Cytochalasans Act as Inhibitors of Biofilm Formation of Staphylococcus aureus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents, Media Ingredients and Solvents
2.2. Microorganisms
2.3. Fungal Specimens and Cultures Used in the Current Study and Origin of Reference Compounds
2.4. Purification of the Compounds
2.5. Spectral Data
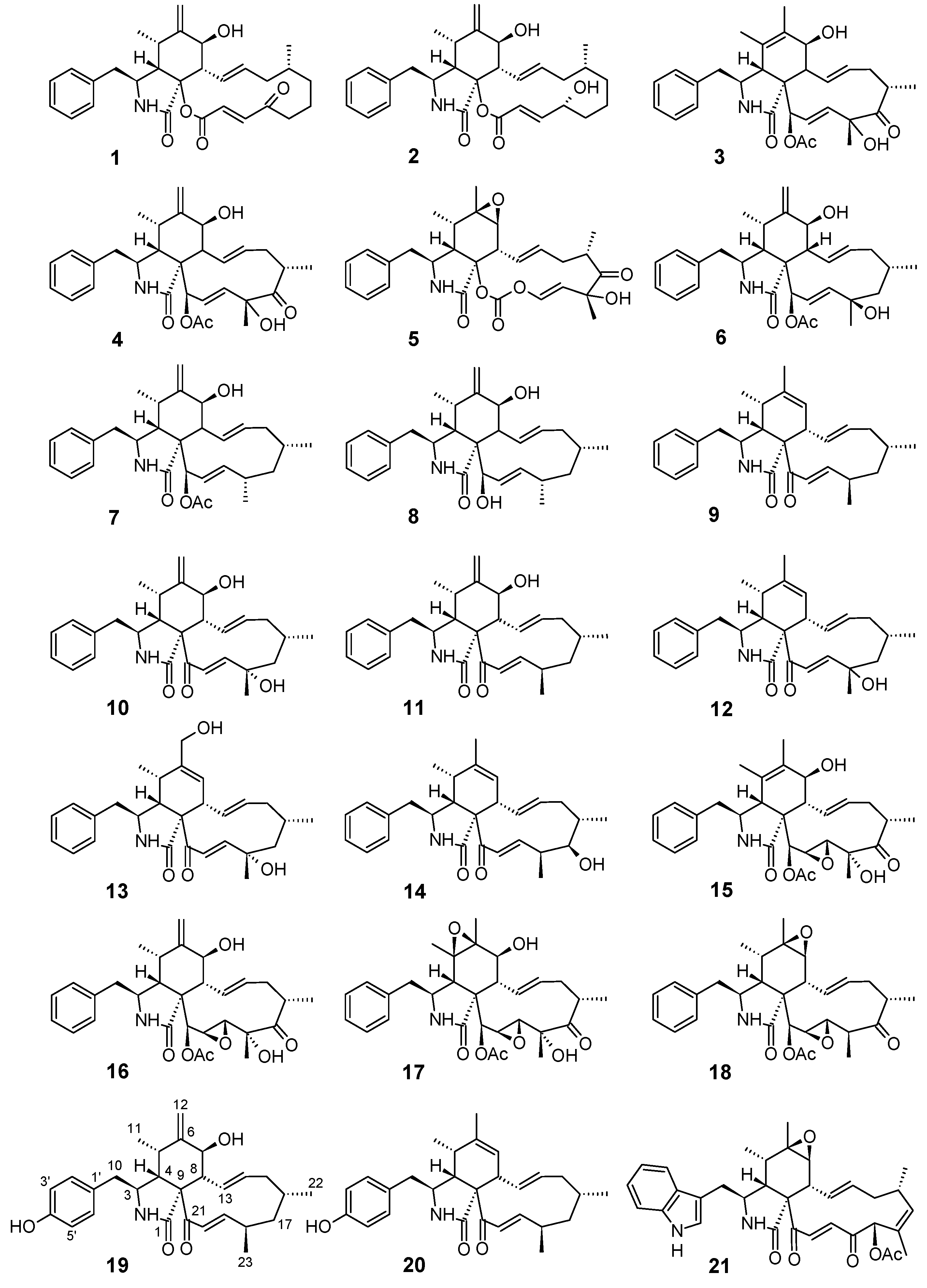
2.5.1. Phenochalasin C (19)
2.5.2. Phenochalasin D (20)
2.6. Bioassays
3. Results
3.1. Structure Elucidation of the New Compounds
3.2. Anti-Biofilm Activities of the Tested Cytochalasins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Klevens, R.M.; Edwards, J.R.; Richards, C.L.; Horan, T.C.; Gaynes, R.P.; Pollock, D.A.; Cardo, D.M. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep. 2007, 122, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef] [PubMed]
- Archer, N.K.; Mazaitis, M.J.; Costerton, J.W.; Leid, J.G.; Powers, M.E.; Shirtliff, M.E. Staphylococcus aureus biofilms: Properties, regulation and roles in human disease. Virulence 2011, 2, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Chepkirui, C.; Yuyama, K.; Wanga, L.; Decock, C.; Matasyoh, J.; Abraham, W.-R.; Stadler, M. Microporenic acids A-G, biofilm inhibitors and antimicrobial agents from the basidiomycete Microporus sp. J. Nat. Prod. 2018, 81, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Phukhamsakda, C.; Macabeo, A.P.G.; Yuyama, K.T.; Hyde, K.D.; Stadler, M. Biofilm inhibitory abscisic acid derivatives from the plant-associated Dothideomycete fungus, Roussoella sp. Molecules 2018, 23, 2190. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, M.P.; Gulotta, G.; do Amaral, M.W.; Lünsdorf, H.; Sasse, F.; Abraham, W.-R. Coprinuslactone protects the edible mushroom Coprinus comatus against biofilm infections by blocking both quorum-sensing and MurA. Environ. Microbiol. 2016, 18, 4254–4264. [Google Scholar] [CrossRef] [PubMed]
- Yuyama, K.T.; Chepkirui, C.; Wendt, L.; Fortkamp, D.; Stadler, M.; Abraham, W.R. Bioactive compounds produced by Hypoxylon fragiforme against Staphylococcus aureus biofilms. Microorganisms 2017, 5, 80. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-H.; Tian, X. Quorum sensing and bacterial social interactions in biofilms. Sensors 2012, 12, 2519–2538. [Google Scholar] [CrossRef] [PubMed]
- Wendt, L.; Sir, E.B.; Kuhnert, E.; Heitkämper, S.; Lambert, C.; Hladki, A.I.; Romero, A.I.; Luangsa-Ard, J.J.; Srikitikulchai, P.; Peršoh, D.; et al. Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycol. Prog. 2018, 17, 115–154. [Google Scholar] [CrossRef]
- Helaly, S.E.; Thongbai, B.; Stadler, M. Diversity of biologically active secondary metabolites from endophytic and saprotrophic fungi of the ascomycete order Xylariales. Nat. Prod. Rep. 2018. [Google Scholar] [CrossRef] [PubMed]
- Stadler, M. Importance of secondary metabolites in the Xylariaceae as parameters for assessment of their taxonomy, phylogeny, and functional biodiversity. Curr. Res. Environ. Appl. Mycol. 2011, 1, 75–133. [Google Scholar] [CrossRef]
- Stadler, M.; Quang, D.N.; Tomita, A.; Hashimoto, T.; Asakawa, Y. Changes in secondary metabolism during stromatal ontogeny of Hypoxylon fragiforme. Mycol. Res. 2006, 110, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.R.; Edwards, R.L.; Whalley, A.J.S. Metabolites of the higher fungi. Part 21. 3-Methyl-3,4-dihydroisocoumarins and related compounds from the Ascomycete family Xylariaceae. J. Chem. Soc. Perkin Trans. 1983, 2165–2192. [Google Scholar] [CrossRef]
- Edwards, R.L.; Fawcett, V.; Maitland, D.J.; Nettleton, R.; Shields, L.; Whalley, A.J.S. Hypoxyxylerone. A novel green pigment from the fungus Hypoxylon fragiforme. J. Chem. Soc. Chem. Commun. 1991, 1009–1010. [Google Scholar] [CrossRef]
- Surup, F.; Narmani, A.; Becker, K.; Wendt, L.; Pfütze, S.; Kretz, R.; Menbrivès, C.; Giosa, A.; Elliott, M.; Petit, C.; et al. Identification of fungal fossils and novel azaphilone pigments in ancient carbonised specimens of Hypoxylon fragiforme from forest soils of Châtillon-sur-Seine (Burgundy). Fungal Divers. 2018, 92, 345–356. [Google Scholar] [CrossRef]
- Stadler, M.; Læssøe, T.; Fournier, J.; Decock, C.; Schmieschek, B.; Tichy, H.V.; Peršoh, D. A polyphasic taxonomy of Daldinia (Xylariaceae). Stud. Mycol. 2014, 77, 1–143. [Google Scholar] [CrossRef] [PubMed]
- Petrini, L.E. Rosellinia—A World Monograph; Bibliotheca Mycologica: Stuttgart, Germany, 2013. [Google Scholar]
- Ashrafi, S.; Helaly, S.E.; Schroers, H.J.; Stadler, M.; Richert-Poeggeler, K.R.; Dababat, A.A.; Maier, W. Ijuhya vitellina sp. nov., a novel source for chaetoglobosin A, is a destructive parasite of the cereal cyst nematode Heterodera filipjevi. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Narmani, A.; Pichai, S.; Palani, P.; Arzanlou, M.; Surup, F.; Stadler, M. Daldinia sacchari (Hypoxylaceae) from India produces the new cytochalasins saccalasins A and B and belongs to the D. eschscholtzii species complex. Mycol. Progr. 2018. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, 47. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, M.S.; Hashimoto, T.; Asakawa, Y. Cytochalasins from a Daldinia sp. of fungus. Phytochemistry 1996, 41, 821–828. [Google Scholar] [CrossRef]
- Buchanan, M.S.; Hashimoto, T.; Asakawa, Y. Five 10-phenyl-[11]-cytochalasans from a Daldinia fungal species. Phytochemistry 1995, 40, 135–140. [Google Scholar] [CrossRef]
- Tomoda, H.; Namatame, I.; Tabata, N.; Kawaguchi, K.; Si, S.; Omura, S. Structure elucidation of fungal phenochalasins, novel inhibitors of lipid droplet formation in mouse macrophages. J. Antibiot. 1999, 52, 857–861. [Google Scholar] [CrossRef] [PubMed]
- Binder, M.; Tamm, C.; Turner, W.B.; Minato, H. Nomenclature of a class of biologically active mould metabolites: The cytochalasins, phomins, and zygosporins. J. Chem. Soc. Perkin 1973, 11, 1146–1147. [Google Scholar] [CrossRef]
- Tao, Y.; Zeng, X.; Mou, C.; Li, J.; Cai, X.; She, Z.; Zhou, S.; Lin, Y. 1H and 13C NMR assignments of three nitrogen containing compounds from the mangrove endophytic fungus (ZZF08). Magn. Reson. Chem. 2008, 46, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Ondeyka, J.; Hensens, O.D.; Zink, D.; Ball, R.; Lingham, R.B.; Bills, G.; Dombrowski, A.; Goetz, M. L-696, 474, a novel cytochalasin as inhibitor of HIV-1 protease. J. Antibiot. 1992, 45, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Elsässer, B.; Krohn, K.; Flörke, U.; Root, N.; Aust, H.-J.; Draeger, S.; Schulz, B.; Antus, S.; Kurtán, T. X-ray structure determination, absolute configuration and biological activity of phomoxanthone A. Eur. J. Org. Chem. 2005, 2005, 4563–4570. [Google Scholar] [CrossRef]
- Espada, A.; Rivera-Sagredo, A.; de la Fuente, J.M.; Hueso-Rodriguez, J.A.; Elson, S.W. New cytochalasins from the fungus Xylaria hypoxylon. Tetrahedron 1997, 53, 6485–6492. [Google Scholar] [CrossRef]
- Yuyama, K.T.; da Costa Neves, T.; Memória, M.; Tartuci, I.; Abraham, W.R. Aurantiogliocladin inhibits biofilm formation at subtoxic concentrations. AIMS Microbiol. 2017, 3, 50–60. [Google Scholar] [CrossRef]
- Skellam, E. The biosynthesis of cytochalasans. Nat. Prod. Rep. 2017, 34, 1252–1263. [Google Scholar] [CrossRef] [PubMed]
- Aldrigde, D.C.; Armstrong, J.J.; Speake, R.N.; Turner, W.B. The cytochalasins, a new class of biologically active mould metabolites. J. Chem. Soc. Chem. Commun. 1967, 3, 26–27. [Google Scholar]
- Rothweiler, W.; Tamm, C. Isolation and structure of Phomin. Experientia 1966, 22, 750–753. [Google Scholar] [CrossRef]
- Jouda, J.B.; Mbazoa, C.D.; Douanla-Meli, C.; Sarkar, P.; Bag, P.K.; Wandji, J. Antibacterial and cytotoxic cytochalasins from the endophytic fungus Phomopsis sp. harbored in Garcinia kola (Heckel) Nut. BMC Complement Altern. Med. 2016, 16, 462. [Google Scholar] [CrossRef] [PubMed]
- Betina, V.; Micekova, D.; Nemec, P. Antimicrobial properties of cytochalasins and their alteration of fungal morphology. Microbiology 1972, 71, 343–349. [Google Scholar] [CrossRef]
- Dagne, E.; Gunatilaka, A.A.L.; Asmellash, S.; Abate, D.; Kingston, D.G.I.; Hofmann, G.A.; Johnson, R.K. Two new cytotoxic cytochalasins from Xylaria obovata. Tetrahedron 1994, 50, 5615–5620. [Google Scholar] [CrossRef]
- Wagenaar, M.M.; Corwin, J.; Strobel, G.; Clardy, J. Three new cytochalasins produced by an endophytic fungus in the genus Rhinocladiella. J. Nat. Prod. 2000, 63, 1692–1695. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xiao, J.; Gao, Y.-Q.; Tang, J.-J.; Zhang, A.-L.; Gao, J.-M. Chaetoglobosins from Chaetomium globosum, an endophytic fungus in Ginkgo biloba, and their phytotoxic and cytotoxic activities. J. Agric. Food Chem. 2014, 62, 3734–3741. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.B. Effects of cytochalasins on mammalian cells. Int. J. Sci. Nat. 1967, 213, 261–264. [Google Scholar] [CrossRef]
- Cooper, J.A. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 1987, 105, 1473–1478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flanagan, M.D.; Lin, S. Cytochalasins block actin filament elongation by binding to high affinity sites associated with F-actin. J. Biol. Chem. 1980, 255, 835–838. [Google Scholar] [PubMed]
- Brown, S.S.; Spudich, J.A. Mechanism of action of cytochalasin: Evidence that it binds to actin filament ends. J. Cell Biol. 1981, 88, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Goietsenoven, G.V.; Mathieu, V.; Andolfi, A.; Cimmino, A.; Lefranc, F.; Kiss, R.; Evidente, A. In vitro growth inhibitory effects of cytochalasins and derivatives in cancer cells. Planta Med. 2011, 77, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.T.; Chou, C.T.; Chen, I.S.; Yu, C.C.; Lu, T.; Hsu, S.S.; Shieh, P.; Jan, C.R.; Liang, W.Z. Mechanisms underlying effect of the mycotoxin cytochalasin B on induction of cytotoxicity, modulation of cell cycle, Ca2+ homeostasis and ROS production in human breast cells. Toxicology 2016, 370, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Trendowski, M. Using cytochalasins to improve current chemotherapeutic approaches. anti-cancer agents in medicinal chemistry. Anticancer Agents Med. Chem. 2015, 15, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Gov, Y.; Bitler, A.; Dell’Acqua, G.; Torres, J.V.; Balaban, N. RNAIII inhibiting peptide (RIP), a global inhibitor of Staphylococcus aureus pathogenesis: Structure and function analysis. Peptides 2001, 22, 1609–1620. [Google Scholar] [CrossRef]
- Francolini, I.; Norris, P.; Piozzi, A.; Donelli, G.; Stoodley, P. Usnic acid, a natural antimicrobial agent able to inhibit bacterial biofilm formation on polymer surfaces. Antimicrob. Agents Chemother. 2004, 48, 4360–4365. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Lim, A.; Lee, J.; Chen, S.; An, S.; Dong, Y.-H.; Zhang, L.-H. Diffusible signal factor (DSF) quorum sensing signal and structurally related molecules enhance the antimicrobial efficacy of antibiotics against some bacterial pathogens. BMC Microbiol. 2014, 14, 51. [Google Scholar] [CrossRef] [PubMed]
- Yuyama, K.T.; Abraham, W.-R. cis-2-alkenoic acids as promising drugs for the control of biofilm infections. Med. Chem. 2016, 13, 3–12. [Google Scholar] [CrossRef] [PubMed]
| 19 a | 20 b | |||
|---|---|---|---|---|
| δC, Mult. | δH, Mult. | δC, Mult. | δH, Mult. | |
| 1 | 173.6, C | 174.3, C | ||
| 2 | 5.49, br s | 5.42, br s | ||
| 3 | 53.3, CH | 3.25, m | 55.0, CH | 3.20, m |
| 4 | 44.8, CH | 3.30, dd (5.7, 2.4) | 48.4, CH | 3.20, m |
| 5 | 31.7, CH | 2.79, m | 34.8, CH | 2.43, m |
| 6 | 148.6, C | 140.2, C | ||
| 7 | 71.6, CH | 4.10, d (10.1) | 125.7, CH | 5.48, m |
| 8 | 51.8, CH | 2.44, m | 49.7, CH | 2.58, d (9.6) |
| 9 | 63.4, C | 68.4, C | ||
| 10 | 43.3, CH2 | 2.63, dd (13.4, 5.2) 2.43, m | 44.2, CH2 | 2.73, dd (13.7, 4.4) 2.41, dd (13.7, 8.8) |
| 11 | 13.2, CH3 | 1.02, m | 13.5, CH3 | 1.18, br d (7.3) |
| 12 | 114.1, CH2 | 5.28, br s 5.09, br s | 20.0, CH3 | 1.75, q (1.3) |
| 13 | 126.9, CH | 5.85, dd (15.6, 9.8) | 128.1, CH | 5.85, ddd (15.5, 9.6, 1.3) |
| 14 | 138.7, CH | 5.22, ddd (15.6, 10.9, 4.8) | 135.9, CH | 5.22, ddd (15.5, 10.9, 4.8) |
| 15 | 42.9, CH2 | 2.02, m 1.81, m | 42.7, CH2 | 2.01, m 1.78, m |
| 16 | 28.7, CH | 1.45, m | 28.7, CH | 1.48, m |
| 17 | 46.7, CH2 | 1.94, m 1.55, m | 46.4, CH2 | 1.95, m 1.52, m |
| 18 | 34.3, CH | 2.63, m | 34.4, CH | 2.63, m |
| 19 | 154.75, CH | 6.52, dd (15.9, 6.9) | 153.2, CH | 6.46, dd (15.9, 6.9) |
| 20 | 132.1, CH | 7.05, br d (15.9) | 132.9, CH | 7.12, dd (15.9, 1.4) |
| 21 | 196.6 | 198.1, C | ||
| 22 | 26.2, CH3 | 1.04, m | 26.1, CH3 | 1.02, d (6.9) |
| 23 | 17.6, CH3 | 1.14, d (7.0) | 17.6, CH3 | 1.13, d (6.9) |
| 1′ | 129.2, C | 129.5, C | ||
| 2′/6′ | 130.3, CH | 6.99, br d (8.4) | 130.3, CH | 6.99, br d (8.4) |
| 3′/5′ | 115.6, CH | 6.77, br d (8.4) | 115.6, CH | 6.77, br d (8.4) |
| 4′ | 154.69, C | OH: 5.41, s | 154.6, C | OH: 5.09, s |
| Compound | Source, Producing Fungus/Strain | MIC (μg mL−1) | Inhibition of Biofilm Formation (%) | Potency of Biofilm Inhibition 1 |
|---|---|---|---|---|
| Cytochalasin A (1) | Drechslera dermatioidea (Sigma) | 32S | 91 ± 1.4 (16 μg mL−1) | +++ |
| Cytochalasin B (2) | Drechslera dermatioidea (Sigma) | >256 | - | - |
| Cytochalasin C (3) | Metarrhizium anisopliae (Sigma) | >256 | 42 ± 6.2 (256 μg mL−1) 27 ± 2.9 (128 μg mL−1) 21 ± 2.3 (64 μg mL−1) | ++ |
| Cytochalasin D (4) | Zygosporium mansonii (Sigma) | >256 | - | - |
| Cytochalasin E (5) | Aspergillus clavatus (Sigma) | >256 | - | |
| Cytochalasin H (6) [12,25] | Hypoxylon fragiforme (cultures) | >256 | - | - |
| L-696,474 (7) [26] | Hypoxylon fragiforme (cultures) | >256 | 44 ± 0.02 (64 μg mL−1) 46 ± 1.2 (32 μg mL−1) 44 ± 0.05 (16 μg mL−1) | ++ |
| 21-O-Deacyl-l-696,474 (8) [27] | Hypoxylon fragiforme (stromata) | >256 | 33 ± 9.1 (256 μg mL−1) 28 ± 15.3 (64 μg mL−1) | + |
| Saccalasin A (9) [19] | Daldinia bambusicola BCC 42280Daldinia sacchari | >256 | 36 ± 8.8 (256 μg mL−1) 33 ± 4.3 (128 μg mL−1) 14 ± 1.8 (32 μg mL−1) | + |
| 10 [21] | Daldinia eschscholtzii BBH 42278 | >256 | 85 ± 5.4 (256 μg mL−1) 54 ± 6.0 (128 μg mL−1) | +++ |
| 11 [22] | Daldinia eschscholtzii BBH 42278 | >256 | - | - |
| 12 [23] | Daldinia eschscholtzii BBH 42278 | >256 | 76 ± 10.8 (256 μg mL−1) 51 ± 4.8 (128 μg mL−1) | +++ |
| 13 [23] | Daldinia eschscholtzii BBH 42278 | >256 | 73.7 ± 16.8 (256 μg mL−1) 44.8 ± 17.0 (128 μg mL−1) 30.6 ± 15.6 (64 μg mL−1) 26.8 ± 13.5 (4 μg mL−1) | +++ |
| 14 [23] | Daldinia eschscholtzii BBH 42278 | >256 | 32 ± 0.7 (256 μg mL−1) | + |
| 19,20-Epoxycytochalasin C (15) [28] | Rosellinia rickii (culture) | >256 | 40 ± 6.0 (256 μg mL−1) 22 ± 12.6 (128 μg mL−1) | ++ |
| 16–18 [28] | Rosellinia rickii (culture) | >256 | - | - |
| Phenochalasin C (19) | Hypoxylon cf. kretzschmarioides BBH 42276 | >256 | 31 ± 6.4 (256 μg mL−1) 14 ± 1.7 (128 μg mL−1) | + |
| Phenochalasin D (20) | Hypoxylon cf. kretzschmarioides BBH 42276 | >256 | 43 ± 4.0 (256 μg mL−1)46 ± 1.5 (64 μg mL−1) 33 ± 6.7 (32 μg mL−1) 15 ± 2.0 (16 μg mL−1) | ++ |
| Chaetoglobosin A (21) [18] | Ijuhya vitellina (culture) | 256 | 87.3 ± 4.4 (128 μg mL−1) 85.1 ± 4.7 (64 μg mL−1) 61.2 ± 1.4 (32 μg mL−1) 17.8 ± 1.1 (8 μg mL−1) | +++ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuyama, K.T.; Wendt, L.; Surup, F.; Kretz, R.; Chepkirui, C.; Wittstein, K.; Boonlarppradab, C.; Wongkanoun, S.; Luangsa-ard, J.; Stadler, M.; et al. Cytochalasans Act as Inhibitors of Biofilm Formation of Staphylococcus aureus. Biomolecules 2018, 8, 129. https://doi.org/10.3390/biom8040129
Yuyama KT, Wendt L, Surup F, Kretz R, Chepkirui C, Wittstein K, Boonlarppradab C, Wongkanoun S, Luangsa-ard J, Stadler M, et al. Cytochalasans Act as Inhibitors of Biofilm Formation of Staphylococcus aureus. Biomolecules. 2018; 8(4):129. https://doi.org/10.3390/biom8040129
Chicago/Turabian StyleYuyama, Kamila Tomoko, Lucile Wendt, Frank Surup, Robin Kretz, Clara Chepkirui, Kathrin Wittstein, Chollaratt Boonlarppradab, Sarunyou Wongkanoun, Jennifer Luangsa-ard, Marc Stadler, and et al. 2018. "Cytochalasans Act as Inhibitors of Biofilm Formation of Staphylococcus aureus" Biomolecules 8, no. 4: 129. https://doi.org/10.3390/biom8040129
APA StyleYuyama, K. T., Wendt, L., Surup, F., Kretz, R., Chepkirui, C., Wittstein, K., Boonlarppradab, C., Wongkanoun, S., Luangsa-ard, J., Stadler, M., & Abraham, W.-R. (2018). Cytochalasans Act as Inhibitors of Biofilm Formation of Staphylococcus aureus. Biomolecules, 8(4), 129. https://doi.org/10.3390/biom8040129








