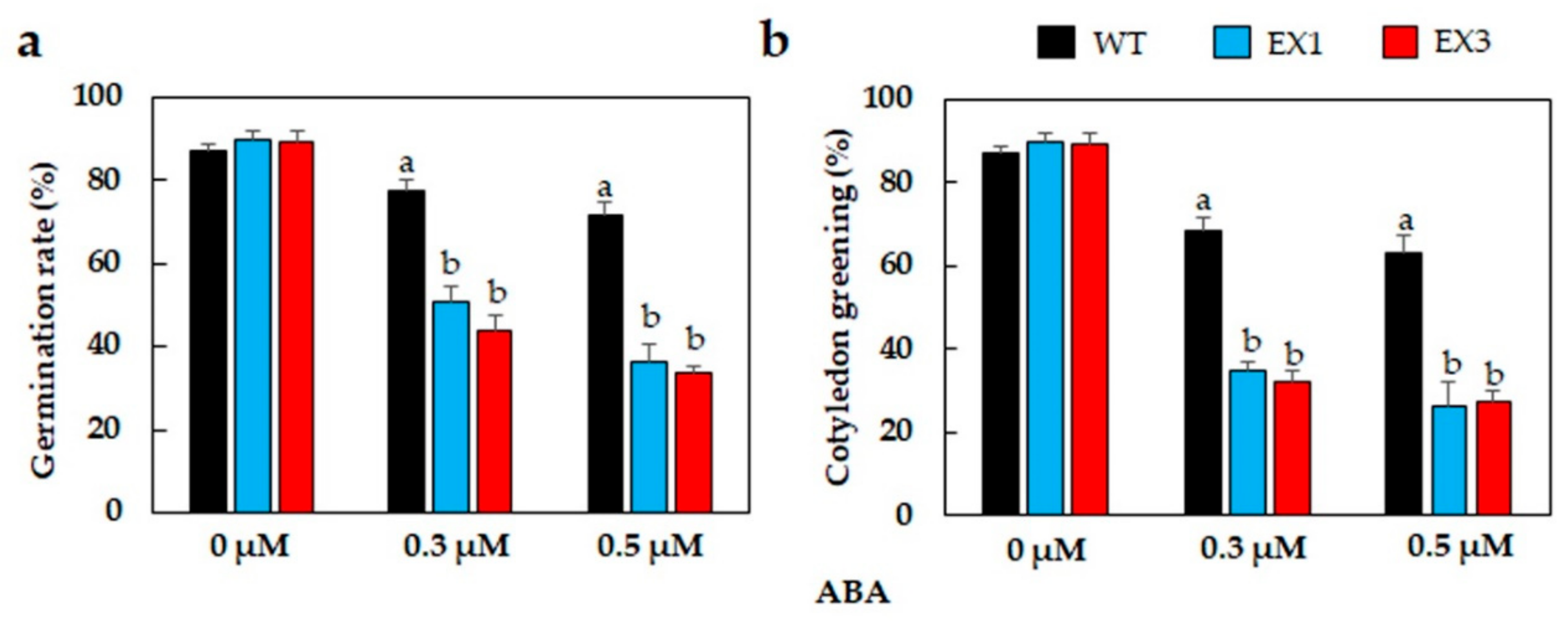
3.3. ABA Hypersensitivity of GmNAC109-EX Lines
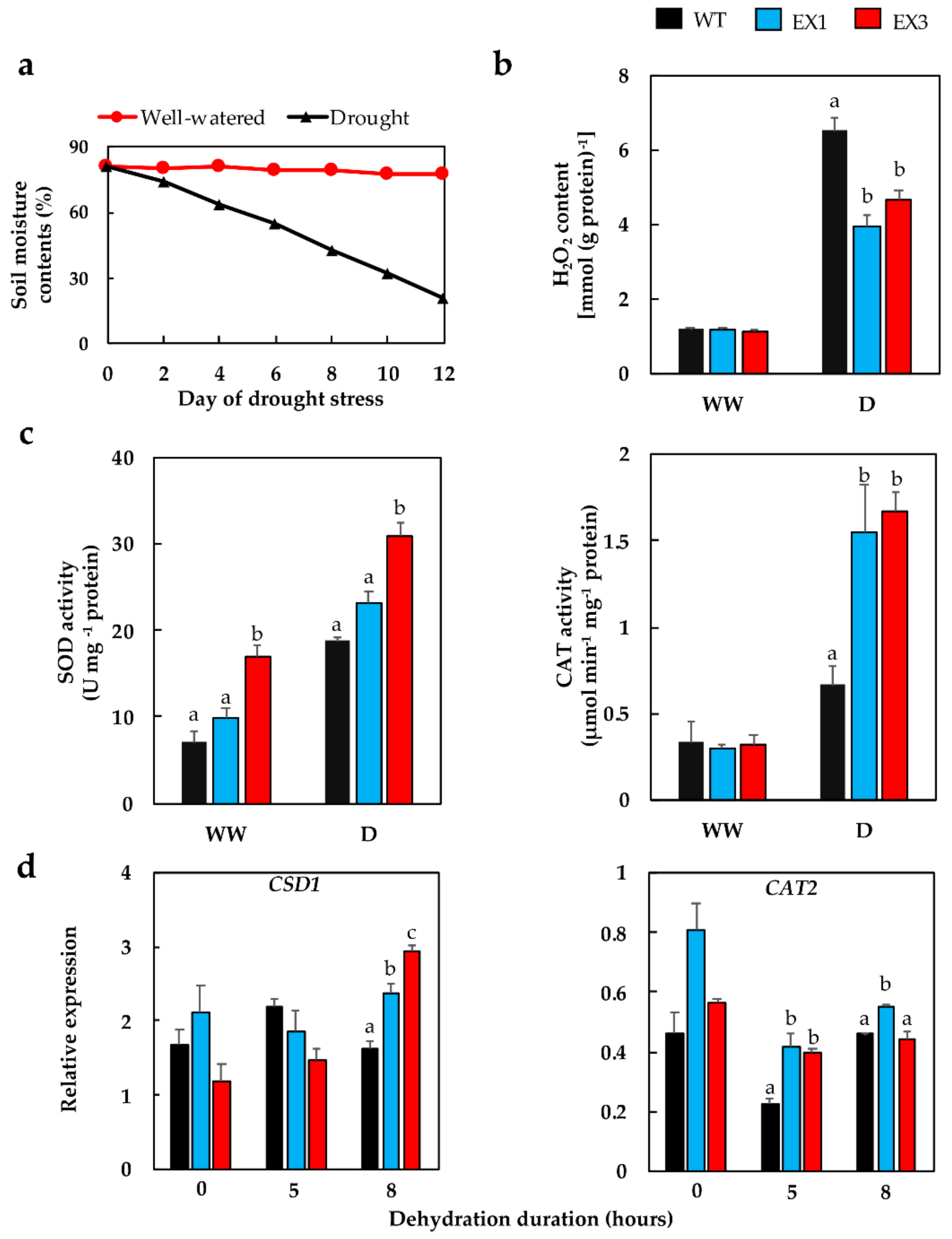
3.4. Enhanced Antioxidant Enzyme Activities in the GmNAC109-EX Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shao, H.; Wang, H.; Tang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef] [PubMed]
- Yadav, N.S.; Shukla, P.S.; Jha, A.; Agarwal, P.K.; Jha, B. The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na+ loading in xylem and confers salt tolerance in transgenic tobacco. BMC Plant Biol. 2012, 12, 188. [Google Scholar] [CrossRef] [PubMed]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Mancosu, N.; Snyder, R.; Kyriakakis, G.; Spano, D. Water scarcity and future challenges for food production. Water 2015, 7, 975–992. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Vardharajula, S.; Shrivastava, M.; Skz, A. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Farooq, M.; Basra, S.M.A.; Wahid, A.; Cheema, Z.A.; Cheema, M.A.; Khaliq, A. Physiological role of exogenously applied glycinebetaine to improve drought tolerance in fine grain aromatic rice (Oryza sativa L.). J. Agron. Crop Sci. 2008, 194, 325–333. [Google Scholar] [CrossRef]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.-S.P. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef]
- Fang, Y.; Liao, K.; Du, H.; Xu, Y.; Song, H.; Li, X.; Xiong, L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015, 66, 6803–6817. [Google Scholar] [CrossRef]
- Bartels, D.; Sunkar, R. Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2006, 58, 221–227. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Hoang, X.L.T.; Nhi, D.N.H.; Thu, N.B.A.; Thao, N.P.; Tran, L.-S.P. Transcription factors and their roles in signal transduction in plants under abiotic stresses. Curr. Genom. 2017, 18, 483–497. [Google Scholar] [CrossRef]
- Hu, W.E.I.; Huang, C.; Deng, X.; Zhou, S.; Chen, L.; Li, Y.I.N.; Wang, C.; Ma, Z.; Yuan, Q.; Wang, Y.A.N.; et al. TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Environ. 2013, 36, 1449–1464. [Google Scholar] [CrossRef] [PubMed]
- Thao, N.P.; Thu, N.B.A.; Hoang, X.L.T.; Van Ha, C.; Tran, L.-S.P. Differential expression analysis of a subset of drought-responsive GmNAC genes in two soybean cultivars differing in drought tolerance. Int. J. Mol. Sci. 2013, 14, 23828–23841. [Google Scholar] [CrossRef]
- Thu, N.B.A.; Hoang, X.L.T.; Doan, H.; Nguyen, T.-H.; Bui, D.; Thao, N.P.; Phan Tran, L.-S. Differential expression analysis of a subset of GmNAC genes in shoots of two contrasting drought-responsive soybean cultivars DT51 and MTD720 under normal and drought conditions. Mol. Biol. Rep. 2014, 41, 5563–5569. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef]
- Guan, H.; Liu, X.; Niu, F.; Zhao, Q.; Fan, N.; Cao, D.; Meng, D.; He, W.; Guo, B.; Wei, Y.; et al. OoNAC72, a NAC-type Oxytropis ochrocephala transcription factor, conferring enhanced drought and salt stress tolerance in Arabidopsis. Front. Plant Sci. 2019, 10, 890. [Google Scholar] [CrossRef]
- Tran, L.; Nakashima, K.; Sakuma, Y.; Simpson, S.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef]
- Quach, T.N.; Tran, L.-S.P.; Valliyodan, B.; Nguyen, H.T.M.; Kumar, R.; Neelakandan, A.K.; Guttikonda, S.K.; Sharp, R.E.; Nguyen, H.T. Functional analysis of water stress-responsive soybean GmNAC003 and GmNAC004 transcription factors in lateral root development in Arabidopsis. PLoS ONE 2014, 9, e84886. [Google Scholar] [CrossRef]
- Hoang, X.L.T.; Thu, N.B.A.; Thao, N.P.; Tran, L.-S.P. Transcription factors in abiotic stress responses: Their potentials in crop improvement. In Improvement of Crops in the Era of Climatic Changes, 1st ed.; Ahmad, P., Wani, M.R., Azooz, M.M., Tran, L.-S.P., Eds.; Springer: New York, NY, USA, 2014; Volume 2, pp. 337–366. [Google Scholar]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Peng, H.; Liu, Y.; Zhang, Y.; Shu, Y.; Chen, Q.; Shi, S.; Ma, L.; Ma, H.; Zhang, H. CarNAC2, a novel NAC transcription factor in chickpea (Cicer arietinum L.), is associated with drought-response and various developmental processes in transgenic Arabidopsis. J. Plant Biol. 2014, 57, 55–66. [Google Scholar] [CrossRef]
- Wu, D.; Sun, Y.; Wang, H.; Shi, H.; Su, M.; Shan, H.; Li, T.; Li, Q. The SlNAC8 gene of the halophyte Suaeda liaotungensis enhances drought and salt stress tolerance in transgenic Arabidopsis thaliana. Gene 2018, 662, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.S.; Kim, Y.S.; Redillas, M.C.F.R.; Jang, G.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.-H.; Reuzeau, C.; Kim, J.-K. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 2013, 11, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Rahman, H.; Ramanathan, V.; Nallathambi, J.; Duraialagaraja, S.; Muthurajan, R. Over-expression of a NAC 67 transcription factor from finger millet (Eleusine coracana L.) confers tolerance against salinity and drought stress in rice. BMC Biotechnol. 2016, 16 (Suppl. 1), 35. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, S.; Ma, X.; Wang, Y.; Kong, F.; Meng, Q. A stress-associated NAC transcription factor (SlNAC35) from tomato plays a positive role in biotic and abiotic stresses. Physiol. Plant. 2016, 158, 45–64. [Google Scholar] [CrossRef] [PubMed]
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018, 16, 354–366. [Google Scholar] [CrossRef]
- Tak, H.; Negi, S.; Ganapathi, T.R. Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma 2017, 254, 803–816. [Google Scholar] [CrossRef]
- Liu, G.; Li, X.; Jin, S.; Liu, X.; Zhu, L.; Nie, Y.; Zhang, X. Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS ONE 2014, 9, e86895. [Google Scholar] [CrossRef]
- Saad, A.S.I.; Li, X.; Li, H.-P.; Huang, T.; Gao, C.-S.; Guo, M.-W.; Cheng, W.; Zhao, G.-Y.; Liao, Y.-C. A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses. Plant Sci. 2013, 203–204, 33–40. [Google Scholar] [CrossRef]
- Xue, G.-P.; Way, H.M.; Richardson, T.; Drenth, J.; Joyce, P.A.; McIntyre, C.L. Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 2011, 4, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Mochida, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011, 18, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Hussain, R.M.; Ali, M.; Feng, X.; Li, X. The essence of NAC gene family to the cultivation of drought-resistant soybean (Glycine max L. Merr.) cultivars. BMC Plant Biol. 2017, 17, 55. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.-S.P.; Quach, T.N.; Guttikonda, S.K.; Aldrich, D.L.; Kumar, R.; Neelakandan, A.; Valliyodan, B.; Nguyen, H.T. Molecular characterization of stress-inducible GmNAC genes in soybean. Mol. Genet. Genom. 2009, 281, 647–664. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.H.; Mostofa, M.G.; Watanabe, Y.; Tran, C.D.; Rahman, M.M.; Tran, L.-S.P. Overexpression of GmNAC085 enhances drought tolerance in Arabidopsis by regulating glutathione biosynthesis, redox balance and glutathione-dependent detoxification of reactive oxygen species and methylglyoxal. Environ. Exp. Bot. 2019, 161, 242–254. [Google Scholar] [CrossRef]
- Nguyen, K.H.; Mostofa, M.G.; Li, W.; Van Ha, C.; Watanabe, Y.; Le, D.T.; Thao, N.P.; Tran, L.-S.P. The soybean transcription factor GmNAC085 enhances drought tolerance in Arabidopsis. Environ. Exp. Bot. 2018, 151, 12–20. [Google Scholar] [CrossRef]
- So, H.-A.; Lee, J.-H. NAC transcription factors from soybean (Glycine max L.) differentially regulated by abiotic stress. J. Plant Biol. 2019, 62, 147–160. [Google Scholar] [CrossRef]
- Hellens, R.P.; Edwards, E.A.; Leyland, N.R.; Bean, S.; Mullineaux, P.M. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000, 42, 819–832. [Google Scholar] [CrossRef]
- Qin, F.; Sakuma, Y.; Tran, L.-S.P.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.-I.; et al. DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress–responsive gene expression. Plant Cell 2008, 20, 1693–1707. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Tizaoui, K.; Kchouk, M.E. Genetic approaches for studying transgene inheritance and genetic recombination in three successive generations of transformed tobacco. Genet. 2012, 35, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Li, Y.; Varala, K.; Moose, S.P.; Hudson, M.E. The inheritance pattern of 24 nt siRNA clusters in Arabidopsis hybrids is influenced by proximity to transposable elements. PLoS ONE 2012, 7, e47043. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Jia, F.; Jiao, Z.; Wang, J.; Xia, X.; Yin, W. Ectopic expression of secretory peptide PdEPF3 in Arabidopsis confers drought tolerance with reduced stomatal density. Acta Soc. Bot. Pol. 2019, 88. [Google Scholar] [CrossRef]
- Ding, S.; Cai, Z.; Du, H.; Wang, H. Genome-wide analysis of TCP family genes in Zea mays L. identified a role for ZmTCP42 in drought tolerance. Int. J. Mol. Sci. 2019, 20, 2762. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Xia, C.; Zhao, G.; Jia, J.; Kong, X. The novel wheat transcription factor TaNAC47 enhances multiple abiotic stress tolerances in transgenic plants. Front. Plant Sci. 2016, 6, 1174. [Google Scholar] [CrossRef]
- Patterson, B.D.; MacRae, E.A.; Ferguson, I.B. Estimation of hydrogen peroxide in plant extracts using titanium(IV). Anal. Biochem. 1984, 139, 487–492. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein of protein utlilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide dismutase: I. Occurence in higher plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef]
- Wang, C.-J.; Yang, W.; Wang, C.; Gu, C.; Niu, D.-D.; Liu, H.-X.; Wang, Y.-P.; Guo, J.-H. Induction of drought tolerance in cucumber plants by a consortium of three plant growth-promoting Rhizobacterium strains. PLoS ONE 2012, 7, e52565. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2-ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Sun, M.-M.; Ye, Q.; Wu, X.-Q.; Wu, W.-H.; Chen, Y.-F. Abscisic acid modulates seed germination via ABA INSENSITIVE5-mediated PHOSPHATE1. Plant Physiol. 2017, 175, 1661–1668. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Manghwar, H.; Shaban, M.; Khan, A.H.; Akbar, A.; Ali, U.; Ali, E.; Fahad, S. Phytohormones enhanced drought tolerance in plants: a coping strategy. Environ. Sci. Pollut. Res. Int. 2018, 25, 33103–33118. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.A.D.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R.O.N. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, S.; Bharti, S.; Khanna-Chopra, R. Superoxide dismutase and abiotic stress tolerance. Physiol. Mol. Biol. Plants 2005, 11, 187–198. [Google Scholar]
- Hossain, M.A.; Bhattacharjee, S.; Armin, S.-M.; Qian, P.; Xin, W.; Li, H.-Y.; Burritt, D.J.; Fujita, M.; Tran, L.-S.P. Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: insights from ROS detoxification and scavenging. Front. Plant Sci. 2015, 6, 420. [Google Scholar] [CrossRef]
- Al-Abdallat, A.M.; Ali-Sheikh-Omar, M.A.; Alnemer, L.M. Overexpression of two ATNAC3-related genes improves drought and salt tolerance in tomato (Solanum lycopersicum L.). Plant Cell Tiss. Org. 2015, 120, 989–1001. [Google Scholar] [CrossRef]
- An, X.; Liao, Y.; Zhang, J.; Dai, L.; Zhang, N.; Wang, B.; Liu, L.; Peng, D. Overexpression of rice NAC gene SNAC1 in ramie improves drought and salt tolerance. Plant Growth Regul. 2015, 76, 211–223. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef]
- Cao, H.; Wang, L.; Nawaz, M.A.; Niu, M.; Sun, J.; Xie, J.; Kong, Q.; Huang, Y.; Cheng, F.; Bie, Z. Ectopic expression of pumpkin NAC transcription factor CmNAC1 improves multiple abiotic stress tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 2052. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, Y.; Li, B.; Chang, J.; Chen, M.; Li, K.; Yang, G.; He, G. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015, 15, 268. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Jiang, Q.; van Nocker, S.; Gong, X.; Ma, F. An apple (Malus domestica) NAC transcription factor enhances drought tolerance in transgenic apple plant. Plant Physiol. Biochem. 2019, 139, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Li, K.-Q.; Xu, X.-Y.; Zhang, H.-P.; Chen, H.-X.; Chen, Y.-H.; Hao, J.; Wang, Y.; Huang, X.-S.; Zhang, S.-L. A novel NAC transcription factor, PbeNAC1, of Pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Front. Plant Sci. 2017, 8, 1049. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-K.; Chung, P.J.; Jeong, J.S.; Jang, G.; Bang, S.W.; Jung, H.; Kim, Y.S.; Ha, S.-H.; Choi, Y.D.; Kim, J.-K. The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol. J. 2017, 15, 754–764. [Google Scholar] [CrossRef]
- Mao, H.; Yu, L.; Han, R.; Li, Z.; Liu, H. ZmNAC55, a maize stress-responsive NAC transcription factor, confers drought resistance in transgenic Arabidopsis. Plant Physiol. Biochem. 2016, 105, 55–66. [Google Scholar] [CrossRef]
- Huang, L.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 2016, 16, 203. [Google Scholar] [CrossRef]
- Farber, M.; Attia, Z.; Weiss, D. Cytokinin activity increases stomatal density and transpiration rate in tomato. J. Exp. Bot. 2016, 67, 6351–6362. [Google Scholar] [CrossRef]
- Pospíšilová, H.; Jiskrová, E.; Vojta, P.; Mrízová, K.; Kokáš, F.; Čudejková, M.M.; Bergougnoux, V.; Plíhal, O.; Klimešová, J.; Novák, O.; et al. Transgenic barley overexpressing a cytokinin dehydrogenase gene shows greater tolerance to drought stress. New Biotechnol. 2016, 33, 692–705. [Google Scholar] [CrossRef]
- Nishiyama, R.; Watanabe, Y.; Fujita, Y.; Le, D.T.; Kojima, M.; Werner, T.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Kakimoto, T.; et al. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 2011, 23, 2169–2183. [Google Scholar] [CrossRef]
- Duval, M.; Hsieh, T.; Kim, S.; Thomas, T. Molecular characterization of AtNAM: A member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 2002, 50, 237–248. [Google Scholar] [CrossRef]
- Willemsen, V.; Bauch, M.; Bennett, T.; Campilho, A.; Wolkenfelt, H.; Xu, J.; Haseloff, J.; Scheres, B. The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev. Cell 2008, 15, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Ying, S.; Zhang, D.-F.; Shi, Y.-S.; Song, Y.-C.; Wang, T.-Y.; Li, Y. A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep. 2012, 31, 1701–1711. [Google Scholar] [CrossRef]
- Yang, X.; Wang, X.; Ji, L.; Yi, Z.; Fu, C.; Ran, J.; Hu, R.; Zhou, G. Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Rep. 2015, 34, 943–958. [Google Scholar] [CrossRef]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef]
- Xie, L.-N.; Chen, M.; Min, D.H.; Feng, L.; Xu, Z.-S.; Zhou, Y.-B.; Xu, D.-B.; Li, L.-C.; Ma, Y.-Z.; Zhang, X.-H. The NAC-like transcription factor SiNAC110 in foxtail millet (Setaria italica L.) confers tolerance to drought and high salt stress through an ABA independent signaling pathway. J. Integr. Agric. 2017, 16, 559–571. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Gómez-Porras, J.L.; Riaño-Pachón, D.M.; Dreyer, I.; Mayer, J.E.; Mueller-Roeber, B. Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genom. 2007, 8, 260. [Google Scholar] [CrossRef]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251. [Google Scholar] [PubMed]
- Daszkowska-Golec, A.; Szarejko, I. Open or close the gate—stomata action under the control of phytohormones in drought stress conditions. Front Plant Sci. 2013, 4, 138. [Google Scholar] [CrossRef] [PubMed]
- Outlaw, J.W.H. Integration of cellular and physiological functions of guard cells. Crit. Rev. Plant Sci. 2003, 22, 503–529. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Hossain, M.A.; Fujita, M.; Tran, L.-S.P. Physiological and biochemical mechanisms associated with trehalose-induced copper-stress tolerance in rice. Sci. Rep. 2015, 5, 11433. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Veljovic-Jovanovic, S.; Driscoll, S.; Novitskaya, L.; Foyer, C.H. Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration? Ann. Bot. 2002, 89, 841–850. [Google Scholar] [CrossRef]
- Ighodaro, O.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. Med. J. 2017, 54. [Google Scholar] [CrossRef]
- Rahman, M.M.; Mostofa, M.G.; Rahman, M.A.; Islam, M.R.; Keya, S.S.; Das, A.K.; Miah, M.G.; Kawser, A.Q.M.R.; Ahsan, S.M.; Hashem, A.; et al. Acetic acid: A cost-effective agent for mitigation of seawater-induced salt toxicity in mung bean. Sci. Rep. 2019, 9, 15186. [Google Scholar] [CrossRef]
- Harb, A.; Awad, D.; Samarah, N. Gene expression and activity of antioxidant enzymes in barley (Hordeum vulgare L.) under controlled severe drought. J. Plant Interact. 2015, 10, 109–116. [Google Scholar] [CrossRef]
- McGrann, G.R.D.; Steed, A.; Burt, C.; Goddard, R.; Lachaux, C.; Bansal, A.; Corbitt, M.; Gorniak, K.; Nicholson, P.; Brown, J.K.M. Contribution of the drought tolerance-related stress-responsive NAC1 transcription factor to resistance of barley to Ramularia leaf spot. Mol. Plant Pathol. 2015, 16, 201–209. [Google Scholar] [CrossRef]
- He, L.; Bian, J.; Xu, J.; Yang, K. Novel maize NAC transcriptional repressor ZmNAC071 confers enhanced sensitivity to ABA and osmotic stress by downregulating stress-responsive genes in transgenic Arabidopsis. J. Agric. Food Chem. 2019, 67, 8905–8918. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, N.C.; Hoang, X.L.T.; Nguyen, Q.T.; Binh, N.X.; Watanabe, Y.; Thao, N.P.; Tran, L.-S.P. Ectopic Expression of Glycine max GmNAC109 Enhances Drought Tolerance and ABA Sensitivity in Arabidopsis. Biomolecules 2019, 9, 714. https://doi.org/10.3390/biom9110714
Nguyen NC, Hoang XLT, Nguyen QT, Binh NX, Watanabe Y, Thao NP, Tran L-SP. Ectopic Expression of Glycine max GmNAC109 Enhances Drought Tolerance and ABA Sensitivity in Arabidopsis. Biomolecules. 2019; 9(11):714. https://doi.org/10.3390/biom9110714
Chicago/Turabian StyleNguyen, Nguyen Cao, Xuan Lan Thi Hoang, Quang Thien Nguyen, Ngo Xuan Binh, Yasuko Watanabe, Nguyen Phuong Thao, and Lam-Son Phan Tran. 2019. "Ectopic Expression of Glycine max GmNAC109 Enhances Drought Tolerance and ABA Sensitivity in Arabidopsis" Biomolecules 9, no. 11: 714. https://doi.org/10.3390/biom9110714
APA StyleNguyen, N. C., Hoang, X. L. T., Nguyen, Q. T., Binh, N. X., Watanabe, Y., Thao, N. P., & Tran, L.-S. P. (2019). Ectopic Expression of Glycine max GmNAC109 Enhances Drought Tolerance and ABA Sensitivity in Arabidopsis. Biomolecules, 9(11), 714. https://doi.org/10.3390/biom9110714




