Antioxidant Barrier, Redox Status, and Oxidative Damage to Biomolecules in Patients with Colorectal Cancer. Can Malondialdehyde and Catalase Be Markers of Colorectal Cancer Advancement?
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Blood Collection
2.3. Determination of Redox Markers
2.4. Enzymatic and Non-Enzymatic Antioxidants
2.5. Total Antioxidant/Oxidant Status
2.6. Oxidative Damage Products
2.7. H + E Staining
2.8. Statistical Analysis
3. Results
3.1. Clinical Findings
3.2. Antioxidant Defence
3.3. Total Antioxidant/Oxidant Status
3.4. Oxidative Damage Products
3.5. ROC Analysis
3.6. Correlations
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Das, V.; Kalita, J.; Pal, M. Predictive and prognostic biomarkers in colorectal cancer: A systematic review of recent advances and challenges. Biomed. Pharmacother. 2017, 87, 8–19. [Google Scholar] [CrossRef]
- Pino, M.S.; Chung, D.C. The chromosomal instability pathway in colon cancer. Gastroenterology 2010, 138, 2059–2072. [Google Scholar] [CrossRef]
- Limoli, C.L.; Giedzinski, E. Induction of Chromosomal Instability by Chronic Oxidative Stress. Neoplasia 2003, 5, 339–346. [Google Scholar] [CrossRef]
- Mena, S.; Ortega, A.; Estrela, J.M. Oxidative stress in environmental-induced carcinogenesis. Mutat. Res. Toxicol. Environ. Mutagen. 2009, 674, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Carini, F.; Mazzola, M.; Rappa, F.; Jurjus, A.; Geagea, A.G.; Al Kattar, S.; Bou-Assi, T.; Jurjus, R.; Damiani, P.; Leone, A.; et al. Colorectal Carcinogenesis: Role of Oxidative Stress and Antioxidants. Anticancer. Res. 2017, 37, 4759–4766. [Google Scholar] [PubMed]
- Mandal, P. Potential biomarkers associated with oxidative stress for risk assessment of colorectal cancer. Naunyn-Schmiedeberg’s Arch. Pharm. 2017, 390, 557–565. [Google Scholar] [CrossRef]
- Poprac, P.; Jomova, K.; Simunkova, M.; Kollar, V.; Rhodes, C.J.; Valko, M. Targeting Free Radicals in Oxidative Stress-Related Human Diseases. Trends Pharmacol. Sci. 2017, 38, 592–607. [Google Scholar] [CrossRef]
- Friguet, B. Oxidized protein degradation and repair in ageing and oxidative stress. FEBS Lett. 2006, 580, 2910–2916. [Google Scholar] [CrossRef]
- Valko, M.; Rhodes, C.; Moncol, J.; Izakovic, M.; Mazur, M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Interact. 2006, 160, 1–40. [Google Scholar] [CrossRef]
- Fejfer, K.; Buczko, P.; Niczyporuk, M.; Ładny, J.R.; Hady, H.R.; Knaś, M.; Waszkiel, D.; Klimiuk, A.; Zalewska, A.; Maciejczyk, M. Oxidative Modification of Biomolecules in the Nonstimulated and Stimulated Saliva of Patients with Morbid Obesity Treated with Bariatric Surgery. BioMed Res. Int. 2017, 2017, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pouvreau, C.; Dayre, A.; Butkowski, E.G.; De Jong, B.; Jelinek, H.F. Inflammation and oxidative stress markers in diabetes and hypertension. J. Inflamm. Res. 2018, 11, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Maciejczyk, M.; Szulimowska, J.; Skutnik, A.; Taranta-Janusz, K.; Wasilewska, A.; Wiśniewska, N.; Zalewska, A. Salivary Biomarkers of Oxidative Stress in Children with Chronic Kidney Disease. J. Clin. Med. 2018, 7, 209. [Google Scholar] [CrossRef] [PubMed]
- Klimiuk, A.; Maciejczyk, M.; Choromańska, M.; Fejfer, K.; Waszkiewicz, N.; Zalewska, A. Salivary Redox Biomarkers in Different Stages of Dementia Severity. J. Clin. Med. 2019, 8, 840. [Google Scholar] [CrossRef] [PubMed]
- Sawczuk, B.; Maciejczyk, M.; Sawczuk-Siemieniuk, M.; Posmyk, R.; Zalewska, A.; Car, H. Salivary Gland Function, Antioxidant Defence and Oxidative Damage in the Saliva of Patients with Breast Cancer: Does the BRCA1 Mutation Disturb the Salivary Redox Profile? Cancers 2019, 11, 1501. [Google Scholar] [CrossRef]
- Bosman, F.; Carneiro, F.; Hruban, R.; Theise, N. WHO Classification of Tumours of the Digestive System, 4th ed.; International World Health Organization: Geneva, Switzerland, 2010. [Google Scholar]
- Amin, M.B.; Edge, S.; Greene, F.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. AJCC Cancer Staging Manual, 8th ed.; Springer: New York, NY, USA, 2017. [Google Scholar]
- Kołodziej, U.; Maciejczyk, M.; Miąsko, A.; Matczuk, J.; Knaś, M.; Żukowski, P.; Żendzian-Piotrowska, M.; Borys, J.; Zalewska, A. Oxidative Modification in the Salivary Glands of High Fat-Diet Induced Insulin Resistant Rats. Front. Physiol. 2017, 8, 736. [Google Scholar] [CrossRef]
- Walker, J.M. The bicinchoninic acid (BCA) assay for protein quantitation. Methods Mol. Biol. 1994, 32, 5–8. [Google Scholar]
- Misra, H.P.; Fridovich, I. The role of superoxide anion in the autoxidation of epinephrine and a simple assay for superoxide dismutase. J. Boil. Chem. 1972, 247, 3170–3175. [Google Scholar]
- Aebi, H. Catalase in vitro. Methods Enzym. 1984, 105, 121–126. [Google Scholar]
- E Paglia, D.; Valentine, W.N. Studies on the quantitative and qualitative characterization of erythrocyte glutathione peroxidase. J. Lab. Clin. Med. 1967, 70, 158–169. [Google Scholar]
- Mize, C.E.; Langdon, R.G. Hepatic glutathione reductase. I. Purification and general kinetic properties. J. Biol. Chem. 1962, 237, 1589–1595. [Google Scholar] [PubMed]
- Griffith, O.W. Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal. Biochem. 1980, 106, 207–212. [Google Scholar] [CrossRef]
- Erel, O. A novel automated direct measurement method for total antioxidant capacity using a new generation, more stable ABTS radical cation. Clin. Biochem. 2004, 37, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Erel, O. A new automated colorimetric method for measuring total oxidant status. Clin. Biochem. 2005, 38, 1103–1111. [Google Scholar] [CrossRef]
- Knaś, M.; Maciejczyk, M.; Daniszewska, I.; Klimiuk, A.; Matczuk, J.; Kołodziej, U.; Waszkiel, D.; Ładny, J.R.; Żendzian-Piotrowska, M.; Zalewska, A. Oxidative Damage to the Salivary Glands of Rats with Streptozotocin-Induced Diabetes-Temporal Study: Oxidative Stress and Diabetic Salivary Glands. J. Diabetes Res. 2016, 2016, 1–13. [Google Scholar] [CrossRef]
- Benzie, I.F.; Strain, J. The Ferric Reducing Ability of Plasma (FRAP) as a Measure of “Antioxidant Power”: The FRAP Assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef]
- Kalousová, M.; Skrha, J.; Zima, T. Advanced glycation end-products and advanced oxidation protein products in patients with diabetes mellitus. Physiol. Res. 2002, 51, 597–604. [Google Scholar]
- Buege, J.A.; Aust, S.D. Microsomal lipid peroxidation. Methods Enzym. 1978, 52, 302–310. [Google Scholar]
- Maciejczyk, M.; Żebrowska, E.; Chabowski, A. Insulin Resistance and Oxidative Stress in the Brain: What’s New? Int. J. Mol. Sci. 2019, 20, 874. [Google Scholar] [CrossRef]
- Gopcevic, K.R.; Rovčanin, B.R.; Tatić, S.B.; Krivokapić, Z.V.; Gajic, M.M.; Dragutinović, V.V. Activity of Superoxide Dismutase, Catalase, Glutathione Peroxidase, and Glutathione Reductase in Different Stages of Colorectal Carcinoma. Dig. Dis. Sci. 2013, 58, 2646–2652. [Google Scholar] [CrossRef]
- Chiang, F.-F.; Wang, H.-M.; Lan, Y.-C.; Yang, M.-H.; Huang, S.-C.; Huang, Y.-C. High homocysteine is associated with increased risk of colorectal cancer independently of oxidative stress and antioxidant capacities. Clin. Nutr. 2014, 33, 1054–1060. [Google Scholar] [CrossRef] [PubMed]
- Lushchak, V.I. Free radicals, reactive oxygen species, oxidative stress and its classification. Chem. Interactions 2014, 224, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Mayo, J.C.; Tan, D.-X.; Sainz, R.M.; Lopez-Burillo, S.; Reiter, R.J. Oxidative damage to catalase induced by peroxyl radicals: Functional protection by melatonin and other antioxidants. Free. Radic. Res. 2003, 37, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Perše, M. Oxidative stress in the pathogenesis of colorectal cancer: Cause or consequence? Biomed Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Perše, M.; Injac, R.; Štrukelj, B.; Cerar, A. High fat mixed lipid diet modifies protective effects of exercise on 1,2 dimethylhydrazine induced colon cancer in rats. Technol. Cancer Res. Treat. 2012, 11, 289–299. [Google Scholar] [CrossRef]
- Perse, M.; Injac, R.; Strukelj, B.; Cerar, A. Effects of high-fat mixed-lipid diet and exercise on the antioxidant system in skeletal and cardiac muscles of rats with colon carcinoma. Pharmacol. Rep. 2009, 61, 909–916. [Google Scholar] [CrossRef]
- Perše, M.; Cerar, A. Morphological and molecular alterations in 1,2 dimethylhydrazine and azoxymethane induced colon carcinogenesis in rats. J. Biomed. Biotechnol. 2011, 2011, 473964. [Google Scholar]
- Zalewska, A.; Ziembicka, D.; Żendzian-Piotrowska, M.; Maciejczyk, M. The Impact of High-Fat Diet on Mitochondrial Function, Free Radical Production, and Nitrosative Stress in the Salivary Glands of Wistar Rats. Oxid. Med. Cell. Longev. 2019, 2019. [Google Scholar] [CrossRef]
- Aleksandrova, K.; Nimptsch, K.; Pischon, T. Influence of Obesity and Related Metabolic Alterations on Colorectal Cancer Risk. Curr. Nutr. Rep. 2013, 2, 1–9. [Google Scholar] [CrossRef][Green Version]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid Peroxidation: Production, Metabolism, and Signaling Mechanisms of Malondialdehyde and 4-Hydroxy-2-Nonenal. Oxidative Med. Cell. Longev. 2014, 2014, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Klaunig, J.E.; Kamendulis, L.M. Therole Ofoxidativestress Incarcinogenesis. Annu. Rev. Pharmacol. Toxicol. 2004, 44, 239–267. [Google Scholar] [CrossRef] [PubMed]
- Adams, S. Reactive carbonyl formation by oxidative and non-oxidative pathways. Front. Biosci. 2001, 6, a17. [Google Scholar] [CrossRef]
- Alderman, C.J.J.; Shah, S.; Foreman, J.C.; Chain, B.M.; Katz, D.R. The role of advanced oxidation protein products in regulation of dendritic cell function. Free. Radic. Boil. Med. 2002, 32, 377–385. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, L.; Rong, S.; Qu, H.; Zhang, Y.; Chang, D.; Pan, H.; Wang, W. Relation between Gastric Cancer and Protein Oxidation, DNA Damage, and Lipid Peroxidation. Oxidative Med. Cell. Longev. 2013, 2013, 1–6. [Google Scholar] [CrossRef]
- Kosova, F.; Çetin, B.; Akıncı, M.; Aslan, S.; Arı, Z.; Sepici, A.; Altan, N.; Cetin, A.; Akinci, M.; Ari, Z. Advanced Oxidation Protein Products, Ferrous Oxidation in Xylenol Orange, and Malondialdehyde Levels in Thyroid Cancer. Ann. Surg. Oncol. 2007, 14, 2616–2620. [Google Scholar] [CrossRef]
- Kiliç, N.; Taslipinar, M.Y.; Guney, Y.; Tekin, E.; Onuk, E. An Investigation into the Serum Thioredoxin, Superoxide Dismutase, Malondialdehyde, and Advanced Oxidation Protein Products in Patients with Breast Cancer. Ann. Surg. Oncol. 2014, 21, 4139–4143. [Google Scholar] [CrossRef]
- Nayyar, A.S.; Khan, M.; Vijayalakshmi, K.R.; Suman, B.; Gayitri, H.C.; Anitha, M. Serum total protein, albumin and advanced oxidation protein products (AOPP)--implications in oral squamous cell carcinoma. Malays. J. Pathol. 2012, 34, 47–52. [Google Scholar]
- Piskounova, E.; Agathocleous, M.; Murphy, M.M.; Hu, Z.; Huddlestun, S.E.; Zhao, Z.; Leitch, A.M.; Johnson, T.M.; DeBerardinis, R.J.; Morrison, S.J. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 2015, 527, 186–191. [Google Scholar] [CrossRef]
- Gao, P.; Zhang, H.; Dinavahi, R.; Li, F.; Xiang, Y.; Raman, V.; Bhujwalla, Z.M.; Felsher, D.W.; Cheng, L.; Pevsner, J.; et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell 2007, 12, 230–238. [Google Scholar] [CrossRef]
- Glasauer, A.; Chandel, N.S. Targeting antioxidants for cancer therapy. Biochem. Pharmacol. 2014, 92, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Glasauer, A.; Sena, L.A.; Diebold, L.P.; Mazar, A.P.; Chandel, N.S. Targeting SOD1 reduces experimental non–small-cell lung cancer. J. Clin. Invest. 2014, 124, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Mysliwiec, P.; Pawlak, K.; Kuklinski, A.; Kędra, B. Combined perioperative plasma endoglin and VEGF--a assessment in colorectal cancer patients. Folia Histochem. Cytobiol. 2009, 47, 231–236. [Google Scholar] [CrossRef]
- Kuklinski, A.; Kamocki, Z.; Koda, M.; Piotrowski, Z.; Sulkowski, S.; Leśniewicz, R.; Pawlak, K.; Myśliwiec, P.; Kedra, B. IGF-IR in patients with advanced colorectal cancer in correlation with certain clinico-morphological factors: Initial report. Oncol. Lett. 2011, 2, 1155–1159. [Google Scholar] [CrossRef] [PubMed]
- Lech, G.; Słotwiński, R.; Słodkowski, M.; Krasnodębski, I.W. Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World J. Gastroenterol. 2016, 22, 1745–1755. [Google Scholar] [CrossRef] [PubMed]
- Pietrucha, B.; Heropolitańska-Pliszka, E.; Maciejczyk, M.; Car, H.; Sawicka-Powierza, J.; Motkowski, R.; Karpinska, J.; Hryniewicka, M.; Zalewska, A.; Pac, M.; et al. Comparison of Selected Parameters of Redox Homeostasis in Patients with Ataxia-Telangiectasia and Nijmegen Breakage Syndrome. Oxidative Med. Cell. Longev. 2017, 2017, 1–8. [Google Scholar] [CrossRef]
- Pallardó, F.V.; Lloret, A.; Lebel, M.; D’Ischia, M.; Cogger, V.C.; Le Couteur, D.G.; Gadaleta, M.N.; Castello, G.; Pagano, G. Mitochondrial dysfunction in some oxidative stress-related genetic diseases: Ataxia-Telangiectasia, Down Syndrome, Fanconi Anaemia and Werner Syndrome. Biogerontology 2010, 11, 401–419. [Google Scholar] [CrossRef]
- Catalán, V.; Frühbeck, G.; Gómez-Ambrosi, J. Inflammatory and Oxidative Stress Markers in Skeletal Muscle of Obese Subjects. In Obesity; Elsevier BV: Amsterdam, The Netherlands, 2018; pp. 163–189. [Google Scholar]
- Świderska, M.; Maciejczyk, M.; Zalewska, A.; Pogorzelska, J.; Flisiak, R.; Chabowski, A. Oxidative stress biomarkers in the serum and plasma of patients with non-alcoholic fatty liver disease (NAFLD). Can plasma AGE be a marker of NAFLD? Free Radic. Res. 2019, 53, 841–851. [Google Scholar] [CrossRef]
- Fortea-Sanchis, C.; Martínez-Ramos, D.; Escrig-Sos, J. The lymph node status as a prognostic factor in colon cancer: Comparative population study of classifications using the logarithm of the ratio between metastatic and nonmetastatic nodes (LODDS) versus the pN-TNM classification and ganglion ratio systems. BMC Cancer 2018, 18, 1208. [Google Scholar] [CrossRef]
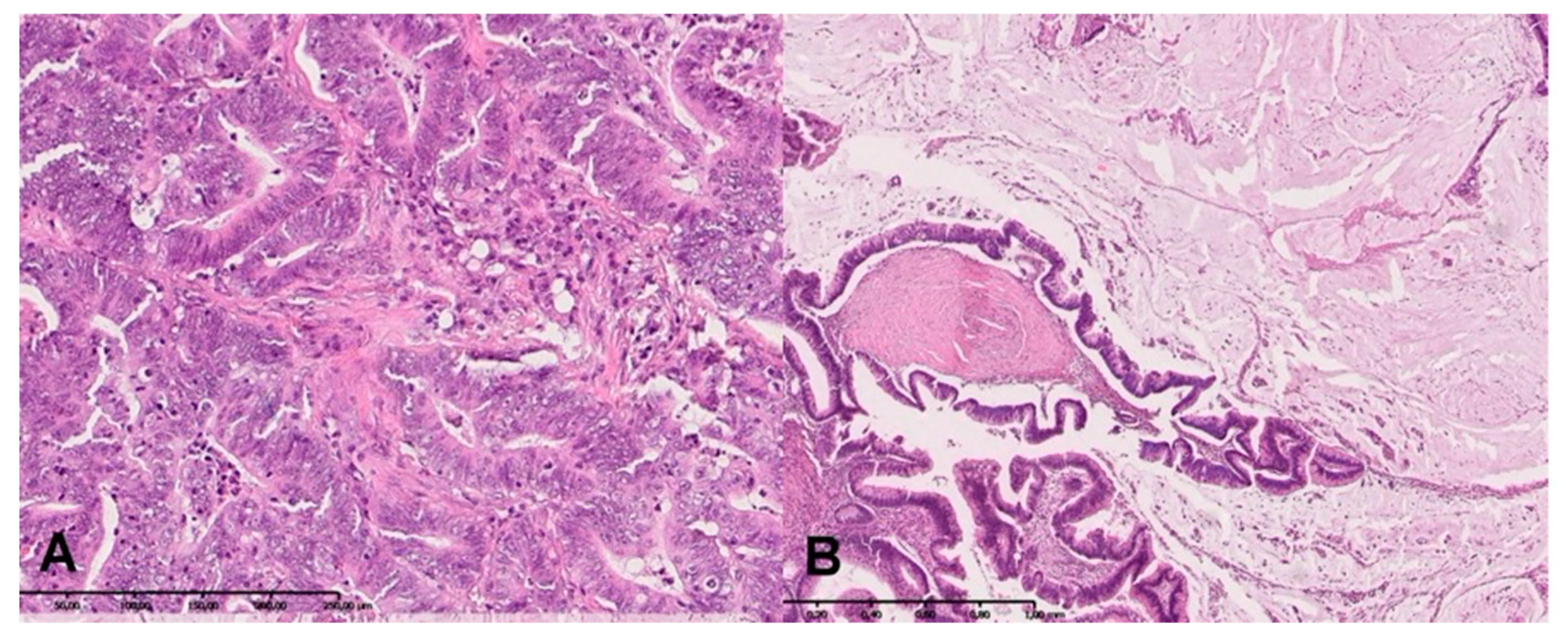
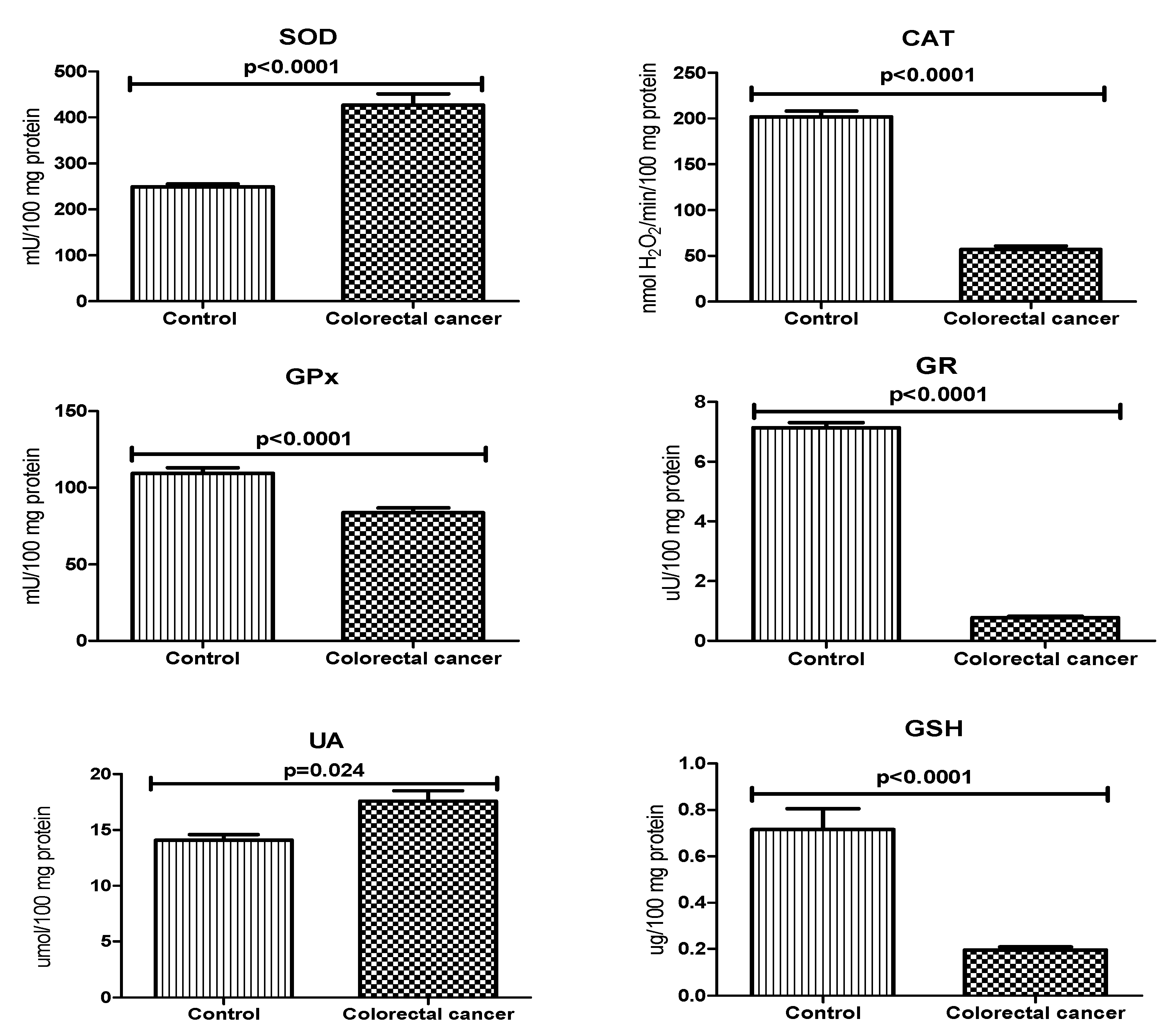
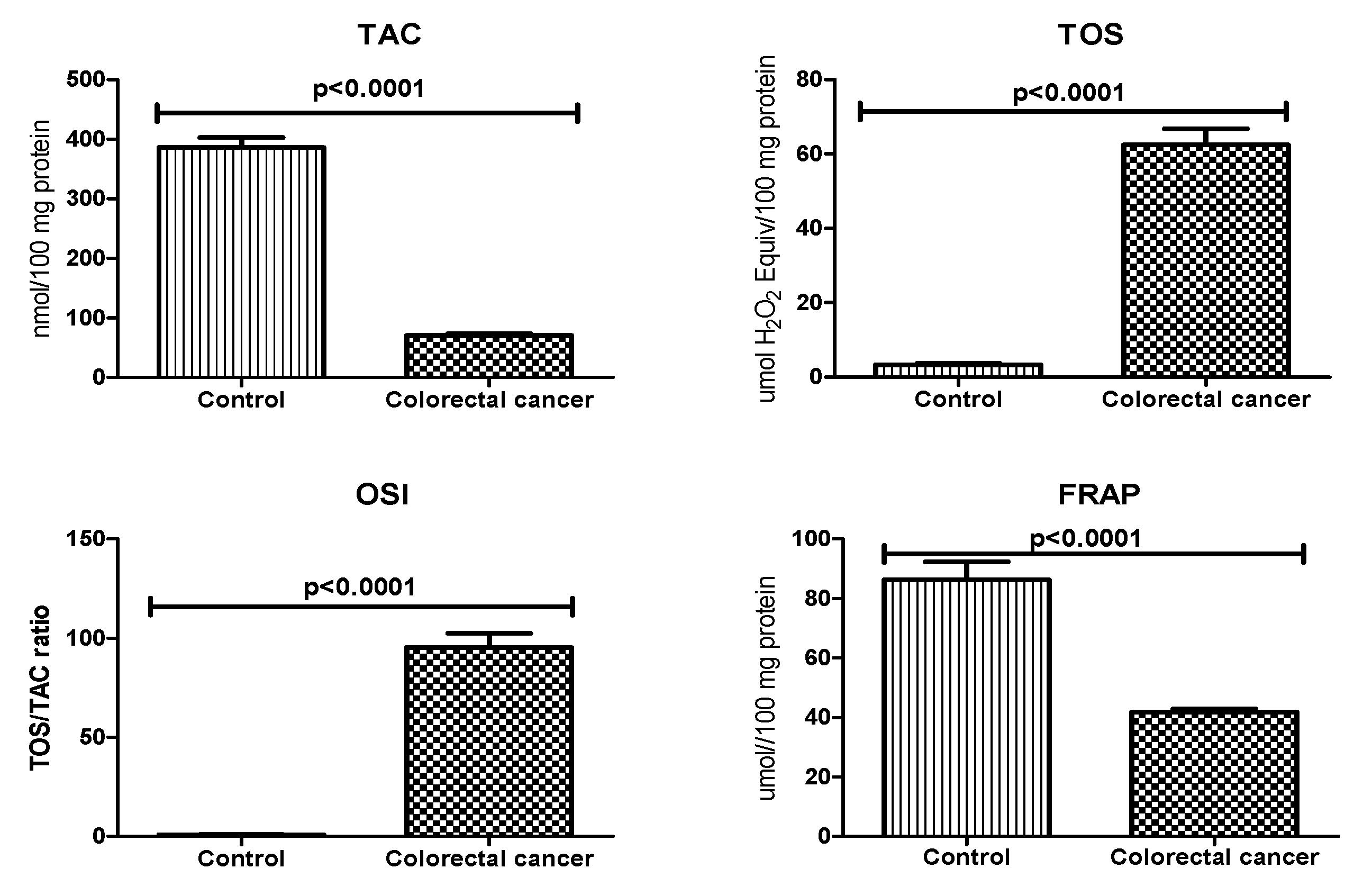
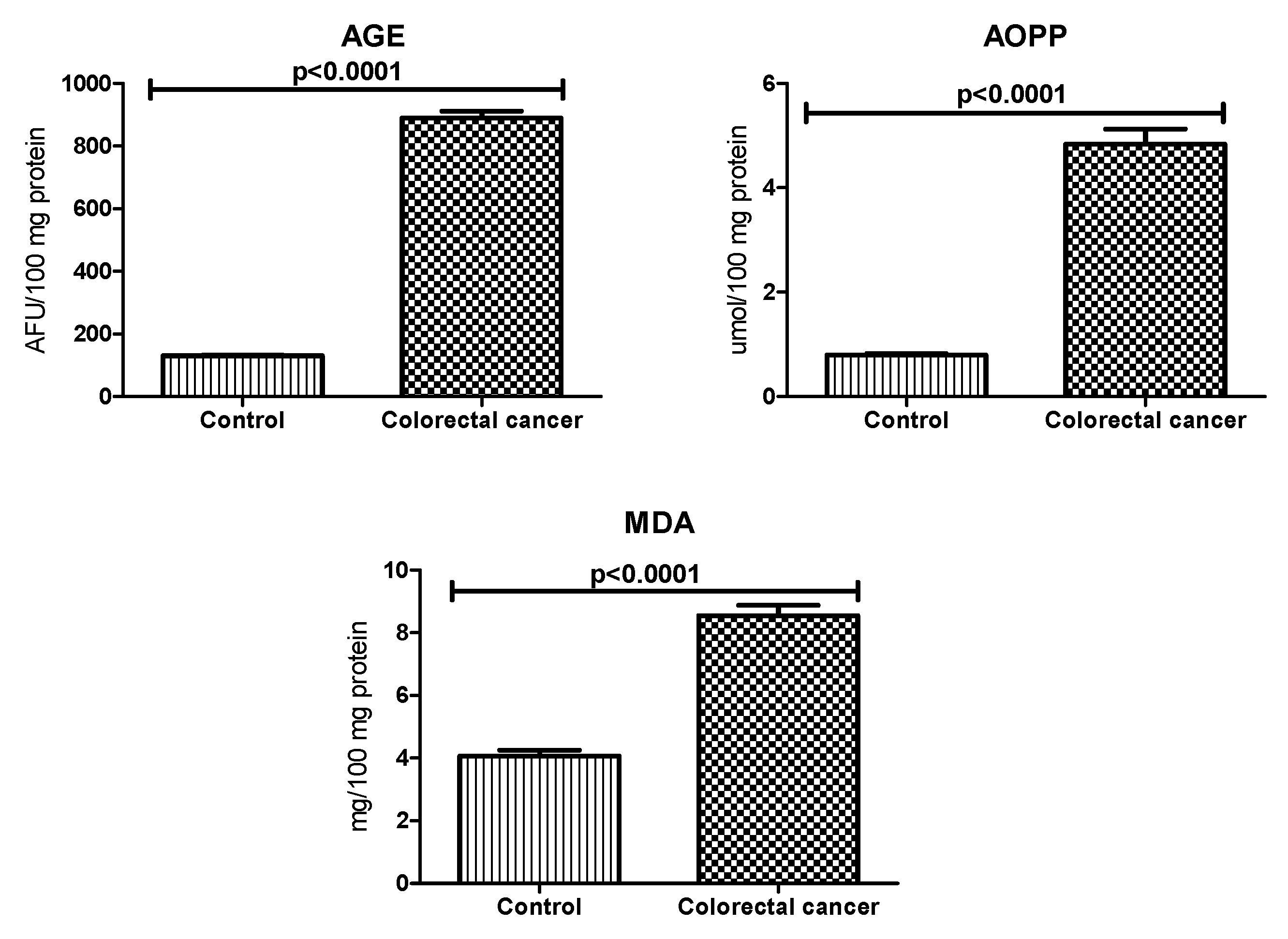
| Parameter | n (%) |
|---|---|
| Age <60 >60 | 11 (22.0%) 39 (78.0%) |
| Sex male female | 31 (62.0%) 19 (38.0%) |
| Histological type adenocarcinoma mucinous adenocarcinoma | 40 (80.0%) 10 (20.0%) |
| Tumour location sigmoid colon rectum cecum ascending colon hepatic fold colon | 15 (30.0%) 15 (30.0%) 7 (14.0%) 6 (12.0%) 5 (10.0%) 2 (4.0%) |
| Tumour’s size <3cm >3cm | 15 (30.0%) 35 (70.0%) |
| pT—depth of invasion T1 T2 T3 T4 | 12 (24.0%) 14 (28.0%) 18 (36.0%) 6 (12.0%) |
| pN—lymph node metastasis N0 N1 N2 | 30 (60.0%) 12 (24.0%) 8 (16.0%) |
| pM—distant metastasis M0 M1 | 44 (88.0%) 6 (12.0%) |
| Stage at diagnosis I II III IV | 11 (22.0%) 16 (32.0%) 18 (36.0%) 5 (10.0%) |
| CEA level (ng/mL) 0–5.0 >5.0 | 36 (72.0%) 14 (28.0%) |
| Parameter | AUC | p-Value | Cut-Off | Sensitivity (%) | Specificity (%) | 95% Confidence Interval |
|---|---|---|---|---|---|---|
| Antioxidant defense | ||||||
| SOD (mU/100 mg protein) | 0.9048 | <0.0001 | >279.7 | 87.80 | 90.48 | 0.8242–0.9854 |
| CAT (nmol H2O2/min/100 mg protein) | 0.9988 | <0.0001 | <123.3 | 97.56 | 97.50 | 0.9955–1.002 |
| GPx (mU/100 mg protein) | 0.9209 | <0.0001 | <99.54 | 90.24 | 93.10 | 0.8387–1.003 |
| GR (uU/100 mg protein) | 1.000 | <0.0001 | <4.205 | 100.0 | 100.0 | 1.000–1.000 |
| UA (umol/100 mg protein) | 0.6880 | 0.002335 | >14.96 | 65.85 | 64.58 | 0.5756–0.8004 |
| GSH (ug/100 mg protein) | 0.9566 | <0.0001 | <0.2986 | 85.37 | 85.19 | 0.9124–1.001 |
| Redox status | ||||||
| TAC (nmol/100 mg protein) | 1.000 | <0.0001 | <168.5 | 100.0 | 100.0 | 1.000–1.000 |
| TOS (umol H2O2 Equiv/100 mg protein) | 1.000 | <0.0001 | >14.27 | 100.0 | 100.0 | 1.000–1.000 |
| OSI (TOS/TAC ratio) | 1.000 | <0.0001 | >15.49 | 100.0 | 100.0 | 1.000–1.000 |
| FRAP (umol/100 mg protein) | 0.9377 | <0.0001 | <49.49 | 90.24 | 89.47 | 0.8726–1.003 |
| Protein and lipid oxidative damage | ||||||
| AGE (AFU/100 mg protein) | 1.000 | <0.0001 | >322.0 | 100.0 | 100.0 | 1.000–1.000 |
| AOPP (umol/100 mg protein) | 1.000 | <0.0001 | >1.637 | 100.0 | 100.0 | 1.000–1.000 |
| MDA (mg/100 mg protein) | 0.9815 | <0.0001 | >5.669 | 92.68 | 92.00 | 0.9561–1.007 |
| Parameter | AUC | p-Value | Cut-Off | Sensitivity (%) | Specificity (%) | 95% Confidence Interval |
|---|---|---|---|---|---|---|
| Antioxidant defense | ||||||
| SOD (mU/100 mg protein) | 0.5507 | 0.6013 | >426.9 | 53.33 | 52.17 | 0.3590–0.7425 |
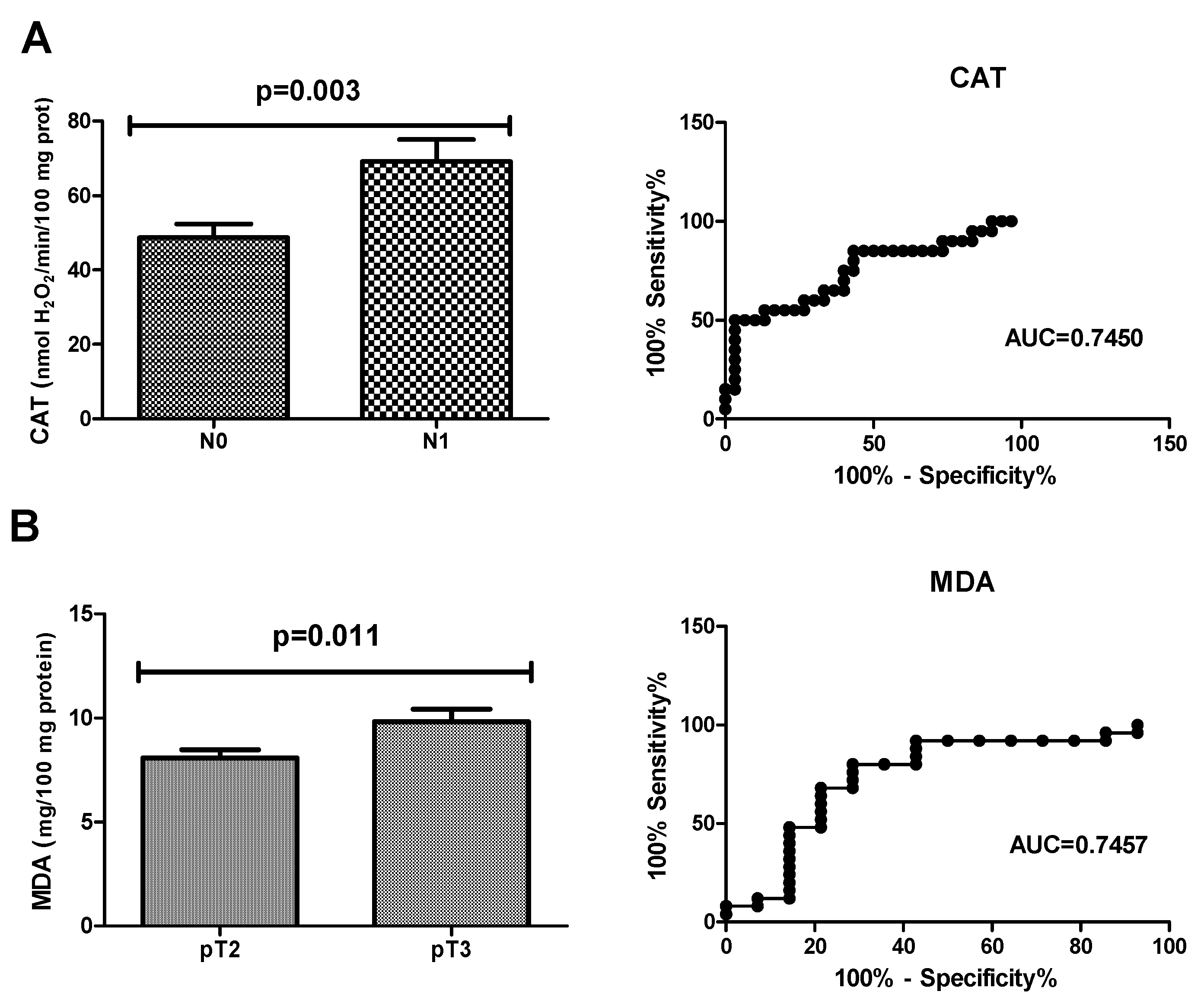
| CAT (nmol H2O2/min/100 mg protein) | 0.7450 | 0.0036 | >61.61 | 65.00 | 66.67 | 0.5989–0.8911 |
| GPx (mU/100 mg protein) | 0.6232 | 0.2044 | >79.95 | 53.33 | 56.52 | 0.4421–0.8043 |
| GR (uU/100 mg protein) | 0.6435 | 0.1394 | >0.7091 | 66.67 | 65.22 | 0.4677–0.8192 |
| UA (umol/100 mg protein) | 0.5681 | 0.4828 | >16.49 | 53.33 | 52.17 | 0.3825–0.7537 |
| GSH (ug/100 mg protein) | 0.6029 | 0.2891 | <0.1736 | 66.67 | 65.22 | 0.4185–0.7873 |
| Redox status | ||||||
| TAC (nmol/100 mg protein) | 0.5318 | 0.7544 | <70.33 | 46.15 | 47.83 | 0.3354–0.7282 |
| TOS (umol H2O2 Equiv/100 mg protein) | 0.5848 | 0.3864 | <48.65 | 53.33 | 54.55 | 0.3931–0.7766 |
| OSI (TOS/TAC ratio) | 0.5245 | 0.8111 | <85.56 | 53.85 | 54.55 | 0.3122–0.7368 |
| FRAP (umol//100 mg protein) | 0.5420 | 0.6650 | <41.71 | 60.00 | 60.87 | 0.3512–0.7329 |
| Protein and lipid oxidative damage | ||||||
| AGE (AFU/100 mg protein) | 0.6087 | 0.2628 | <873.3 | 53.33 | 52.17 | 0.4204–0.7970 |
| AOPP (umol/100 mg protein) | 0.6290 | 0.1839 | <4.388 | 66.67 | 65.22 | 0.4506–0.8074 |
| MDA (mg/100 mg protein) | 0.5188 | 0.8461 | <8.707 | 60.00 | 60.87 | 0.3293–0.7084 |
| Parameter | AUC | p-Value | Cut-Off | Sensitivity (%) | Specificity (%) | 95% Confidence Interval |
|---|---|---|---|---|---|---|
| Antioxidant defense | ||||||
| SOD (mU/100 mg protein) | 0.5500 | 0.6265 | >426.9 | 56.00 | 58.33 | 0.3361–0.7639 |
| CAT (nmol H2O2/min/100 mg protein) | 0.5633 | 0.5376 | >49.17 | 56.00 | 58.33 | 0.3758–0.7509 |
| GPx (mU/100 mg protein) | 0.5600 | 0.5592 | >79.22 | 48.00 | 50.00 | 0.3515–0.7685 |
| GR (uU/100 mg protein) | 0.6833 | 0.0744 | >0.6881 | 68.00 | 66.67 | 0.4925–0.8742 |
| UA (umol/100 mg protein) | 0.6067 | 0.2992 | >15.42 | 42.52 | 41.67 | 0.4228–0.7905 |
| GSH (ug/100 mg protein) | 0.5233 | 0.8203 | >0.1857 | 44.00 | 41.67 | 0.3302–0.7164 |
| Redox status | ||||||
| TAC (nmol/100 mg protein) | 0.5652 | 0.5316 | >70.33 | 56.52 | 58.33 | 0.3342–0.7962 |
| TOS (umol H2O2 Equiv/100 mg protein) | 0.5818 | 0.4397 | >47.87 | 56.00 | 54.55 | 0.3897–0.7739 |
| OSI (TOS/TAC ratio) | 0.5771 | 0.4729 | >85.56 | 52.17 | 54.55 | 0.3704–0.7837 |
| FRAP (umol//100 mg protein) | 0.5033 | 0.9741 | >43.19 | 40.00 | 41.67 | 0.2912–0.7154 |
| Protein and lipid oxidative damage | ||||||
| AGE (AFU/100 mg protein) | 0.5133 | 0.8967 | >873.3 | 48.00 | 50.00 | 0.3288–0.6978 |
| AOPP (umol/100 mg protein) | 0.5067 | 0.9483 | <4.475 | 52.00 | 50.00 | 0.3054–0.7079 |
| MDA (mg/100 mg protein) | 0.7457 | 0.0118 | <9.361 | 72.00 | 71.43 | 0.5678–0.9236 |
| Pair of Variables | r | p |
|---|---|---|
| GPx and GR | 0.575 | <0.0001 |
| FRAP and GSH | 0.575 | <0.0001 |
| UA and AGE | 0.407 | 0.008 |
| CAT and AOPP | −0.341 | 0.029 |
| GSH and AOPP | 0.351 | 0.024 |
| TOS and AOPP | 0.650 | <0.0001 |
| OSI and AOPP | 0.466 | 0.004 |
| UA and CA 19-9 | 0.509 | 0.026 |
| CAT and CA 19-9 | 0.642 | <0.0001 |
| GR and CA19-9 | 0.522 | 0.018 |
| GPx and CEA | −0.448 | 0.036 |
| MDA and CEA | 0.560 | 0.008 |
| UA and α1globulin | 0.547 | 0.028 |
| FRAP and location | 0.332 | 0.045 |
| MDA and pT | 0.460 | 0.008 |
| GPx and vascular invasion | 0.512 | 0.043 |
| MDA and CRP | 0.980 | <0.0001 |
| FRAP and total cholesterol | 0.670 | 0.009 |
| UA and total cholesterol | 0.565 | 0.035 |
| UA and eosinophils | −0.663 | 0.037 |
| SOD and monocytes | 0.745 | 0.013 |
| TOS and basophils | −0.735 | 0.016 |
| AGE and eosinophils | −0.717 | 0.020 |
| AGE and neutrophils | −0.636 | 0.047 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zińczuk, J.; Maciejczyk, M.; Zaręba, K.; Romaniuk, W.; Markowski, A.; Kędra, B.; Zalewska, A.; Pryczynicz, A.; Matowicka-Karna, J.; Guzińska-Ustymowicz, K. Antioxidant Barrier, Redox Status, and Oxidative Damage to Biomolecules in Patients with Colorectal Cancer. Can Malondialdehyde and Catalase Be Markers of Colorectal Cancer Advancement? Biomolecules 2019, 9, 637. https://doi.org/10.3390/biom9100637
Zińczuk J, Maciejczyk M, Zaręba K, Romaniuk W, Markowski A, Kędra B, Zalewska A, Pryczynicz A, Matowicka-Karna J, Guzińska-Ustymowicz K. Antioxidant Barrier, Redox Status, and Oxidative Damage to Biomolecules in Patients with Colorectal Cancer. Can Malondialdehyde and Catalase Be Markers of Colorectal Cancer Advancement? Biomolecules. 2019; 9(10):637. https://doi.org/10.3390/biom9100637
Chicago/Turabian StyleZińczuk, Justyna, Mateusz Maciejczyk, Konrad Zaręba, Wioletta Romaniuk, Adam Markowski, Bogusław Kędra, Anna Zalewska, Anna Pryczynicz, Joanna Matowicka-Karna, and Katarzyna Guzińska-Ustymowicz. 2019. "Antioxidant Barrier, Redox Status, and Oxidative Damage to Biomolecules in Patients with Colorectal Cancer. Can Malondialdehyde and Catalase Be Markers of Colorectal Cancer Advancement?" Biomolecules 9, no. 10: 637. https://doi.org/10.3390/biom9100637
APA StyleZińczuk, J., Maciejczyk, M., Zaręba, K., Romaniuk, W., Markowski, A., Kędra, B., Zalewska, A., Pryczynicz, A., Matowicka-Karna, J., & Guzińska-Ustymowicz, K. (2019). Antioxidant Barrier, Redox Status, and Oxidative Damage to Biomolecules in Patients with Colorectal Cancer. Can Malondialdehyde and Catalase Be Markers of Colorectal Cancer Advancement? Biomolecules, 9(10), 637. https://doi.org/10.3390/biom9100637







