The Mechanisms Underlying PTEN Loss in Human Tumors Suggest Potential Therapeutic Opportunities
Abstract
1. PI3K Mutations in Human Cancers
2. Prevalence of PTEN Mutations in Cancer
3. Epigenetic, Transcriptional, and Post-Transcriptional Regulation of PTEN
3.1. Transcriptional Regulation of PTEN
3.2. Epigenetic Regulation of PTEN Expression
3.3. miRNA and PTEN Expression
3.4. Long Non-Coding RNA and PTEN Expression
4. Post-Translational Modifications of PTEN
4.1. Phosphorylation
4.2. Ubiquitination
4.3. Oxidation
4.4. Acetylation
4.5. S-Nitrosylation
4.6. Ribosylation
4.7. Sumoylation
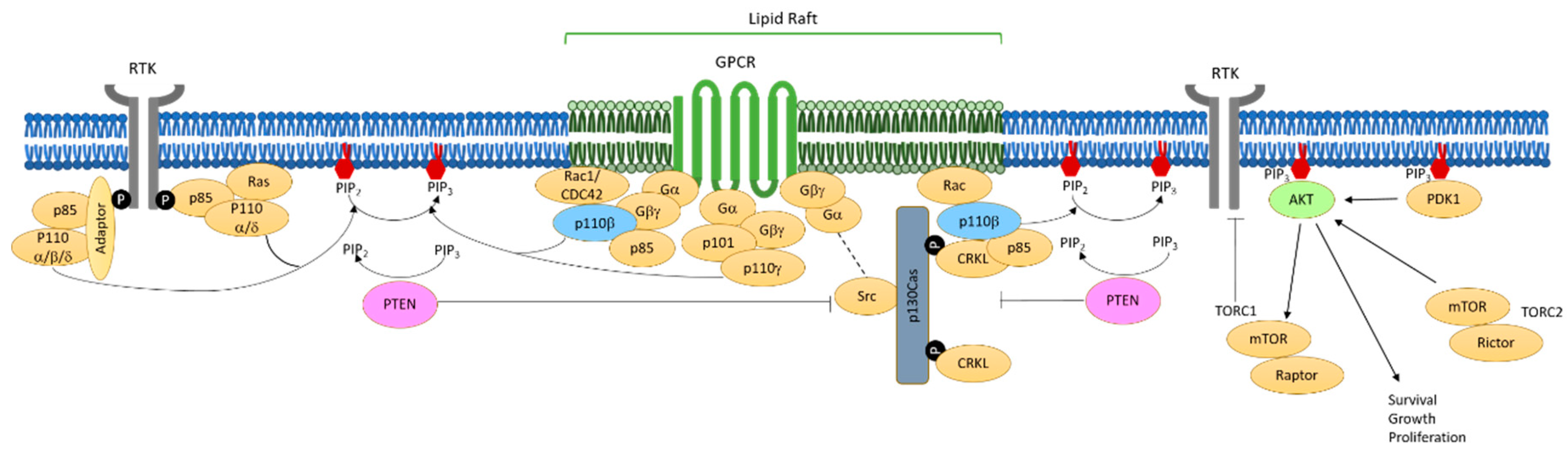
5. Mechanisms of PI3K Activation in PTEN-Null Tumor Cells
6. Turning Mechanistic Insights into Possible Therapies
6.1. Combining p110β Inhbition and Immunotherapies
6.2. PTEN-Deficient Synthetic Lethality
6.3. Reactivation of PTEN
7. Conclusions and Perspective
Funding
Conflicts of Interest
References
- Engelman, J.A.; Luo, J.; Cantley, L.C. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006, 7, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Cheng, H.; Roberts, T.M.; Zhao, J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009, 8, 627–644. [Google Scholar] [CrossRef] [PubMed]
- Chantry, D.; Vojtek, A.; Kashishian, A.; Holtzman, D.A.; Wood, C.; Gray, P.W.; Cooper, J.A.; Hoekstra, M.F. p110delta, a novel phosphatidylinositol 3-kinase catalytic subunit that associates with p85 and is expressed predominantly in leukocytes. J. Biol. Chem. 1997, 272, 19236–19241. [Google Scholar] [CrossRef] [PubMed]
- Vanhaesebroeck, B.; Welham, M.J.; Kotani, K.; Stein, R.; Warne, P.H.; Zvelebil, M.J.; Higashi, K.; Volinia, S.; Downward, J.; Waterfield, M.D. P110delta, a novel phosphoinositide 3-kinase in leukocytes. Proc. Natl. Acad. Sci. USA 1997, 94, 4330–4335. [Google Scholar] [CrossRef]
- Kaneda, M.M.; Messer, K.S.; Ralainirina, N.; Li, H.; Leem, C.J.; Gorjestani, S.; Woo, G.; Nguyen, A.V.; Figueiredo, C.C.; Foubert, P.; et al. PI3Kγ is a molecular switch that controls immune suppression. Nature 2016, 539, 437–442. [Google Scholar] [CrossRef]
- Li, J.; Yen, C.; Liaw, D.; Podsypanina, K.; Bose, S.; Wang, S.I.; Puc, J.; Miliaresis, C.; Rodgers, L.; McCombie, R.; et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997, 275, 1943–1947. [Google Scholar] [CrossRef]
- Liaw, D.; Marsh, D.J.; Li, J.; Dahia, P.L.; Wang, S.I.; Zheng, Z.; Bose, S.; Call, K.M.; Tsou, H.C.; Peacocke, M.; et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Gene. 1997, 16, 64–67. [Google Scholar] [CrossRef]
- Steck, P.A.; Pershouse, M.A.; Jasser, S.A.; Yung, W.K.; Lin, H.; Ligon, A.H.; Langford, L.A.; Baumgard, M.L.; Hattier, T.; Davis, T.; et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Gene. 1997, 15, 356–362. [Google Scholar] [CrossRef]
- Zhang, X.C.; Piccini, A.; Myers, M.P.; Van Aelst, L.; Tonks, N.K. Functional analysis of the protein phosphatase activity of PTEN. Biochem. J. 2012, 444, 457–464. [Google Scholar] [CrossRef]
- Leslie, N.R.; Downes, C.P. PTEN: The down side of PI 3-kinase signalling. Cell Signal. 2002, 14, 285–295. [Google Scholar] [CrossRef]
- Planchon, S.M.; Waite, K.A.; Eng, C. The nuclear affairs of PTEN. J. Cell Sci. 2008, 121, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Gil, A.; Andres-Pons, A.; Fernandez, E.; Valiente, M.; Torres, J.; Cervera, J.; Pulido, R. Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: Involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs. Mol. Biol. Cell 2006, 17, 4002–4013. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Shen, W.H. PTEN: A new guardian of the genome. Oncogene 2008, 27, 5443–5453. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, L.M.; Yuzugullu, H.; Zhao, J.J. PI3K in cancer: Divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 2015, 15, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Leslie, N.R.; Downes, C.P. PTEN function: How normal cells control it and tumour cells lose it. Biochem. J. 2004, 382, 1–11. [Google Scholar] [CrossRef]
- Berns, K.; Horlings, H.M.; Hennessy, B.T.; Madiredjo, M.; Hijmans, E.M.; Beelen, K.; Linn, S.C.; Gonzalez-Angulo, A.M.; Stemke-Hale, K.; Hauptmann, M.; et al. A Functional Genetic Approach Identifies the PI3K Pathway as a Major Determinant of Trastuzumab Resistance in Breast Cancer. Cancer Cell 2007, 12, 395–402. [Google Scholar] [CrossRef]
- Utermark, T.; Rao, T.; Cheng, H.; Wang, Q.; Lee, S.H.; Wang, Z.C.; Iglehart, J.D.; Roberts, T.M.; Muller, W.J.; Zhao, J.J. The p110alpha and p110beta isoforms of PI3K play divergent roles in mammary gland development and tumorigenesis. Genes Dev. 2012, 26, 1573–1586. [Google Scholar] [CrossRef]
- Tsolakos, N.; Durrant, T.N.; Chessa, T.; Suire, S.M.; Oxley, D.; Kulkarni, S.; Downward, J.; Perisic, O.; Williams, R.L.; Stephens, L.; et al. Quantitation of class IA PI3Ks in mice reveals p110-free-p85s and isoform-selective subunit associations and recruitment to receptors. Proc. Natl. Acad. Sci. USA 2018, 115, 12176–12181. [Google Scholar] [CrossRef]
- Wee, S.; Wiederschain, D.; Maira, S.M.; Loo, A.; Miller, C.; de Beaumont, R.; Stegmeier, F.; Yao, Y.M.; Lengauer, C. PTEN-deficient cancers depend on PIK3CB. Proc. Natl. Acad. Sci. USA 2008, 105, 13057–13062. [Google Scholar] [CrossRef]
- Jia, S.; Liu, Z.; Zhang, S.; Liu, P.; Zhang, L.; Lee, S.H.; Zhang, J.; Signoretti, S.; Loda, M.; Roberts, T.M.; et al. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature 2008, 454, 776–779. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Millis, S.Z.; Ikeda, S.; Reddy, S.; Gatalica, Z.; Kurzrock, R. Landscape of Phosphatidylinositol-3-Kinase Pathway Alterations Across 19784 Diverse Solid Tumors. JAMA Oncol. 2016, 2, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Schade, B.; Rao, T.; Dourdin, N.; Lesurf, R.; Hallett, M.; Cardiff, R.D.; Muller, W.J. PTEN deficiency in a luminal ErbB-2 mouse model results in dramatic acceleration of mammary tumorigenesis and metastasis. J. Biol. Chem. 2009, 284, 19018–19026. [Google Scholar] [CrossRef]
- Chalhoub, N.; Baker, S.J. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 2009, 4, 127–150. [Google Scholar] [CrossRef]
- Salmena, L.; Carracedo, A.; Pandolfi, P.P. Tenets of PTEN tumor suppression. Cell 2008, 133, 403–414. [Google Scholar] [CrossRef]
- Bignell, G.R.; Greenman, C.D.; Davies, H.; Butler, A.P.; Edkins, S.; Andrews, J.M.; Buck, G.; Chen, L.; Beare, D.; Latimer, C.; et al. Signatures of mutation and selection in the cancer genome. Nature 2010, 463, 893–898. [Google Scholar] [CrossRef]
- Song, M.S.; Salmena, L.; Pandolfi, P.P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012, 13, 283–296. [Google Scholar] [CrossRef]
- Bonneau, D.; Longy, M. Mutations of the human PTEN gene. Hum. Mutat. 2000, 16, 109–122. [Google Scholar] [CrossRef]
- Georgescu, M.M.; Kirsch, K.H.; Kaloudis, P.; Yang, H.; Pavletich, N.P.; Hanafusa, H. Stabilization and productive positioning roles of the C2 domain of PTEN tumor suppressor. Cancer Res. 2000, 60, 7033–7038. [Google Scholar]
- Di Cristofano, A.; Pesce, B.; Cordon-Cardo, C.; Pandolfi, P.P. Pten is essential for embryonic development and tumour suppression. Nat. Gene. 1998, 19, 348–355. [Google Scholar] [CrossRef]
- Podsypanina, K.; Ellenson, L.H.; Nemes, A.; Gu, J.; Tamura, M.; Yamada, K.M.; Cordon-Cardo, C.; Catoretti, G.; Fisher, P.E.; Parsons, R. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl. Acad. Sci. USA 1999, 96, 1563–1568. [Google Scholar] [CrossRef]
- Alimonti, A.; Carracedo, A.; Clohessy, J.G.; Trotman, L.C.; Nardella, C.; Egia, A.; Salmena, L.; Sampieri, K.; Haveman, W.J.; Brogi, E.; et al. Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 2010, 42, 454–458. [Google Scholar] [CrossRef]
- Trotman, L.C.; Niki, M.; Dotan, Z.A.; Koutcher, J.A.; Di Cristofano, A.; Xiao, A.; Khoo, A.S.; Roy-Burman, P.; Greenberg, N.M.; Van Dyke, T.; et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003, 1, 59. [Google Scholar] [CrossRef]
- Kwabi-Addo, B.; Giri, D.; Schmidt, K.; Podsypanina, K.; Parsons, R.; Greenberg, N.; Ittmann, M. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proc. Natl. Acad. Sci. USA 2001, 98, 11563–11568. [Google Scholar] [CrossRef]
- Zhou, X.P.; Loukola, A.; Salovaara, R.; Nystrom-Lahti, M.; Peltomaki, P.; de la Chapelle, A.; Aaltonen, L.A.; Eng, C. PTEN mutational spectra, expression levels, and subcellular localization in microsatellite stable and unstable colorectal cancers. Am. J. Pathol. 2002, 161, 439–447. [Google Scholar] [CrossRef]
- Ali, I.U.; Schriml, L.M.; Dean, M. Mutational spectra of PTEN/MMAC1 gene: A tumor suppressor with lipid phosphatase activity. J. Natl. Cancer Inst. 1999, 91, 1922–1932. [Google Scholar] [CrossRef]
- Bubien, V.; Bonnet, F.; Brouste, V.; Hoppe, S.; Barouk-Simonet, E.; David, A.; Edery, P.; Bottani, A.; Layet, V.; Caron, O.; et al. High cumulative risks of cancer in patients with PTEN hamartoma tumour syndrome. J. Med. Genet. 2013, 50, 255–263. [Google Scholar] [CrossRef]
- Knobbe, C.B.; Lapin, V.; Suzuki, A.; Mak, T.W. The roles of PTEN in development, physiology and tumorigenesis in mouse models: A tissue-by-tissue survey. Oncogene 2008, 27, 5398–5415. [Google Scholar] [CrossRef]
- Suzuki, A.; de la Pompa, J.L.; Stambolic, V.; Elia, A.J.; Sasaki, T.; del Barco Barrantes, I.; Ho, A.; Wakeham, A.; Itie, A.; Khoo, W.; et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr. Biol. 1998, 8, 1169–1178. [Google Scholar] [CrossRef]
- Stambolic, V.; MacPherson, D.; Sas, D.; Lin, Y.; Snow, B.; Jang, Y.; Benchimol, S.; Mak, T.W. Regulation of PTEN transcription by p53. Mol. Cell 2001, 8, 317–325. [Google Scholar] [CrossRef]
- Wang, J.; Ouyang, W.; Li, J.; Wei, L.; Ma, Q.; Zhang, Z.; Tong, Q.; He, J.; Huang, C. Loss of tumor suppressor p53 decreases PTEN expression and enhances signaling pathways leading to activation of activator protein 1 and nuclear factor kappaB induced by UV radiation. Cancer Res. 2005, 65, 6601–6611. [Google Scholar] [CrossRef]
- Virolle, T.; Adamson, E.D.; Baron, V.; Birle, D.; Mercola, D.; Mustelin, T.; de Belle, I. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat. Cell Biol. 2001, 3, 1124–1128. [Google Scholar] [CrossRef]
- Ferraro, B.; Bepler, G.; Sharma, S.; Cantor, A.; Haura, E.B. EGR1 predicts PTEN and survival in patients with non-small-cell lung cancer. J. Clin. Oncol. 2005, 23, 1921–1926. [Google Scholar] [CrossRef]
- Baron, V.; Adamson, E.D.; Calogero, A.; Ragona, G.; Mercola, D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006, 13, 115–124. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, S.S.; Saito, K.; Williams, S.; Arimura, Y.; Ma, Y.; Ke, Y.; Baron, V.; Mercola, D.; Feng, G.S.; et al. PTEN regulation by Akt-EGR1-ARF-PTEN axis. EMBO J. 2009, 28, 21–33. [Google Scholar] [CrossRef]
- Yamamoto, C.; Basaki, Y.; Kawahara, A.; Nakashima, K.; Kage, M.; Izumi, H.; Kohno, K.; Uramoto, H.; Yasumoto, K.; Kuwano, M.; et al. Loss of PTEN expression by blocking nuclear translocation of EGR1 in gefitinib-resistant lung cancer cells harboring epidermal growth factor receptor-activating mutations. Cancer Res. 2010, 70, 8715–8725. [Google Scholar] [CrossRef]
- Su, L.; Cheng, H.; Sampaio, A.V.; Nielsen, T.O.; Underhill, T.M. EGR1 reactivation by histone deacetylase inhibitors promotes synovial sarcoma cell death through the PTEN tumor suppressor. Oncogene 2010, 29, 4352–4361. [Google Scholar] [CrossRef]
- Lau, M.T.; Klausen, C.; Leung, P.C. E-cadherin inhibits tumor cell growth by suppressing PI3K/Akt signaling via beta-catenin-Egr1-mediated PTEN expression. Oncogene 2011, 30, 2753–2766. [Google Scholar] [CrossRef]
- Kim, J.; Kang, H.S.; Lee, Y.J.; Lee, H.J.; Yun, J.; Shin, J.H.; Lee, C.W.; Kwon, B.M.; Hong, S.H. EGR1-dependent PTEN upregulation by 2-benzoyloxycinnamaldehyde attenuates cell invasion and EMT in colon cancer. Cancer Lett. 2014, 349, 35–44. [Google Scholar] [CrossRef]
- Chappell, W.H.; Green, T.D.; Spengeman, J.D.; McCubrey, J.A.; Akula, S.M.; Bertrand, F.E. Increased protein expression of the PTEN tumor suppressor in the presence of constitutively active Notch-1. Cell Cycle 2005, 4, 1389–1395. [Google Scholar] [CrossRef] [PubMed]
- Whelan, J.T.; Forbes, S.L.; Bertrand, F.E. CBF-1 (RBP-J kappa) binds to the PTEN promoter and regulates PTEN gene expression. Cell Cycle 2007, 6, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Whelan, J.T.; Kellogg, A.; Shewchuk, B.M.; Hewan-Lowe, K.; Bertrand, F.E. Notch-1 signaling is lost in prostate adenocarcinoma and promotes PTEN gene expression. J. Cell Biochem. 2009, 107, 992–1001. [Google Scholar] [CrossRef]
- Patel, L.; Pass, I.; Coxon, P.; Downes, C.P.; Smith, S.A.; Macphee, C.H. Tumor suppressor and anti-inflammatory actions of PPARgamma agonists are mediated via upregulation of PTEN. Curr. Biol. 2001, 11, 764–768. [Google Scholar] [CrossRef]
- Farrow, B.; Evers, B.M. Activation of PPARgamma increases PTEN expression in pancreatic cancer cells. Biochem. Bio. Res. Commun. 2003, 301, 50–53. [Google Scholar] [CrossRef]
- Lee, K.S.; Park, S.J.; Hwang, P.H.; Yi, H.K.; Song, C.H.; Chai, O.H.; Kim, J.S.; Lee, M.K.; Lee, Y.C. PPAR-gamma modulates allergic inflammation through up-regulation of PTEN. FASEB J. 2005, 19, 1033–1035. [Google Scholar] [CrossRef]
- Teresi, R.E.; Shaiu, C.W.; Chen, C.S.; Chatterjee, V.K.; Waite, K.A.; Eng, C. Increased PTEN expression due to transcriptional activation of PPARgamma by Lovastatin and Rosiglitazone. Int. J. Cancer 2006, 118, 2390–2398. [Google Scholar] [CrossRef]
- Vasudevan, K.M.; Gurumurthy, S.; Rangnekar, V.M. Suppression of PTEN expression by NF-kappa B prevents apoptosis. Mol. Cell Biol. 2004, 24, 1007–1021. [Google Scholar] [CrossRef]
- Xia, D.; Srinivas, H.; Ahn, Y.H.; Sethi, G.; Sheng, X.; Yung, W.K.; Xia, Q.; Chiao, P.J.; Kim, H.; Brown, P.H.; et al. Mitogen-activated protein kinase kinase-4 promotes cell survival by decreasing PTEN expression through an NF kappa B-dependent pathway. J. Boil. Chem. 2007, 282, 3507–3519. [Google Scholar] [CrossRef]
- Ghosh-Choudhury, N.; Mandal, C.C.; Ghosh-Choudhury, N.; Ghosh Choudhury, G. Simvastatin induces derepression of PTEN expression via NFkappaB to inhibit breast cancer cell growth. Cell Signal. 2010, 22, 749–758. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J.; Gao, G.; Li, J.; Huang, H.; Jin, H.; Zhu, J.; Che, X.; Huang, C. Tumor-suppressor NFkappaB2 p100 interacts with ERK2 and stabilizes PTEN mRNA via inhibition of miR-494. Oncogene 2016, 35, 4080–4090. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Zhang, L.; Youker, K.; Zhang, M.X.; Wang, J.; LeMaire, S.A.; Coselli, J.S.; Shen, Y.H. Free fatty acids inhibit insulin signaling-stimulated endothelial nitric oxide synthase activation through upregulating PTEN or inhibiting Akt kinase. Diabetes 2006, 55, 2301–2310. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.H.; Zhang, L.; Gan, Y.; Wang, X.; Wang, J.; LeMaire, S.A.; Coselli, J.S.; Wang, X.L. Up-regulation of PTEN (phosphatase and tensin homolog deleted on chromosome ten) mediates p38 MAPK stress signal-induced inhibition of insulin signaling. A cross-talk between stress signaling and insulin signaling in resistin-treated human endothelial cells. J. Biol. Chem. 2006, 281, 7727–7736. [Google Scholar] [CrossRef]
- Hettinger, K.; Vikhanskaya, F.; Poh, M.K.; Lee, M.K.; de Belle, I.; Zhang, J.T.; Reddy, S.A.; Sabapathy, K. c-Jun promotes cellular survival by suppression of PTEN. Cell Death Differ. 2007, 14, 218–229. [Google Scholar] [CrossRef]
- Song, L.B.; Li, J.; Liao, W.T.; Feng, Y.; Yu, C.P.; Hu, L.J.; Kong, Q.L.; Xu, L.H.; Zhang, X.; Liu, W.L.; et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J. Clin. Invest. 2009, 119, 3626–3636. [Google Scholar] [CrossRef]
- Escriva, M.; Peiro, S.; Herranz, N.; Villagrasa, P.; Dave, N.; Montserrat-Sentis, B.; Murray, S.A.; Franci, C.; Gridley, T.; Virtanen, I.; et al. Repression of PTEN phosphatase by Snail1 transcriptional factor during gamma radiation-induced apoptosis. Mol. Cell Biol. 2008, 28, 1528–1540. [Google Scholar] [CrossRef]
- Garcia, J.M.; Silva, J.; Pena, C.; Garcia, V.; Rodriguez, R.; Cruz, M.A.; Cantos, B.; Provencio, M.; Espana, P.; Bonilla, F. Promoter methylation of the PTEN gene is a common molecular change in breast cancer. Genes Chrom. Cancer 2004, 41, 117–124. [Google Scholar] [CrossRef]
- Goel, A.; Arnold, C.N.; Niedzwiecki, D.; Carethers, J.M.; Dowell, J.M.; Wasserman, L.; Compton, C.; Mayer, R.J.; Bertagnolli, M.M.; Boland, C.R. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res. 2004, 64, 3014–3021. [Google Scholar] [CrossRef]
- Khan, S.; Kumagai, T.; Vora, J.; Bose, N.; Sehgal, I.; Koeffler, P.H.; Bose, S. PTEN promoter is methylated in a proportion of invasive breast cancers. Int. J. Cancer 2004, 112, 407–410. [Google Scholar] [CrossRef]
- Phuong, N.T.; Kim, S.K.; Lim, S.C.; Kim, H.S.; Kim, T.H.; Lee, K.Y.; Ahn, S.G.; Yoon, J.H.; Kang, K.W. Role of PTEN promoter methylation in tamoxifen-resistant breast cancer cells. Breast Cancer Res. Treat. 2011, 130, 73–83. [Google Scholar] [CrossRef]
- Soria, J.C.; Lee, H.Y.; Lee, J.I.; Wang, L.; Issa, J.P.; Kemp, B.L.; Liu, D.D.; Kurie, J.M.; Mao, L.; Khuri, F.R. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin. Cancer Res. 2002, 8, 1178–1184. [Google Scholar]
- Maeda, M.; Murakami, Y.; Watari, K.; Kuwano, M.; Izumi, H.; Ono, M. CpG hypermethylation contributes to decreased expression of PTEN during acquired resistance to gefitinib in human lung cancer cell lines. Lung Cancer 2015, 87, 265–271. [Google Scholar] [CrossRef]
- Ho, C.M.; Lin, M.C.; Huang, S.H.; Huang, C.J.; Lai, H.C.; Chien, T.Y.; Chang, S.F. PTEN promoter methylation and LOH of 10q22-23 locus in PTEN expression of ovarian clear cell adenocarcinomas. Gynecol. Oncol. 2009, 112, 307–313. [Google Scholar] [CrossRef]
- Qi, Q.; Ling, Y.; Zhu, M.; Zhou, L.; Wan, M.; Bao, Y.; Liu, Y. Promoter region methylation and loss of protein expression of PTEN and significance in cervical cancer. Biomed. Rep. 2014, 2, 653–658. [Google Scholar] [CrossRef]
- Huhns, M.; Salem, T.; Schneider, B.; Krohn, M.; Linnebacher, M.; Prall, F. PTEN mutation, loss of heterozygosity, promoter methylation and expression in colorectal carcinoma: Two hits on the gene? Oncol. Rep. 2014, 31, 2236–2244. [Google Scholar] [CrossRef]
- Alvarez-Nunez, F.; Bussaglia, E.; Mauricio, D.; Ybarra, J.; Vilar, M.; Lerma, E.; de Leiva, A.; Matias-Guiu, X.; Thyroid Neoplasia Study Group. PTEN promoter methylation in sporadic thyroid carcinomas. Thyroid 2006, 16, 17–23. [Google Scholar] [CrossRef]
- Hashemi, M.; Rezaei, H.; Eskandari-Nasab, E.; Kaykhaei, M.A.; Taheri, M. Association of promoter methylation and 32-bp deletion of the PTEN gene with susceptibility to metabolic syndrome. Mol. Med. Rep. 2013, 7, 342–346. [Google Scholar] [CrossRef]
- Hino, R.; Uozaki, H.; Murakami, N.; Ushiku, T.; Shinozaki, A.; Ishikawa, S.; Morikawa, T.; Nakaya, T.; Sakatani, T.; Takada, K.; et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009, 69, 2766–2774. [Google Scholar] [CrossRef]
- Lu, J.; Jeong, H.W.; Kong, N.; Yang, Y.; Carroll, J.; Luo, H.R.; Silberstein, L.E.; Yupoma; Chai, L. Stem cell factor SALL4 represses the transcriptions of PTEN and SALL1 through an epigenetic repressor complex. PLoS ONE 2009, 4, 5577. [Google Scholar] [CrossRef]
- Liu, C.; Wu, H.; Li, Y.; Shen, L.; Yu, R.; Yin, H.; Sun, T.; Sun, C.; Zhou, Y.; Du, Z. SALL4 suppresses PTEN expression to promote glioma cell proliferation via PI3K/AKT signaling pathway. J. Neur. 2017, 135, 263–272. [Google Scholar] [CrossRef]
- Yoshimi, A.; Goyama, S.; Watanabe-Okochi, N.; Yoshiki, Y.; Nannya, Y.; Nitta, E.; Arai, S.; Sato, T.; Shimabe, M.; Nakagawa, M.; et al. Evi1 represses PTEN expression and activates PI3K/AKT/mTOR via interactions with polycomb proteins. Blood 2011, 117, 3617–3628. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Wehbe-Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef]
- Yang, S.M.; Huang, C.; Li, X.F.; Yu, M.Z.; He, Y.; Li, J. miR-21 confers cisplatin resistance in gastric cancer cells by regulating PTEN. Toxicology 2013, 306, 162–168. [Google Scholar] [CrossRef]
- Zhang, W.; Bai, W.; Zhang, W. MiR-21 suppresses the anticancer activities of curcumin by targeting PTEN gene in human non-small cell lung cancer A549 cells. Clin. Transl. Oncol. 2014, 16, 708–713. [Google Scholar] [CrossRef]
- He, C.; Dong, X.; Zhai, B.; Jiang, X.; Dong, D.; Li, B.; Jiang, H.; Xu, S.; Sun, X. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget 2015, 6, 28867–28881. [Google Scholar] [CrossRef]
- Song, L.; Liu, S.; Zhang, L.; Yao, H.; Gao, F.; Xu, D.; Li, Q. MiR-21 modulates radiosensitivity of cervical cancer through inhibiting autophagy via the PTEN/Akt/HIF-1alpha feedback loop and the Akt-mTOR signaling pathway. Tumour Biol. 2016, 37, 12161–12168. [Google Scholar] [CrossRef]
- Tumaneng, K.; Schlegelmilch, K.; Russell, R.C.; Yimlamai, D.; Basnet, H.; Mahadevan, N.; Fitamant, J.; Bardeesy, N.; Camargo, F.D.; Guan, K.L. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat. Cell Biol. 2012, 14, 1322–1329. [Google Scholar] [CrossRef]
- Shen, H.; Li, L.; Yang, S.; Wang, D.; Zhong, S.; Zhao, J.; Tang, J. MicroRNA-29a contributes to drug-resistance of breast cancer cells to adriamycin through PTEN/AKT/GSK3beta signaling pathway. Gene 2016, 593, 84–90. [Google Scholar] [CrossRef]
- Zheng, J.; Wu, C.; Lin, Z.; Guo, Y.; Shi, L.; Dong, P.; Lu, Z.; Gao, S.; Liao, Y.; Chen, B.; et al. Curcumin up-regulates phosphatase and tensin homologue deleted on chromosome 10 through microRNA-mediated control of DNA methylation--a novel mechanism suppressing liver fibrosis. FEBS J. 2014, 281, 88–103. [Google Scholar] [CrossRef]
- Yang, Y.L.; Wang, F.S.; Li, S.C.; Tiao, M.M.; Huang, Y.H. MicroRNA-29a Alleviates Bile Duct Ligation Exacerbation of Hepatic Fibrosis in Mice through Epigenetic Control of Methyltransferases. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef]
- Chang, R.M.; Xu, J.F.; Fang, F.; Yang, H.; Yang, L.Y. MicroRNA-130b promotes proliferation and EMT-induced metastasis via PTEN/p-AKT/HIF-1alpha signaling. Tumour Biol. 2016, 37, 10609–10619. [Google Scholar] [CrossRef] [PubMed]
- Egawa, H.; Jingushi, K.; Hirono, T.; Ueda, Y.; Kitae, K.; Nakata, W.; Fujita, K.; Uemura, M.; Nonomura, N.; Tsujikawa, K. The miR-130 family promotes cell migration and invasion in bladder cancer through FAK and Akt phosphorylation by regulating PTEN. Sci. Rep. 2016, 6, 20574. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Cui, R.; Bahr, J.; Zanesi, N.; Luo, Z.; Meng, W.; Liang, G.; Croce, C.M. miR-130a Deregulates PTEN and Stimulates Tumor Growth. Cancer Res. 2017, 77, 6168–6178. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, N.; Wang, Z.; Ai, D.M.; Cao, Z.Y.; Pan, H.P. Pseudogene PTENP1 Functions as a Competing Endogenous RNA (ceRNA) to Regulate PTEN Expression by Sponging miR-499-5p. Biochemistry 2016, 81, 739–747. [Google Scholar] [CrossRef]
- Guo, X.; Deng, L.; Deng, K.; Wang, H.; Shan, T.; Zhou, H.; Liang, Z.; Xia, J.; Li, C. Pseudogene PTENP1 Suppresses Gastric Cancer Progression by Modulating PTEN. Anticancer Agents Med. Chem. 2016, 16, 456–464. [Google Scholar] [CrossRef]
- Li, R.K.; Gao, J.; Guo, L.H.; Huang, G.Q.; Luo, W.H. PTENP1 acts as a ceRNA to regulate PTEN by sponging miR-19b and explores the biological role of PTENP1 in breast cancer. Cancer Gene Ther. 2017, 24, 309–315. [Google Scholar] [CrossRef]
- Qian, Y.Y.; Li, K.; Liu, Q.Y.; Liu, Z.S. Long non-coding RNA PTENP1 interacts with miR-193a-3p to suppress cell migration and invasion through the PTEN pathway in hepatocellular carcinoma. Oncotarget 2017, 8, 107859–107869. [Google Scholar] [CrossRef]
- Chen, S.; Wang, Y.; Zhang, J.H.; Xia, Q.J.; Sun, Q.; Li, Z.K.; Zhang, J.G.; Tang, M.S.; Dong, M.S. Long non-coding RNA PTENP1 inhibits proliferation and migration of breast cancer cells via AKT and MAPK signaling pathways. Oncol. Lett. 2017, 14, 4659–4662. [Google Scholar] [CrossRef]
- Zhang, R.; Guo, Y.; Ma, Z.; Ma, G.; Xue, Q.; Li, F.; Liu, L. Long non-coding RNA PTENP1 functions as a ceRNA to modulate PTEN level by decoying miR-106b and miR-93 in gastric cancer. Oncotarget 2017, 8, 26079–26089. [Google Scholar] [CrossRef]
- Gao, L.; Ren, W.; Zhang, L.; Li, S.; Kong, X.; Zhang, H.; Dong, J.; Cai, G.; Jin, C.; Zheng, D.; et al. PTENp1, a natural sponge of miR-21, mediates PTEN expression to inhibit the proliferation of oral squamous cell carcinoma. Mol. Carcinog. 2017, 56, 1322–1334. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Song, W.Q.; Sun, P.; Jin, L.; Dai, H.Y. LncRNA-GAS5 induces PTEN expression through inhibiting miR-103 in endometrial cancer cells. J. Biomed. Sci. 2015, 22, 100. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Chen, S.; Jiang, Z.; Shao, Y.; Jiang, X.; Li, P.; Xiao, B.; Guo, J. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci. Rep. 2015, 5, 13445. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Q.; Li, H. LncRNA FER1L4 suppresses cancer cell proliferation and cycle by regulating PTEN expression in endometrial carcinoma. Biochem. Biophys. Res. Commun. 2016, 478, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Chen, B.; Wang, X.; Wu, K.; Sun, Y. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer 2017, 17, 248. [Google Scholar] [CrossRef]
- Gu, S.; Xie, R.; Liu, X.; Shou, J.; Gu, W.; Che, X. Long Coding RNA XIST Contributes to Neuronal Apoptosis through the Downregulation of AKT Phosphorylation and Is Negatively Regulated by miR-494 in Rat Spinal Cord Injury. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef]
- Li, Y.; Gu, J.; Lu, H. The GAS5/miR-222 Axis Regulates Proliferation of Gastric Cancer Cells Through the PTEN/Akt/mTOR Pathway. Dig. Dis. Sci. 2017, 62, 3426–3437. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Yao, G.; Liu, B.; Ma, T.; Xia, Y.; Wei, K.; Wang, J.; Xu, J.; Chen, L.; Chen, Y. Long Noncoding RNA FAL1 Promotes Cell Proliferation, Invasion and Epithelial-Mesenchymal Transition Through the PTEN/AKT Signaling Axis in Non-Small Cell Lung Cancer. Cell Physiol. Biochem. 2017, 43, 339–352. [Google Scholar] [CrossRef]
- Qin, N.; Tong, G.F.; Sun, L.W.; Xu, X.L. Long Noncoding RNA MEG3 Suppresses Glioma Cell Proliferation, Migration, and Invasion by Acting as a Competing Endogenous RNA of miR-19a. Oncol. Res. 2017, 25, 1471–1478. [Google Scholar] [CrossRef]
- Tao, H.; Zhang, J.G.; Qin, R.H.; Dai, C.; Shi, P.; Yang, J.J.; Deng, Z.Y.; Shi, K.H. LncRNA GAS5 controls cardiac fibroblast activation and fibrosis by targeting miR-21 via PTEN/MMP-2 signaling pathway. Toxicology 2017, 386, 11–18. [Google Scholar] [CrossRef]
- Wu, Z.; He, Y.; Li, D.; Fang, X.; Shang, T.; Zhang, H.; Zheng, X. Long noncoding RNA MEG3 suppressed endothelial cell proliferation and migration through regulating miR-21. Am. J. Transl. Res. 2017, 9, 3326–3335. [Google Scholar] [PubMed]
- Yang, C.; Wang, G.; Yang, J.; Wang, L. Long noncoding RNA NBAT1 negatively modulates growth and metastasis of osteosarcoma cells through suppression of miR-21. Am. J. Cancer Res. 2017, 7, 2009–2019. [Google Scholar] [PubMed]
- Zhu, B.; Gong, Y.; Yan, G.; Wang, D.; Qiao, Y.; Wang, Q.; Liu, B.; Hou, J.; Li, R.; Tang, C. Down-regulation of lncRNA MEG3 promotes hypoxia-induced human pulmonary artery smooth muscle cell proliferation and migration via repressing PTEN by sponging miR-21. Biochem. Biophys Res. Commun. 2018, 495, 2125–2132. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Liu, J. LncRNA-p21 inhibited the proliferation of osteosarcoma cells via the miR-130b/PTEN/AKT signaling pathway. Biomed. Pharm. 2018, 97, 911–918. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, H.; Jin, L.; Li, G.; Hu, S.; Ning, C.; Guo, J.; Shuai, S.; Li, X.; Li, M. Long Noncoding RNA GAS5 Suppresses 3T3-L1 Cells Adipogenesis Through miR-21a-5p/PTEN Signal Pathway. DNA Cell Biol. 2018, 37, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Lin, Z.; Xu, J.; Lu, Y.; Meng, Q.; Wang, C.; Yang, Y.; Xin, X.; Li, X.; Pu, H.; et al. Long noncoding RNA MEG3 suppresses liver cancer cells growth through inhibiting beta-catenin by activating PKM2 and inactivating PTEN. Cell Death Dis. 2018, 9, 253. [Google Scholar] [CrossRef]
- Li, D.; Feng, J.; Wu, T.; Wang, Y.; Sun, Y.; Ren, J.; Liu, M. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am. J. Pathol. 2013, 182, 64–70. [Google Scholar] [CrossRef]
- Lai, Y.; He, S.; Ma, L.; Lin, H.; Ren, B.; Ma, J.; Zhu, X.; Zhuang, S. HOTAIR functions as a competing endogenous RNA to regulate PTEN expression by inhibiting miR-19 in cardiac hypertrophy. Mol. Cell Biochem. 2017, 432, 179–187. [Google Scholar] [CrossRef]
- Yu, F.; Chen, B.; Dong, P.; Zheng, J. HOTAIR Epigenetically Modulates PTEN Expression via MicroRNA-29b: A Novel Mechanism in Regulation of Liver Fibrosis. Mol. Ther. 2017, 25, 205–217. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, G.; Yang, Y.; Jiang, Z.; Cai, J.; Zhang, Z.; Hu, H. Long noncoding RNA HOTAIRM1 inhibits cell progression by regulating miR-17-5p/ PTEN axis in gastric cancer. J. Cell Biochem. 2018, 120, 4952–4965. [Google Scholar] [CrossRef]
- Vazquez, F.; Ramaswamy, S.; Nakamura, N.; Sellers, W.R. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell Biol. 2000, 20, 5010–5018. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.; Pulido, R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J. Biol. Chem. 2001, 276, 993–998. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.; Rodriguez, J.; Myers, M.P.; Valiente, M.; Graves, J.D.; Tonks, N.K.; Pulido, R. Phosphorylation-regulated cleavage of the tumor suppressor PTEN by caspase-3: Implications for the control of protein stability and PTEN-protein interactions. J. Biol. Chem. 2003, 278, 30652–30660. [Google Scholar] [CrossRef] [PubMed]
- Maccario, H.; Perera, N.M.; Davidson, L.; Downes, C.P.; Leslie, N.R. PTEN is destabilized by phosphorylation on Thr366. Biochem. J. 2007, 405, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Tolkacheva, T.; Boddapati, M.; Sanfiz, A.; Tsuchida, K.; Kimmelman, A.C.; Chan, A.M. Regulation of PTEN binding to MAGI-2 by two putative phosphorylation sites at threonine 382 and 383. Cancer Res. 2001, 61, 4985–4989. [Google Scholar] [PubMed]
- Yang, Z.; Xie, C.; Xu, W.; Liu, G.; Cao, X.; Li, W.; Chen, J.; Zhu, Y.; Luo, S.; Luo, Z.; et al. Phosphorylation and inactivation of PTEN at residues Ser380/Thr382/383 induced by Helicobacter pylori promotes gastric epithelial cell survival through PI3K/Akt pathway. Oncotarget 2015, 6, 31916–31926. [Google Scholar] [CrossRef] [PubMed]
- Pua, T.L.; Wang, F.Q.; Fishman, D.A. Roles of LPA in ovarian cancer development and progression. Future Oncol. 2009, 5, 1659–1673. [Google Scholar] [CrossRef]
- Rahdar, M.; Inoue, T.; Meyer, T.; Zhang, J.; Vazquez, F.; Devreotes, P.N. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc. Natl. Acad. Sci. USA 2009, 106, 480–485. [Google Scholar] [CrossRef]
- Trotman, L.C.; Wang, X.; Alimonti, A.; Chen, Z.; Teruya-Feldstein, J.; Yang, H.; Pavletich, N.P.; Carver, B.S.; Cordon-Cardo, C.; Erdjument-Bromage, H.; et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 2007, 128, 141–156. [Google Scholar] [CrossRef]
- Yan, Y.; Li, Z.; Zeng, S.; Wang, X.; Gong, Z.; Xu, Z. FGFR2-mediated phosphorylation of PTEN at tyrosine 240 contributes to the radioresistance of glioma. J. Cell Commun. Signal. 2019, 13, 279–280. [Google Scholar] [CrossRef]
- Fenton, T.R.; Nathanson, D.; Ponte de Albuquerque, C.; Kuga, D.; Iwanami, A.; Dang, J.; Yang, H.; Tanaka, K.; Oba-Shinjo, S.M.; Uno, M.; et al. Resistance to EGF receptor inhibitors in glioblastoma mediated by phosphorylation of the PTEN tumor suppressor at tyrosine 240. Proc. Natl. Acad. Sci. USA 2012, 109, 14164–14169. [Google Scholar] [CrossRef]
- Wang, X.; Trotman, L.C.; Koppie, T.; Alimonti, A.; Chen, Z.; Gao, Z.; Wang, J.; Erdjument-Bromage, H.; Tempst, P.; Cordon-Cardo, C.; et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 2007, 128, 129–139. [Google Scholar] [CrossRef]
- Wang, X.; Shi, Y.; Wang, J.; Huang, G.; Jiang, X. Crucial role of the C-terminus of PTEN in antagonizing NEDD4-1-mediated PTEN ubiquitination and degradation. Biochem. J. 2008, 414, 221–229. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Deb, S.; Paul, I.; Chatterjee, A.; Mandal, T.; Chatterjee, U.; Ghosh, M.K. The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J. Biol. Chem. 2012, 287, 15996–16006. [Google Scholar] [CrossRef]
- Li, N.; Zhang, Y.; Han, X.; Liang, K.; Wang, J.; Feng, L.; Wang, W.; Songyang, Z.; Lin, C.; Yang, L.; et al. Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth. Genes Dev. 2015, 29, 157–170. [Google Scholar] [CrossRef]
- Maddika, S.; Kavela, S.; Rani, N.; Palicharla, V.R.; Pokorny, J.L.; Sarkaria, J.N.; Chen, J. WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 2011, 13, 728–733. [Google Scholar] [CrossRef]
- Li, H.; Zhang, P.; Zhang, Q.; Li, C.; Zou, W.; Chang, Z.; Cui, C.P.; Zhang, L. WWP2 is a physiological ubiquitin ligase for phosphatase and tensin homolog (PTEN) in mice. J. Biol. Chem. 2018, 293, 8886–8899. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, Z.; Ma, Z.; Liu, H.; Wu, Y.; Zhang, Q. WWP1 as a potential tumor oncogene regulates PTEN-Akt signaling pathway in human gastric carcinoma. Tumour Biol. 2015, 36, 787–798. [Google Scholar] [CrossRef]
- Lee, Y.R.; Chen, M.; Lee, J.D.; Zhang, J.; Lin, S.Y.; Fu, T.M.; Chen, H.; Ishikawa, T.; Chiang, S.Y.; Katon, J.; et al. Reactivation of PTEN tumor suppressor for cancer treatment through inhibition of a MYC-WWP1 inhibitory pathway. Science 2019, 364, 159. [Google Scholar] [CrossRef]
- Van Themsche, C.; Leblanc, V.; Parent, S.; Asselin, E. X-linked inhibitor of apoptosis protein (XIAP) regulates PTEN ubiquitination, content, and compartmentalization. J. Biol. Chem. 2009, 284, 20462–20466. [Google Scholar] [CrossRef]
- Lee, S.R.; Yang, K.S.; Kwon, J.; Lee, C.; Jeong, W.; Rhee, S.G. Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 2002, 277, 20336–20342. [Google Scholar] [CrossRef]
- Kwon, J.; Lee, S.R.; Yang, K.S.; Ahn, Y.; Kim, Y.J.; Stadtman, E.R.; Rhee, S.G. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc. Natl. Acad. Sci. USA 2004, 101, 16419–16424. [Google Scholar] [CrossRef]
- Okumura, K.; Mendoza, M.; Bachoo, R.M.; DePinho, R.A.; Cavenee, W.K.; Furnari, F.B. PCAF modulates PTEN activity. J. Biol. Chem. 2006, 281, 26562–26568. [Google Scholar] [CrossRef]
- Ikenoue, T.; Inoki, K.; Zhao, B.; Guan, K.L. PTEN acetylation modulates its interaction with PDZ domain. Cancer Res. 2008, 68, 6908–6912. [Google Scholar] [CrossRef]
- Meng, Z.; Jia, L.F.; Gan, Y.H. PTEN activation through K163 acetylation by inhibiting HDAC6 contributes to tumour inhibition. Oncogene 2016, 35, 2333–2344. [Google Scholar] [CrossRef]
- Numajiri, N.; Takasawa, K.; Nishiya, T.; Tanaka, H.; Ohno, K.; Hayakawa, W.; Asada, M.; Matsuda, H.; Azumi, K.; Kamata, H.; et al. On-off system for PI3-kinase-Akt signaling through S-nitrosylation of phosphatase with sequence homology to tensin (PTEN). Proc. Natl. Acad. Sci. USA 2011, 108, 10349–10354. [Google Scholar] [CrossRef]
- Huang, J.; Yan, J.; Zhang, J.; Zhu, S.; Wang, Y.; Shi, T.; Zhu, C.; Chen, C.; Liu, X.; Cheng, J.; et al. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat. Commun. 2012, 3, 911. [Google Scholar] [CrossRef]
- Fouladkou, F.; Landry, T.; Kawabe, H.; Neeb, A.; Lu, C.; Brose, N.; Stambolic, V.; Rotin, D. The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proc. Natl. Acad. Sci. USA 2008, 105, 8585–8590. [Google Scholar] [CrossRef]
- Covey, T.M.; Edes, K.; Fitzpatrick, F.A. Akt activation by arachidonic acid metabolism occurs via oxidation and inactivation of PTEN tumor suppressor. Oncogene 2007, 26, 5784–5792. [Google Scholar] [CrossRef]
- Hui, S.T.; Andres, A.M.; Miller, A.K.; Spann, N.J.; Potter, D.W.; Post, N.M.; Chen, A.Z.; Sachithanantham, S.; Jung, D.Y.; Kim, J.K.; et al. Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proc. Natl. Acad. Sci. USA 2008, 105, 3921–3926. [Google Scholar] [CrossRef]
- Shen, S.M.; Guo, M.; Xiong, Z.; Yu, Y.; Zhao, X.Y.; Zhang, F.F.; Chen, G.Q. AIF inhibits tumor metastasis by protecting PTEN from oxidation. EMBO Rep. 2015, 16, 1563–1580. [Google Scholar] [CrossRef] [PubMed]
- Pei, D.S.; Sun, Y.F.; Song, Y.J. S-nitrosylation of PTEN Invovled in ischemic brain injury in rat hippocampal CA1 region. Neurochem Res. 2009, 34, 1507–1512. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Anjomani-Virmouni, S.; Koundouros, N.; Dimitriadi, M.; Choo-Wing, R.; Valle, A.; Zheng, Y.; Chiu, Y.H.; Agnihotri, S.; Zadeh, G.; et al. PARK2 Depletion Connects Energy and Oxidative Stress to PI3K/Akt Activation via PTEN S-Nitrosylation. Mol. Cell 2017, 65, 999–1013. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Anjomani-Virmouni, S.; Koundouros, N.; Poulogiannis, G. PARK2 loss promotes cancer progression via redox-mediated inactivation of PTEN. Mol. Cell Oncol. 2017, 4, 1329692. [Google Scholar] [CrossRef]
- Zhu, L.; Li, L.; Zhang, Q.; Yang, X.; Zou, Z.; Hao, B.; Marincola, F.M.; Liu, Z.; Zhong, Z.; Wang, M.; et al. NOS1 S-nitrosylates PTEN and inhibits autophagy in nasopharyngeal carcinoma cells. Cell Death Discov. 2017, 3, 17011. [Google Scholar] [CrossRef]
- Zhao, J.J.; Cheng, H.; Jia, S.; Wang, L.; Gjoerup, O.V.; Mikami, A.; Roberts, T.M. The p110α isoform of PI3K is essential for proper growth factor signaling and oncogenic transformation. Proc. Natl. Acad. Sci. USA 2006, 103, 16296–16300. [Google Scholar] [CrossRef]
- Gupta, S.; Ramjaun, A.R.; Haiko, P.; Wang, Y.; Warne, P.H.; Nicke, B.; Nye, E.; Stamp, G.; Alitalo, K.; Downward, J. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell 2007, 129, 957–968. [Google Scholar] [CrossRef]
- Jiang, X.; Chen, S.; Asara, J.M.; Balk, S.P. Phosphoinositide 3-kinase pathway activation in phosphate and tensin homolog (PTEN)-deficient prostate cancer cells is independent of receptor tyrosine kinases and mediated by the p110beta and p110delta catalytic subunits. J. Biol. Chem. 2010, 285, 14980–14989. [Google Scholar] [CrossRef]
- Cizmecioglu, O.; Ni, J.; Xie, S.; Zhao, J.J.; Roberts, T.M. Rac1-mediated membrane raft localization of PI3K/p110beta is required for its activation by GPCRs or PTEN loss. eLife 2016, 5, 17635. [Google Scholar] [CrossRef]
- Lasserre, R.; Guo, X.J.; Conchonaud, F.; Hamon, Y.; Hawchar, O.; Bernard, A.M.; Soudja, S.M.; Lenne, P.F.; Rigneault, H.; Olive, D.; et al. Raft nanodomains contribute to Akt/PKB plasma membrane recruitment and activation. Nat. Chem. Biol. 2008, 4, 538–547. [Google Scholar] [CrossRef]
- Fritsch, R.; de Krijger, I.; Fritsch, K.; George, R.; Reason, B.; Kumar, M.S.; Diefenbacher, M.; Stamp, G.; Downward, J. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell 2013, 153, 1050–1063. [Google Scholar] [CrossRef]
- Welch, H.C.; Coadwell, W.J.; Stephens, L.R.; Hawkins, P.T. Phosphoinositide 3-kinase-dependent activation of Rac. FEBS Lett. 2003, 546, 93–97. [Google Scholar] [CrossRef]
- Yuzugullu, H.; Baitsch, L.; Von, T.; Steiner, A.; Tong, H.; Ni, J.; Clayton, L.K.; Bronson, R.; Roberts, T.M.; Gritsman, K.; et al. A PI3K p110beta-Rac signalling loop mediates Pten-loss-induced perturbation of haematopoiesis and leukaemogenesis. Nat. Commun. 2015, 6, 8501. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, X.; Schmit, F.; Adelmant, G.; Eck, M.J.; Marto, J.A.; Zhao, J.J.; Roberts, T.M. CRKL Mediates p110beta-Dependent PI3K Signaling in PTEN-Deficient Cancer Cells. Cell Rep. 2017, 20, 549–557. [Google Scholar] [CrossRef]
- Sharma, A.; Mayer, B.J. Phosphorylation of p130Cas initiates Rac activation and membrane ruffling. BMC Cell Biol. 2008, 9, 50. [Google Scholar] [CrossRef]
- Wozniak, M.A.; Modzelewska, K.; Kwong, L.; Keely, P.J. Focal adhesion regulation of cell behavior. Biochim. Bio. Acta. 2004, 1692, 103–119. [Google Scholar] [CrossRef]
- Lin, Z.; Zhou, P.; von Gise, A.; Gu, F.; Ma, Q.; Chen, J.; Guo, H.; van Gorp, P.R.; Wang, D.Z.; Pu, W.T. Pi3kcb links Hippo-YAP and PI3K-AKT signaling pathways to promote cardiomyocyte proliferation and survival. Circ. Res. 2015, 116, 35–45. [Google Scholar] [CrossRef]
- Jang, J.W.; Kim, M.K.; Lee, Y.S.; Lee, J.W.; Kim, D.M.; Song, S.H.; Lee, J.Y.; Choi, B.Y.; Min, B.; Chi, X.Z.; et al. RAC-LATS1/2 signaling regulates YAP activity by switching between the YAP-binding partners TEAD4 and RUNX3. Oncogene 2017, 36, 999–1011. [Google Scholar] [CrossRef]
- Sethi, I.; Cai, Z.; Roberts, T.M.; Yuan, G.C. Molecular Profiling Establishes Genetic Features Predictive of the Efficacy of the p110beta Inhibitor KIN-193. Cancer Res. 2019, 79, 4524–4531. [Google Scholar] [CrossRef]
- Rodon, J.; Dienstmann, R.; Serra, V.; Tabernero, J. Development of PI3K inhibitors: Lessons learned from early clinical trials. Nat. Rev. Clin. Oncol. 2013, 10, 143–153. [Google Scholar] [CrossRef]
- Yap, T.A.; Bjerke, L.; Clarke, P.A.; Workman, P. Drugging PI3K in cancer: Refining targets and therapeutic strategies. Curr. Opin. Pharm. 2015, 23, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Ganji, G.; Lemech, C.; Burris, H.A.; Han, S.W.; Swales, K.; Decordova, S.; DeYoung, M.P.; Smith, D.A.; Kalyana-Sundaram, S.; et al. A First-Time-in-Human Study of GSK2636771, a Phosphoinositide 3 Kinase Beta-Selective Inhibitor, in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2017, 23, 5981–5992. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.R.; Shapiro, G.; Do, K.T.; Kumar, R.; Martin-Liberal, J.; Higano, C.S.; Wisinski, K.B.; Dean, E.J.; Heath, E.I.; Rathkopf, D.E.; et al. A first in human phase I study of AZD8186, a potent and selective inhibitor of PI3K in patients with advanced solid tumours as monotherapy and in combination with the dual mTORC1/2 inhibitor vistusertib (AZD2014) or abiraterone acetate. J. Clin. Oncol. 2017, 35, 2570. [Google Scholar] [CrossRef]
- Saura, C.; Roda, D.; Rosello, S.; Oliveira, M.; Macarulla, T.; Perez-Fidalgo, J.A.; Morales-Barrera, R.; Sanchis-Garcia, J.M.; Musib, L.; Budha, N.; et al. A First-in-Human Phase I Study of the ATP-Competitive AKT Inhibitor Ipatasertib Demonstrates Robust and Safe Targeting of AKT in Patients with Solid Tumors. Cancer Discov. 2017, 7, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Foukas, L.C.; Claret, M.; Pearce, W.; Okkenhaug, K.; Meek, S.; Peskett, E.; Sancho, S.; Smith, A.J.; Withers, D.J.; Vanhaesebroeck, B. Critical role for the p110alpha phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature 2006, 441, 366–370. [Google Scholar] [CrossRef]
- Mulholland, D.J.; Tran, L.M.; Li, Y.; Cai, H.; Morim, A.; Wang, S.; Plaisier, S.; Garraway, I.P.; Huang, J.; Graeber, T.G.; et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 2011, 19, 792–804. [Google Scholar] [CrossRef]
- Rescigno, P.; de Bono, J.; Aparicio, A.; Chowdhury, S.; Twardowski, P.; Dawson, N.; Vaishampayan, U.; Pantuck, A.J.; Zhou, Y.; Fecteau, D.; et al. 792PDPhase I, open-label, dose-finding study of GSK2636771, a phosphoinositide 3-kinase (PI3K)β inhibitor, in combination with enzalutamide in male subjects with metastatic castration-resistant prostate cancer (mCRPC). Annals Oncol. 2017, 28, 5. [Google Scholar] [CrossRef]
- Suzuki, A.; Yamaguchi, M.T.; Ohteki, T.; Sasaki, T.; Kaisho, T.; Kimura, Y.; Yoshida, R.; Wakeham, A.; Higuchi, T.; Fukumoto, M.; et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity 2001, 14, 523–534. [Google Scholar] [CrossRef]
- Peng, W.; Chen, J.Q.; Liu, C.; Malu, S.; Creasy, C.; Tetzlaff, M.T.; Xu, C.; McKenzie, J.A.; Zhang, C.; Liang, X.; et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov. 2016, 6, 202–216. [Google Scholar] [CrossRef]
- Peng, W.; Xu, C.; Melendez, B.; Jackson, H.; McKenzi, J.A.; Williams, L.J.; Chen, Y.; Mbofung, R.M.; Leahey, S.E.L.E.; Lizee, G.; et al. Abstract 4938: OX40 agonist antibody-based combination therapy with PI3Kβ selective inhibitor enhances T cell immunity. Cancer Res. 2018, 78, 4938. [Google Scholar] [CrossRef]
- Tawbi, H.A.-H.; Peng, W.; Milton, D.; Amaria, R.N.; Glitza, I.C.; Hwu, W.-J.; Patel, S.P.; Wong, M.K.K.; Woodman, S.E.; Yee, C.; et al. Phase I/II study of the PI3Kβ inhibitor GSK2636771 in combination with pembrolizumab (P) in patients (pts) with PD-1 refractory metastatic melanoma (MM) and PTEN loss. J. Clin. Oncol. 2018, 36, 9596. [Google Scholar] [CrossRef]
- Kaelin, W.G., Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer 2005, 5, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Mendes-Pereira, A.M.; Martin, S.A.; Brough, R.; McCarthy, A.; Taylor, J.R.; Kim, J.S.; Waldman, T.; Lord, C.J.; Ashworth, A. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol. Med. 2009, 1, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Lu, X.; Wang, G.; Lan, Z.; Liao, W.; Li, J.; Liang, X.; Chen, J.R.; Shah, S.; Shang, X.; et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature 2017, 542, 484–488. [Google Scholar] [CrossRef]
- Turchick, A.; Liu, Y.; Zhao, W.; Cohen, I.; Glazer, P.M. Synthetic lethality of a cell-penetrating anti-RAD51 antibody in PTEN-deficient melanoma and glioma cells. Oncotarget 2019, 10, 1272–1283. [Google Scholar] [CrossRef]
- Ren, W.; Joshi, R.; Mathew, P. Synthetic Lethality in PTEN-Mutant Prostate Cancer Is Induced by Combinatorial PI3K/Akt and BCL-XL Inhibition. Mol. Cancer Res. 2016, 14, 1176–1181. [Google Scholar] [CrossRef]
- Li, K.; Yan, H.; Guo, W.; Tang, M.; Zhao, X.; Tong, A.; Peng, Y.; Li, Q.; Yuan, Z. ATM inhibition induces synthetic lethality and enhances sensitivity of PTEN-deficient breast cancer cells to cisplatin. Exp. Cell Res. 2018, 366, 24–33. [Google Scholar] [CrossRef]
- Ding, Y.; Li, N.; Dong, B.; Guo, W.; Wei, H.; Chen, Q.; Yuan, H.; Han, Y.; Chang, H.; Kan, S.; et al. Chromatin remodeling ATPase BRG1 and PTEN are synthetic lethal in prostate cancer. J. Clin. Invest. 2019, 129, 759–773. [Google Scholar] [CrossRef]
- Chen, P.; Zhao, D.; Li, J.; Liang, X.; Li, J.; Chang, A.; Henry, V.K.; Lan, Z.; Spring, D.J.; Rao, G.; et al. Symbiotic Macrophage-Glioma Cell Interactions Reveal Synthetic Lethality in PTEN-Null Glioma. Cancer Cell 2019, 35, 868–884. [Google Scholar] [CrossRef]
- Chatterjee, N.; Pazarentzos, E.; Mayekar, M.K.; Gui, P.; Allegakoen, D.V.; Hrustanovic, G.; Olivas, V.; Lin, L.; Verschueren, E.; Johnson, J.R.; et al. Synthetic Essentiality of Metabolic Regulator PDHK1 in PTEN-Deficient Cells and Cancers. Cell Rep. 2019, 28, 2317–2330. [Google Scholar] [CrossRef]
- Schwertassek, U.; Haque, A.; Krishnan, N.; Greiner, R.; Weingarten, L.; Dick, T.P.; Tonks, N.K. Reactivation of oxidized PTP1B and PTEN by thioredoxin 1. FEBS J. 2014, 281, 3545–3558. [Google Scholar] [CrossRef] [PubMed]
| Modification | Modifier | Site |
|---|---|---|
| phosphorylation | CK2 [121,122,123] | S370, S385 [121,122,123] |
| GSK3β [124] | T366 [124] | |
| T382, T383 [125] S380, T382, T383 [126] S380, T382, T383, T385 [127,128] | ||
| FGFR,SRC family kinases [129] FGFR2 [130] | Y240 [130,131] | |
| Ubiquitination | Need4-1 [129,132,133] CHIP [134] NRF146 [135] WWP2 [136,137] WWP1 [138,139] XIAP [140] | K13, K289 K48 [134] K27 [139] |
| Oxidation | ROS [141,142] Thioreductase [142] | C71, C124 [141,142] |
| Acetylation | PCAF [143] | K125, K128 [143] |
| CBP [144] | K402 [144] | |
| HDAC6 [145] | K163 [145] | |
| S-nitrosylation | NO [146] | C83 [146] |
| Ribosylation | TNKS1, TNKS2 [135] | |
| Sumoylation | SUMO1 [147] | K266 [147] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, H.; Cai, Z.; Roberts, T.M. The Mechanisms Underlying PTEN Loss in Human Tumors Suggest Potential Therapeutic Opportunities. Biomolecules 2019, 9, 713. https://doi.org/10.3390/biom9110713
Chang H, Cai Z, Roberts TM. The Mechanisms Underlying PTEN Loss in Human Tumors Suggest Potential Therapeutic Opportunities. Biomolecules. 2019; 9(11):713. https://doi.org/10.3390/biom9110713
Chicago/Turabian StyleChang, Hyeyoun, Zhenying Cai, and Thomas M. Roberts. 2019. "The Mechanisms Underlying PTEN Loss in Human Tumors Suggest Potential Therapeutic Opportunities" Biomolecules 9, no. 11: 713. https://doi.org/10.3390/biom9110713
APA StyleChang, H., Cai, Z., & Roberts, T. M. (2019). The Mechanisms Underlying PTEN Loss in Human Tumors Suggest Potential Therapeutic Opportunities. Biomolecules, 9(11), 713. https://doi.org/10.3390/biom9110713




