The Production of Curli Amyloid Fibers Is Deeply Integrated into the Biology of Escherichia coli
Abstract
1. Introduction
2. Results and Discussion
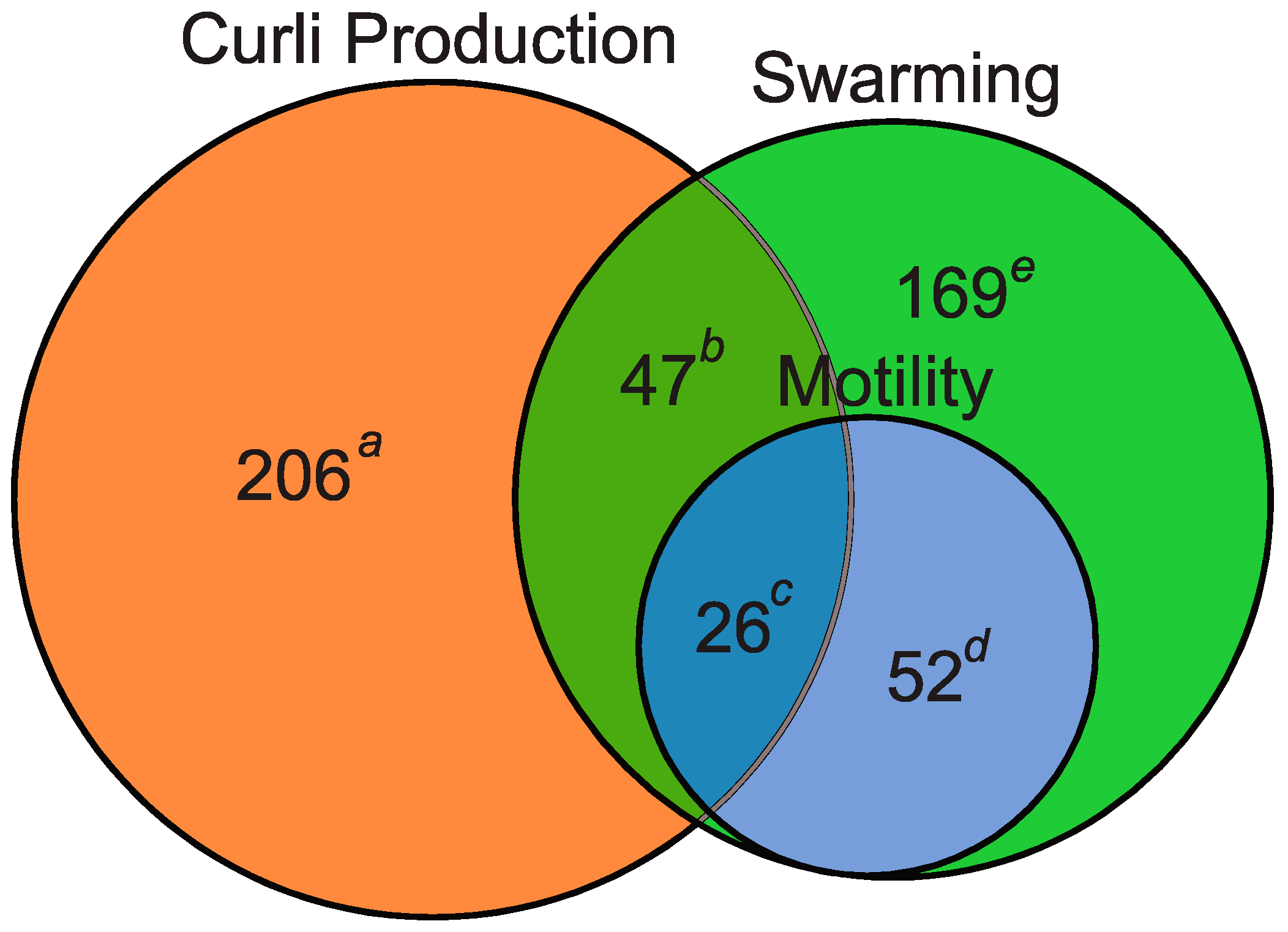
2.1. Forward Genetic Screen to Identify Genes Involved in Curli Production
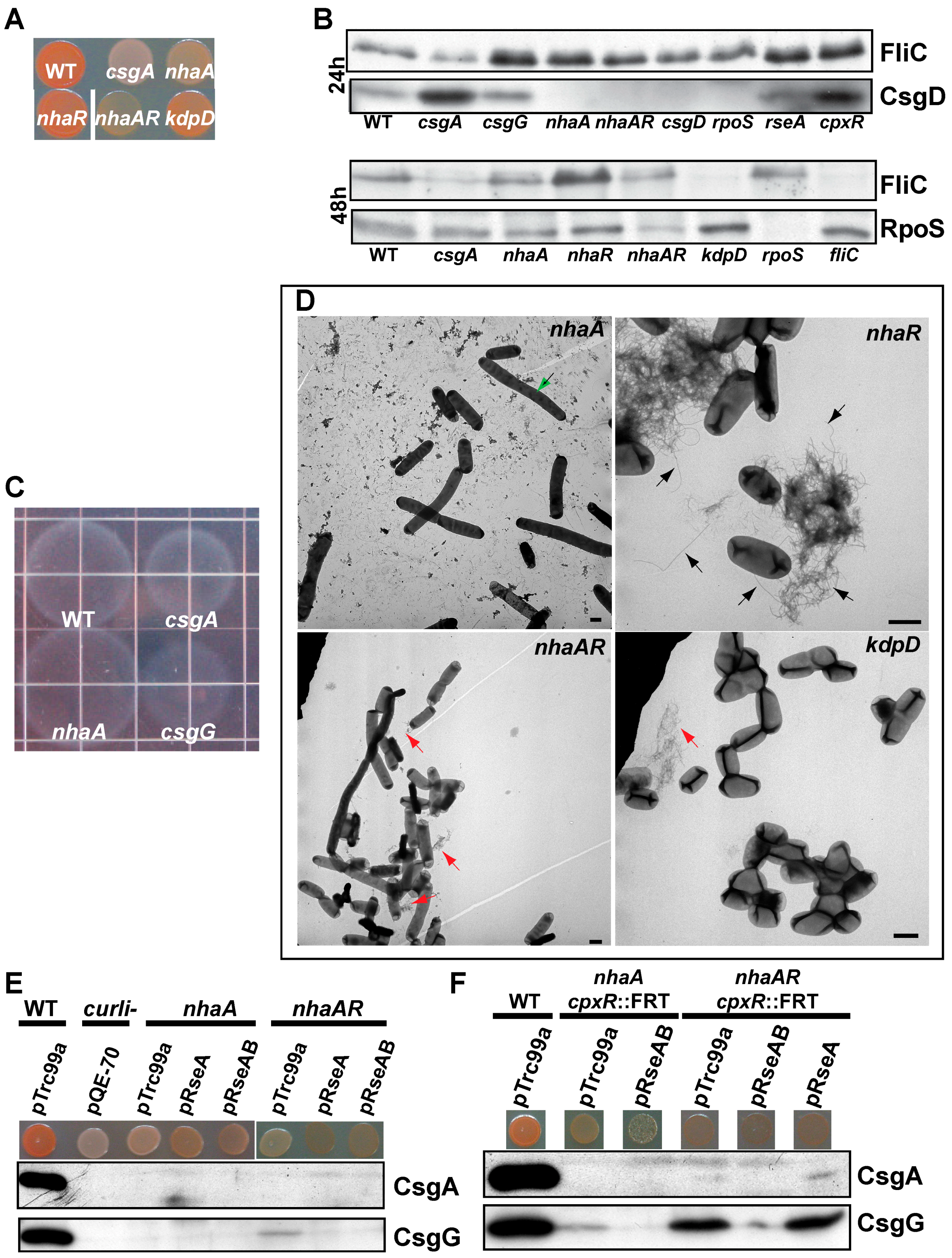
2.2. Cell Envelope
2.3. Carbohydrate Metabolism, Energy Production, and Gluconeogenesis
2.4. Multiple Regulatory Networks Control Curli Gene Expression
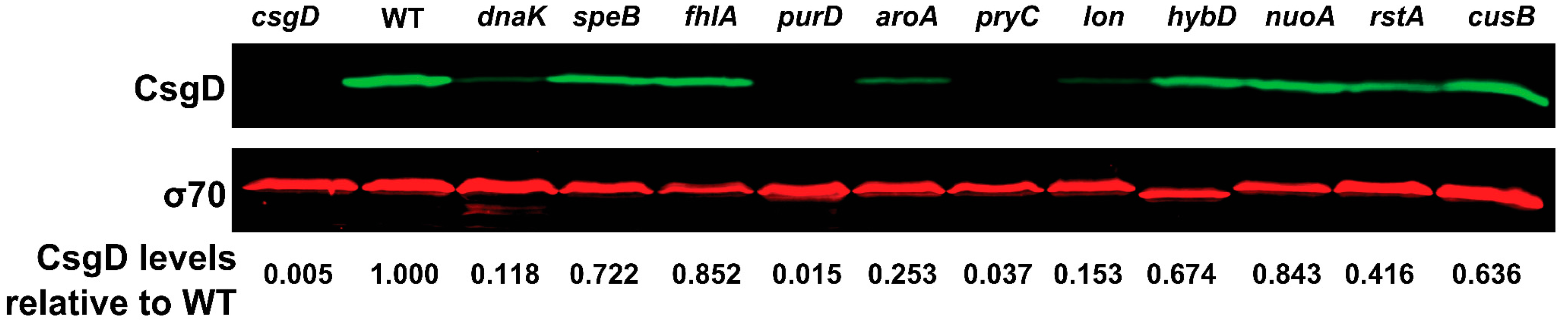
2.5. CsgD Transcript and Protein Levels Are Altered in Several Strains with CR Phenotypes
3. Materials and Methods
3.1. Bacterial Strains and Growth Conditions
3.2. Western Blotting and LPS Silver Staining
3.3. RNA Extraction
3.4. Reverse Transcriptase Complementary DNA Synthesis and Real-Time Quantitative PCR
3.5. Quantitative Real Time-PCR Analysis
3.6. Motility Assay
3.7. Electron Microscopy
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chapman, M.R.; Robinson, L.S.; Pinkner, J.S.; Roth, R.; Heuser, J.; Hammar, M.; Normark, S.; Hultgren, S.J. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 2002, 295, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Collinson, S.K.; Emody, L.; Muller, K.H.; Trust, T.J.; Kay, W.W. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 1991, 173, 4773–4781. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Mizunoe, Y.; Takade, A.; Naito, S.; Yoshida, S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 2005, 49, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Uhlich, G.A.; Cooke, P.H.; Solomon, E.B. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl. Environ. Microbiol. 2006, 72, 2564–2572. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.H.; Beuchat, L.R. Biofilm formation by Escherichia coli O157:H7 on stainless steel: Effect of exopolysaccharide and curli production on its resistance to chlorine. Appl. Environ. Microbiol. 2005, 71, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Jeter, C.; Matthysse, A.G. Characterization of the binding of diarrheagenic strains of E. coli to plant surfaces and the role of curli in the interaction of the bacteria with alfalfa sprouts. Mol. Plant Microbe Interact. 2005, 18, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Barak, J.D.; Gorski, L.; Naraghi-Arani, P.; Charkowski, A.O. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl. Environ. Microbiol. 2005, 71, 5685–5691. [Google Scholar] [CrossRef] [PubMed]
- Vidal, O.; Longin, R.; Prigent-Combaret, C.; Dorel, C.; Hooreman, M.; Lejeune, P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: Involvement of a new ompR allele that increases curli expression. J. Bacteriol. 1998, 180, 2442–2449. [Google Scholar] [PubMed]
- White, A.P.; Gibson, D.L.; Kim, W.; Kay, W.W.; Surette, M.G. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J. Bacteriol. 2006, 188, 3219–3227. [Google Scholar] [CrossRef] [PubMed]
- DePas, W.H.; Hufnagel, D.A.; Lee, J.S.; Blanco, L.P.; Bernstein, H.C.; Fisher, S.T.; James, G.A.; Stewart, P.S.; Chapman, M.R. Iron induces bimodal population development by Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 2629–2634. [Google Scholar] [CrossRef] [PubMed]
- Depas, W.H.; Syed, A.K.; Sifuentes, M.; Lee, J.S.; Warshaw, D.; Saggar, V.; Csankovszki, G.; Boles, B.R.; Chapman, M.R. Biofilm formation protects Escherichia coli against killing by Caenorhabditis elegans and Myxococcus xanthus. Appl. Environ. Microbiol. 2014, 80, 7079–7087. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Rochon, M.; Lamprokostopoulou, A.; Lunsdorf, H.; Nimtz, M.; Romling, U. Impact of biofilm matrix components on interaction of commensal Escherichia coli with the gastrointestinal cell line HT-29. Cell. Mol. Life Sci. 2006, 63, 2352–2363. [Google Scholar] [CrossRef] [PubMed]
- Gophna, U.; Barlev, M.; Seijffers, R.; Oelschlager, T.A.; Hacker, J.; Ron, E.Z. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 2001, 69, 2659–2665. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Brauner, A.; Li, Y.; Normark, S. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 2000, 181, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Hammar, M.; Arnqvist, A.; Bian, Z.; Olsen, A.; Normark, S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 1995, 18, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.L.; Chapman, M.R. Curli biogenesis: Order out of disorder. Biochim. Biophys. Acta 2014, 1843, 1551–1558. [Google Scholar] [CrossRef] [PubMed]
- Romling, U.; Bian, Z.; Hammar, M.; Sierralta, W.D.; Normark, S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 1998, 180, 722–731. [Google Scholar] [PubMed]
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M.; et al. String 8—A global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009, 37, D412–D416. [Google Scholar] [CrossRef] [PubMed]
- Romling, U.; Sierralta, W.D.; Eriksson, K.; Normark, S. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 1998, 28, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Normark, S. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 1997, 16, 5827–5836. [Google Scholar] [CrossRef] [PubMed]
- Hammar, M.; Bian, Z.; Normark, S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl. Acad. Sci. USA 1996, 93, 6562–6566. [Google Scholar] [CrossRef] [PubMed]
- Hammer, N.D.; Schmidt, J.C.; Chapman, M.R. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. USA 2007, 104, 12494–12499. [Google Scholar] [CrossRef] [PubMed]
- Goyal, P.; Krasteva, P.V.; Van Gerven, N.; Gubellini, F.; Van den Broeck, I.; Troupiotis-Tsailaki, A.; Jonckheere, W.; Pehau-Arnaudet, G.; Pinkner, J.S.; Chapman, M.R.; et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 2014, 516, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Loferer, H.; Hammar, M.; Normark, S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol. Microbiol. 1997, 26, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Robinson, L.S.; Ashman, E.M.; Hultgren, S.J.; Chapman, M.R. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 2006, 59, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Epstein, E.A.; Reizian, M.A.; Chapman, M.R. Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly. J. Bacteriol. 2009, 191, 608–615. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shu, Q.; Krezel, A.M.; Cusumano, Z.T.; Pinkner, J.S.; Klein, R.; Hultgren, S.J.; Frieden, C. Solution NMR structure of CsgE: Structural insights into a chaperone and regulator protein important for functional amyloid formation. Proc. Natl. Acad. Sci. USA 2016, 113, 7130–7135. [Google Scholar] [CrossRef] [PubMed]
- Nenninger, A.A.; Robinson, L.S.; Hammer, N.D.; Epstein, E.A.; Badtke, M.P.; Hultgren, S.J.; Chapman, M.R. CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol. Microbiol. 2011, 81, 486–499. [Google Scholar] [CrossRef] [PubMed]
- Nenninger, A.A.; Robinson, L.S.; Hultgren, S.J. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc. Natl. Acad. Sci. USA 2009, 106, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.L.; White, A.P.; Rajotte, C.M.; Kay, W.W. AgfC and AgfE facilitate extracellular thin aggregative fimbriae synthesis in Salmonella enteritidis. Microbiology 2007, 153, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.D.; Zhou, Y.; Salgado, P.S.; Patwardhan, A.; McGuffie, M.; Pape, T.; Grabe, G.; Ashman, E.; Constable, S.C.; Simpson, P.J.; et al. Atomic resolution insights into curli fibre biogenesis. Structure 2011, 19, 1307–1316. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.L.; Chorell, E.; Taylor, J.D.; Aden, J.; Gotheson, A.; Li, F.; Koch, M.; Sefer, L.; Matthews, S.J.; Wittung-Stafshede, P.; et al. The bacterial curli system possesses a potent and selective inhibitor of amyloid formation. Mol. Cell 2015, 57, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.L.L.; Kwan, A.H.; Sunde, M. Functional amyloid: Widespread in nature, diverse in purpose. In Amyloids in Health and Disease; Perrett, S., Ed.; Portland Press Ltd.: London, UK, 2014; Volume 56, pp. 207–219. [Google Scholar]
- Ogasawara, H.; Yamada, K.; Kori, A.; Yamamoto, K.; Ishihama, A. Regulation of the E. coli csgD promoter: Interplay between five transcription factors. Microbiology 2010, 156, 2470–2483. [Google Scholar] [CrossRef] [PubMed]
- Barnhart, M.M.; Lynem, J.; Chapman, M.R. GlcNAc-6P levels modulate the expression of curli fibers by Escherichia coli. J. Bacteriol. 2006, 188, 5212–5219. [Google Scholar] [CrossRef] [PubMed]
- Hammar, M. Assembly and Adhesive Properties of Curli. Ph.D. Thesis, Karolinska Institute, Stockholm, Sweden, 1997. [Google Scholar]
- Vogeleer, P.; Tremblay, Y.D.N.; Mafu, A.A.; Jacques, M.; Harel, J. Life on the outside: Role of biofilms in environmental persistence of Shiga-toxin producing Escherichia coli. Front. Microbiol. 2014, 5, 317. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Ksoll, W.B.; Hicks, R.E.; Sadowsky, M.J. Presence and growth of naturalized Escherichia coli in temperate soils from lake superior watersheds. Appl. Environ. Microbiol. 2006, 72, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Hassett, D.J.; Sutton, M.D.; Schurr, M.J.; Herr, A.B.; Caldwell, C.C.; Matu, J.O. Pseudomonas aeruginosa hypoxic or anaerobic biofilm infections within cystic fibrosis airways. Trends Microbiol. 2009, 17, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006–2008. [Google Scholar] [CrossRef] [PubMed]
- Collinson, S.K.; Doig, P.C.; Doran, J.L.; Clouthier, S.; Trust, T.J.; Kay, W.W. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J. Bacteriol. 1993, 175, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.; Szklarczyk, D.; Trachana, K.; Roth, A.; Kuhn, M.; Muller, J.; Arnold, R.; Rattei, T.; Letunic, I.; Doerks, T.; et al. Eggnog v3.0: Orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012, 40, D284–D289. [Google Scholar] [CrossRef] [PubMed]
- Keseler, I.M.; Mackie, A.; Peralta-Gil, M.; Santos-Zavaleta, A.; Gama-Castro, S.; Bonavides-Martinez, C.; Fulcher, C.; Huerta, A.M.; Kothari, A.; Krummenacker, M.; et al. Ecocyc: Fusing model organism databases with systems biology. Nucleic Acids Res. 2013, 41, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015, 43, D261–D269. [Google Scholar] [CrossRef] [PubMed]
- Lipman, D.J.; Souvorov, A.; Koonin, E.V.; Panchenko, A.R.; Tatusova, T.A. The relationship of protein conservation and sequence length. BMC Evol. Biol. 2002, 2, 20. [Google Scholar] [CrossRef]
- Misra, R.V.; Horler, R.S.; Reindl, W.; Goryanin, I.I.; Thomas, G.H. Echobase: An integrated post-genomic database for Escherichia coli. Nucleic Acids Res. 2005, 33, D329–D333. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Fujita, N.; Yamamoto, K.; Ishihama, A. Novel Roles of cAMP Receptor Protein (CRP) in Regulation of Transport and Metabolism of Carbon Sources. PLoS ONE 2011, 6, e20081. [Google Scholar] [CrossRef] [PubMed]
- Barnhart, M.M.; Chapman, M.R. Curli biogenesis and function. Annu. Rev. Microbiol. 2006, 60, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A genomic perspective on protein families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Weiner, J.H.; Li, L. Proteome of the Escherichia coli envelope and technological challenges in membrane proteome analysis. Biochim. Biophys. Acta 2008, 1778, 1698–1713. [Google Scholar] [CrossRef] [PubMed]
- Pradel, E.; Schnaitman, C.A. Effect of RfaH (SfrB) and temperature on expression of Rfa genes of Escherichia coli K-12. J. Bacteriol. 1991, 173, 6428–6431. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]
- Schnaitman, C.A.; Klena, J.D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 1993, 57, 655–682. [Google Scholar] [PubMed]
- Klein, G.; Müller-Loennies, S.; Lindner, B.; Kobylak, N.; Brade, H.; Raina, S. Molecular and Structural Basis of Inner Core Lipopolysaccharide Alterations in Escherichia coli. J. Biol. Chem. 2013, 288, 8111–8127. [Google Scholar] [CrossRef] [PubMed]
- Anriany, Y.; Sahu, S.N.; Wessels, K.R.; McCann, L.M.; Joseph, S.W. Alteration of the rugose phenotype in waaG and ddhC mutants of Salmonella enterica serovar typhimurium dt104 is associated with inverse production of curli and cellulose. Appl. Environ. Microbiol. 2006, 72, 5002–5012. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, Y.H. Escherichia coli O157:H7 adherence to HEp-2 cells is implicated with curli expression and outer membrane integrity. J. Vet. Sci. 2004, 5, 119–124. [Google Scholar] [PubMed]
- Zogaj, X.; Nimtz, M.; Rohde, M.; Bokranz, W.; Romling, U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 2001, 39, 1452–1463. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wood, T.K. OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ. Microbiol. 2009, 11, 2735–2746. [Google Scholar] [CrossRef] [PubMed]
- Coleman, W.G., Jr. The rfaD gene codes for ADP-l-glycero-d-mannoheptose-6-epimerase. An enzyme required for lipopolysaccharide core biosynthesis. J. Biol. Chem. 1983, 258, 1985–1990. [Google Scholar] [PubMed]
- Kneidinger, B.; Marolda, C.; Graninger, M.; Zamyatina, A.; McArthur, F.; Kosma, P.; Valvano, M.A.; Messner, P. Biosynthesis pathway of ADP-l-glycero-beta-d-manno-heptose in Escherichia coli. J. Bacteriol. 2002, 184, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, E.; Proudfoot, M.; Gonzalez, C.F.; Brown, G.; Omelchenko, M.V.; Borozan, I.; Carmel, L.; Wolf, Y.I.; Mori, H.; Savchenko, A.V.; et al. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J. Biol. Chem. 2006, 281, 36149–36161. [Google Scholar] [CrossRef] [PubMed]
- Parker, C.T.; Kloser, A.W.; Schnaitman, C.A.; Stein, M.A.; Gottesman, S.; Gibson, B.W. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 1992, 174, 2525–2538. [Google Scholar] [CrossRef] [PubMed]
- Weissborn, A.C.; Liu, Q.; Rumley, M.K.; Kennedy, E.P. Utp: α-d-glucose-1-phosphate uridylyltransferase of Escherichia coli: Isolation and DNA sequence of the galU gene and purification of the enzyme. J. Bacteriol. 1994, 176, 2611–2618. [Google Scholar] [CrossRef] [PubMed]
- Karp, P.D.; Keseler, I.M.; Shearer, A.; Latendresse, M.; Krummenacker, M.; Paley, S.M.; Paulsen, I.; Collado-Vides, J.; Gama-Castro, S.; Peralta-Gil, M.; et al. Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res. 2007, 35, 7577–7590. [Google Scholar] [CrossRef] [PubMed]
- Boos, W.; Ehmann, U.; Forkl, H.; Klein, W.; Rimmele, M.; Postma, P. Trehalose transport and metabolism in Escherichia coli. J. Bacteriol. 1990, 172, 3450–3461. [Google Scholar] [CrossRef] [PubMed]
- Genevaux, P.; Bauda, P.; DuBow, M.S.; Oudega, B. Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch. Microbiol. 1999, 172, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, M.; Gutnick, D.; Rosenberg, E. Adherence of bacteria to hydrocarbons: A simple method for measuring cell surface hydrophobicity. FEMS Microbiol. Lett. 1980, 9, 29–33. [Google Scholar] [CrossRef]
- Nikaido, H.; Vaara, M. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 1985, 49, 1–32. [Google Scholar] [CrossRef] [PubMed]
- De Cock, H.; Tommassen, J. Lipopolysaccharides and divalent cations are involved in the formation of an assembly-competent intermediate of outer-membrane protein PhoE of E. coli. EMBO J. 1996, 15, 5567–5573. [Google Scholar] [PubMed]
- De Maagd, R.A.; Wientjes, F.B.; Lugtenberg, B.J. Evidence for divalent cation (Ca2+)-stabilized oligomeric proteins and covalently bound protein-peptidoglycan complexes in the outer membrane of Rhizobium leguminosarum. J. Bacteriol. 1989, 171, 3989–3995. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Barr, K.; Rick, P.D. Identification of the structural gene for the TDP-Fuc4NAc:lipid II Fuc4NAc transferase involved in synthesis of enterobacterial common antigen in Escherichia coli K-12. J. Bacteriol. 2001, 183, 6509–6516. [Google Scholar] [CrossRef] [PubMed]
- Danese, P.N.; Oliver, G.R.; Barr, K.; Bowman, G.D.; Rick, P.D.; Silhavy, T.J. Accumulation of the enterobacterial common antigen lipid II biosynthetic intermediate stimulates degP transcription in Escherichia coli. J. Bacteriol. 1998, 180, 5875–5884. [Google Scholar] [PubMed]
- Serra, D.O.; Mika, F.; Richter, A.M.; Hengge, R. The green tea polyphenol EGCG inhibits E. coli biofilm formation by impairing amyloid curli fibre assembly and downregulating the biofilm regulator CsgD via the σE-dependent sRNA RybB. Mol. Microbiol. 2016, 101, 136–151. [Google Scholar] [CrossRef] [PubMed]
- Vines, E.D.; Marolda, C.L.; Balachandran, A.; Valvano, M.A. Defective O-antigen polymerization in tolA and pal mutants of Escherichia coli in response to extracytoplasmic stress. J. Bacteriol. 2005, 187, 3359–3368. [Google Scholar] [CrossRef] [PubMed]
- Chaba, R.; Alba, B.M.; Guo, M.S.; Sohn, J.; Ahuja, N.; Sauer, R.T.; Gross, C.A. Signal integration by DegS and RseB governs the σE-mediated envelope stress response in Escherichia coli. Proc. Natl. Acad. Sci. USA 2011, 108, 2106–2111. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, N.; Korkin, D.; Chaba, R.; Cezairliyan, B.O.; Sauer, R.T.; Kim, K.K.; Gross, C.A. Analyzing the interaction of RseA and RseB, the two negative regulators of the σE envelope stress response, using a combined bioinformatic and experimental strategy. J. Biol. Chem. 2009, 284, 5403–5413. [Google Scholar] [CrossRef] [PubMed]
- Missiakas, D.; Mayer, M.P.; Lemaire, M.; Georgopoulos, C.; Raina, S. Modulation of the Escherichia coli σE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 1997, 24, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Rhodius, V.A.; Suh, W.C.; Nonaka, G.; West, J.; Gross, C.A. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 2006, 4, e2. [Google Scholar] [CrossRef] [PubMed]
- Bury-Mone, S.; Nomane, Y.; Reymond, N.; Barbet, R.; Jacquet, E.; Imbeaud, S.; Jacq, A.; Bouloc, P. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 2009, 5, e1000651. [Google Scholar] [CrossRef] [PubMed]
- Tam, C.; Missiakas, D. Changes in lipopolysaccharide structure induce the σE-dependent response of Escherichia coli. Mol. Microbiol. 2005, 55, 1403–1412. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Stupak, A.; Biernacka, D.; Wojtkiewicz, P.; Lindner, B.; Raina, S. Multiple transcriptional factors regulate transcription of the rpoE gene in Escherichia coli under different growth conditions and when the lipopolysaccharide biosynthesis is defective. J. Biol. Chem. 2016, 291, 22999–23019. [Google Scholar] [CrossRef] [PubMed]
- Missiakas, D.; Betton, J.M.; Raina, S. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol. Microbiol. 1996, 21, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.S.; Lin, T.Y.; Wang, W.B.; Liu, M.C.; Hsueh, P.R.; Liaw, S.J. Characterization of UDP-glucose dehydrogenase and UDP-glucose pyrophosphorylase mutants of Proteus mirabilis: Defectiveness in polymyxin Bb resistance, swarming, and virulence. Antimicrob. Agents Chemother. 2010, 54, 2000–2009. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Lindner, B.; Brabetz, W.; Brade, H.; Raina, S. Escherichia coli K-12 suppressor-free mutants lacking early glycosyltransferases and late acyltransferases: Minimal lipopolysaccharide structure and induction of envelope stress response. J. Biol. Chem. 2009, 284, 15369–15389. [Google Scholar] [CrossRef] [PubMed]
- Dorel, C.; Vidal, O.; Prigent-Combaret, C.; Vallet, I.; Lejeune, P. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 1999, 178, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Jubelin, G.; Vianney, A.; Beloin, C.; Ghigo, J.M.; Lazzaroni, J.C.; Lejeune, P.; Dorel, C. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 2005, 187, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Ishihama, A. Transcriptional response of Escherichia coli to external zinc. J. Bacteriol. 2005, 187, 6333–6340. [Google Scholar] [CrossRef] [PubMed]
- Egler, M.; Grosse, C.; Grass, G.; Nies, D.H. Role of the extracytoplasmic function protein family sigma factor RpoE in metal resistance of Escherichia coli. J. Bacteriol. 2005, 187, 2297–2307. [Google Scholar] [CrossRef] [PubMed]
- De Las Penas, A.; Connolly, L.; Gross, C.A. The σE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of σE. Mol. Microbiol. 1997, 24, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Perrenoud, A.; Sauer, U. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J. Bacteriol. 2005, 187, 3171–3179. [Google Scholar] [CrossRef] [PubMed]
- Nanchen, A.; Schicker, A.; Revelles, O.; Sauer, U. Cyclic AMP-dependent catabolite repression is the dominant control mechanism of metabolic fluxes under glucose limitation in Escherichia coli. J. Bacteriol. 2008, 190, 2323–2330. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H., Jr.; Ramseier, T.M. The catabolite repressor/activator (Cra) protein of enteric bacteria. J. Bacteriol. 1996, 178, 3411–3417. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Constantinidou, C.; Hobman, J.L.; Minchin, S.D. Identification of the Crp regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004, 32, 5874–5893. [Google Scholar] [CrossRef] [PubMed]
- Neidhardt, F.C.; Curtiss, R., III; Ingraham, J.L.; Lin, E.C.C.; Low, K.B., Jr.; Magasanik, B.; Reznikoff, W.S.; Riley, M.; Schaechter, M.; Umbarger, H.E. Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd ed.; ASM Press: Washington, DC, USA, 1996. [Google Scholar]
- Hufnagel, D.A.; Evans, M.L.; Greene, S.E.; Pinkner, J.S.; Hultgren, S.J.; Chapman, M.R. Crp-cAMP regulates csgD and biofilm formation by uropathogenic Escherichia coli. J. Bacteriol. 2016, 198, 3329–3334. [Google Scholar] [CrossRef] [PubMed]
- Ramseier, T.M.; Bledig, S.; Michotey, V.; Feghali, R.; Saier, M.H., Jr. The global regulatory protein FruR modulates the direction of carbon flow in Escherichia coli. Mol. Microbiol. 1995, 16, 1157–1169. [Google Scholar] [CrossRef] [PubMed]
- White, A.P.; Weljie, A.M.; Apel, D.; Zhang, P.; Shaykhutdinov, R.; Vogel, H.J.; Surette, M.G. A global metabolic shift is linked to Salmonella multicellular development. PLoS ONE 2010, 5, e11814. [Google Scholar] [CrossRef] [PubMed]
- Alteri, C.J.; Smith, S.N.; Mobley, H.L. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 2009, 5, e1000448. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.A.; Chowdhury, F.Z.; Fabich, A.J.; Anderson, A.; Schreiner, D.M.; House, A.L.; Autieri, S.M.; Leatham, M.P.; Lins, J.J.; Jorgensen, M.; et al. Respiration of Escherichia coli in the mouse intestine. Infect. Immun. 2007, 75, 4891–4899. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.S.; Hennigan, R.F.; Hilliard, G.M.; Ochsner, U.A.; Parvatiyar, K.; Kamani, M.C.; Allen, H.L.; DeKievit, T.R.; Gardner, P.R.; Schwab, U.; et al. Pseudomonas aeruginosa anaerobic respiration in biofilms. Dev. Cell 2002, 3, 593–603. [Google Scholar] [CrossRef]
- Mangalea, M.R.; Plumley, B.A.; Borlee, B.R. Nitrate sensing and metabolism inhibit biofilm formation in the opportunistic pathogen Burkholderia pseudomallei by reducing the intracellular concentration of c-di-GMP. Front. Microbiol. 2017, 8, 1353. [Google Scholar] [CrossRef] [PubMed]
- Van Alst, N.E.; Picardo, K.F.; Iglewski, B.H.; Haidaris, C.G. Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect. Immun. 2007, 75, 3780–3790. [Google Scholar] [CrossRef] [PubMed]
- Kalamorz, F.; Reichenbach, B.; Marz, W.; Rak, B.; Gorke, B. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol. Microbiol. 2007, 65, 1518–1533. [Google Scholar] [CrossRef] [PubMed]
- Reichenbach, B.; Maes, A.; Kalamorz, F.; Hajnsdorf, E.; Gorke, B. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of GlmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 2008, 36, 2570–2580. [Google Scholar] [CrossRef] [PubMed]
- Gruber, C.C.; Sperandio, V. Global analysis of posttranscriptional regulation by GlmY and GlmZ in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 2015, 83, 1286–1295. [Google Scholar] [CrossRef] [PubMed]
- Urban, J.H.; Vogel, J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008, 6, e64. [Google Scholar] [CrossRef] [PubMed]
- Mengin-Lecreulx, D.; van Heijenoort, J. Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J. Biol. Chem. 1996, 271, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, A.G.; Jensen, L.J.; Brunak, S.; Staerfeldt, H.H.; Ussery, D.W. A DNA structural atlas for Escherichia coli. J. Mol. Biol. 2000, 299, 907–930. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, H.; Yamamoto, K.; Ishihama, A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J. Bacteriol. 2011, 193, 2587–2597. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, J.; Yoshimura, S.H.; Takeyasu, K.; Ishihama, A. A novel nucleoid protein of Escherichia coli induced under anaerobiotic growth conditions. Nucleic Acids Res. 2010, 38, 3605–3618. [Google Scholar] [CrossRef] [PubMed]
- Tamayo, R.; Pratt, J.T.; Camilli, A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 2007, 61, 131–148. [Google Scholar] [CrossRef] [PubMed]
- Hengge-Aronis, R. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 2002, 66, 373–395. [Google Scholar] [CrossRef] [PubMed]
- Bohringer, J.; Fischer, D.; Mosler, G.; Hengge-Aronis, R. UDP-glucose is a potential intracellular signal molecule in the control of expression of σS and σS-dependent genes in Escherichia coli. J. Bacteriol. 1995, 177, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Loewen, P.C.; Hu, B.; Strutinsky, J.; Sparling, R. Regulation in the RpoS regulon of Escherichia coli. Can. J. Microbiol. 1998, 44, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Sevcik, M.; Sebkova, A.; Volf, J.; Rychlik, I. Transcription of arcA and rpoS during growth of Salmonella typhimurium under aerobic and microaerobic conditions. Microbiology 2001, 147, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Lange, R.; Fischer, D.; Hengge-Aronis, R. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 1995, 177, 4676–4680. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.; Gentry, D.; Elliott, T.; Cashel, M. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 2002, 184, 4455–4465. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Sullivan, S.M.; Wout, P.K.; Maddock, J.R. G-protein control of the ribosome-associated stress response protein SpoT. J. Bacteriol. 2007, 189, 6140–6147. [Google Scholar] [CrossRef] [PubMed]
- Kostakioti, M.; Hadjifrangiskou, M.; Pinkner, J.S.; Hultgren, S.J. QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol. Microbiol. 2009, 73, 1020–1031. [Google Scholar] [CrossRef] [PubMed]
- Pesavento, C.; Becker, G.; Sommerfeldt, N.; Possling, A.; Tschowri, N.; Mehlis, A.; Hengge, R. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 2008, 22, 2434–2446. [Google Scholar] [CrossRef] [PubMed]
- Dyszel, J.L.; Soares, J.A.; Swearingen, M.C.; Lindsay, A.; Smith, J.N.; Ahmer, B.M. E. coli K-12 and EHEC genes regulated by SdiA. PLoS ONE 2010, 5, e8946. [Google Scholar] [CrossRef] [PubMed]
- Leaphart, A.B.; Thompson, D.K.; Huang, K.; Alm, E.; Wan, X.-F.; Arkin, A.; Brown, S.D.; Wu, L.; Yan, T.; Liu, X.; et al. Transcriptome profiling of Shewanella oneidensis gene expression following exposure to acidic and alkaline pH. J. Bacteriol. 2006, 188, 1633–1642. [Google Scholar] [CrossRef] [PubMed]
- Popp, D.; Iwasa, M.; Narita, A.; Erickson, H.P.; Maéda, Y. FtsZ condensates: An in vitro electron microscopy study. Biopolymers 2009, 91, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Pogliano, J.; Dong, J.M.; De Wulf, P.; Furlong, D.; Boyd, D.; Losick, R.; Pogliano, K.; Lin, E.C. Aberrant cell division and random FtsZ ring positioning in Escherichia coli cpxA* mutants. J. Bacteriol. 1998, 180, 3486–3490. [Google Scholar] [PubMed]
- Inoue, T.; Shingaki, R.; Hirose, S.; Waki, K.; Mori, H.; Fukui, K. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J. Bacteriol. 2007, 189, 950–957. [Google Scholar] [CrossRef] [PubMed]
- Harshey, R.M. Bacterial motility on a surface: Many ways to a common goal. Annu. Rev. Microbiol. 2003, 57, 249–273. [Google Scholar] [CrossRef] [PubMed]
- Pirooznia, M.; Nagarajan, V.; Deng, Y. GeneVenn—A web application for comparing gene lists using Venn diagrams. Bioinformation 2007, 1, 420–422. [Google Scholar] [CrossRef] [PubMed]
- Hadjifrangiskou, M.; Gu, A.P.; Pinkner, J.S.; Kostakioti, M.; Zhang, E.W.; Greene, S.E.; Hultgren, S.J. Transposon mutagenesis identifies uropathogenic Escherichia coli biofilm factors. J. Bacteriol. 2012, 194, 6195–6205. [Google Scholar] [CrossRef] [PubMed]
- Niba, E.T.; Naka, Y.; Nagase, M.; Mori, H.; Kitakawa, M. A genome-wide approach to identify the genes involved in biofilm formation in E. coli. DNA Res. 2007, 14, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Rohde, M.; Olsén, A.; Normark, S.; Reinköster, J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 2000, 36, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Garavaglia, M.; Rossi, E.; Landini, P. The pyrimidine nucleotide biosynthetic pathway modulates production of biofilm determinants in Escherichia coli. PLoS ONE 2012, 7, e31252. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, C.; Papenfort, K.; Hentrich, K.; Ahmad, I.; Le Guyon, S.; Reimann, R.; Grantcharova, N.; Romling, U. Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica Serovar Typhimurium. RNA Biol. 2012, 9, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Andresen, L.; Sala, E.; Koiv, V.; Mae, A. A role for the Rcs phosphorelay in regulating expression of plant cell wall degrading enzymes in Pectobacterium carotovorum subsp. carotovorum. Microbiology 2010, 156, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Mika, F.; Busse, S.; Possling, A.; Berkholz, J.; Tschowri, N.; Sommerfeldt, N.; Pruteanu, M.; Hengge, R. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol. Microbiol. 2012, 84, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, M.G.; Nielsen, J.S.; Boysen, A.; Franch, T.; Moller-Jensen, J.; Valentin-Hansen, P. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol. Microbiol. 2012, 84, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Schu, D.J.; Tjaden, B.C.; Storz, G.; Gottesman, S. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J. Mol. Biol. 2013, 425, 3678–3697. [Google Scholar] [CrossRef] [PubMed]
- Mika, F.; Hengge, R. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol. 2014, 11, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Tsay, J.T.; Oh, W.; Larson, T.J.; Jackowski, S.; Rock, C.O. Isolation and characterization of the ß-ketoacyl-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J. Biol. Chem. 1992, 267, 6807–6814. [Google Scholar] [PubMed]
- Dehesh, K.; Tai, H.; Edwards, P.; Byrne, J.; Jaworski, J.G. Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol. 2001, 125, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.-Y.; Cronan, J.E. β-ketoacyl-acyl carrier protein synthase III (FabH) is essential for bacterial fatty acid synthesis. J. Biol. Chem. 2003, 278, 51494–51503. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Smith, D.R.; Jones, J.W.; Chapman, M.R. In vitro polymerization of a functional Escherichia coli amyloid protein. J. Biol. Chem. 2007, 282, 3713–3719. [Google Scholar] [CrossRef] [PubMed]
- Marolda, C.L.; Lahiry, P.; Vines, E.; Saldias, S.; Valvano, M.A. Micromethods for the characterization of lipid A-core and O-antigen lipopolysaccharide. Methods Mol. Biol. 2006, 347, 237–252. [Google Scholar] [PubMed]
- Stead, M.B.; Agrawal, A.; Bowden, K.E.; Nasir, R.; Mohanty, B.K.; Meagher, R.B.; Kushner, S.R. Rnasnap: A rapid, quantitative and inexpensive, method for isolating total RNA from bacteria. Nucleic Acids Res. 2012, 40, e156. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
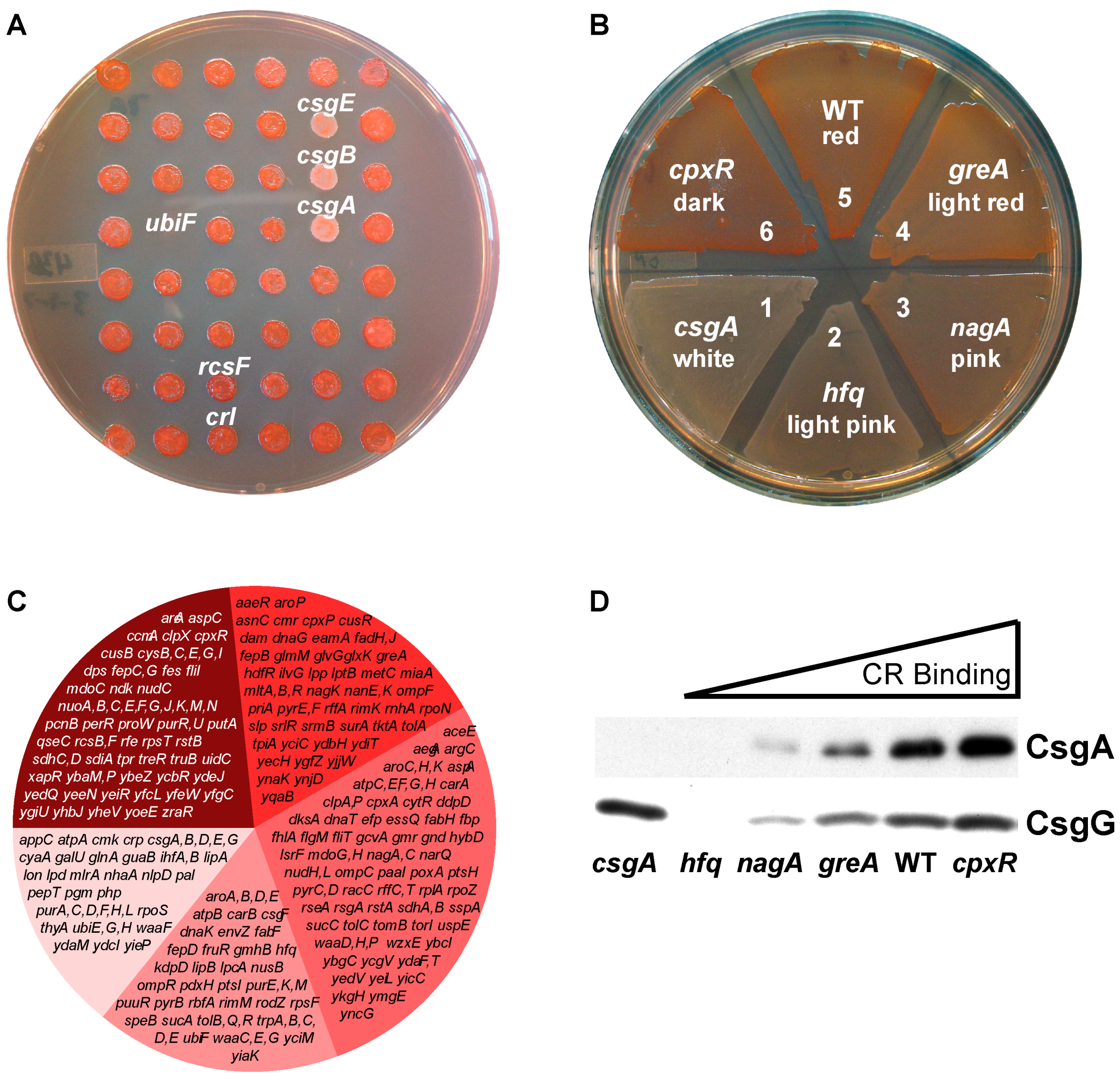
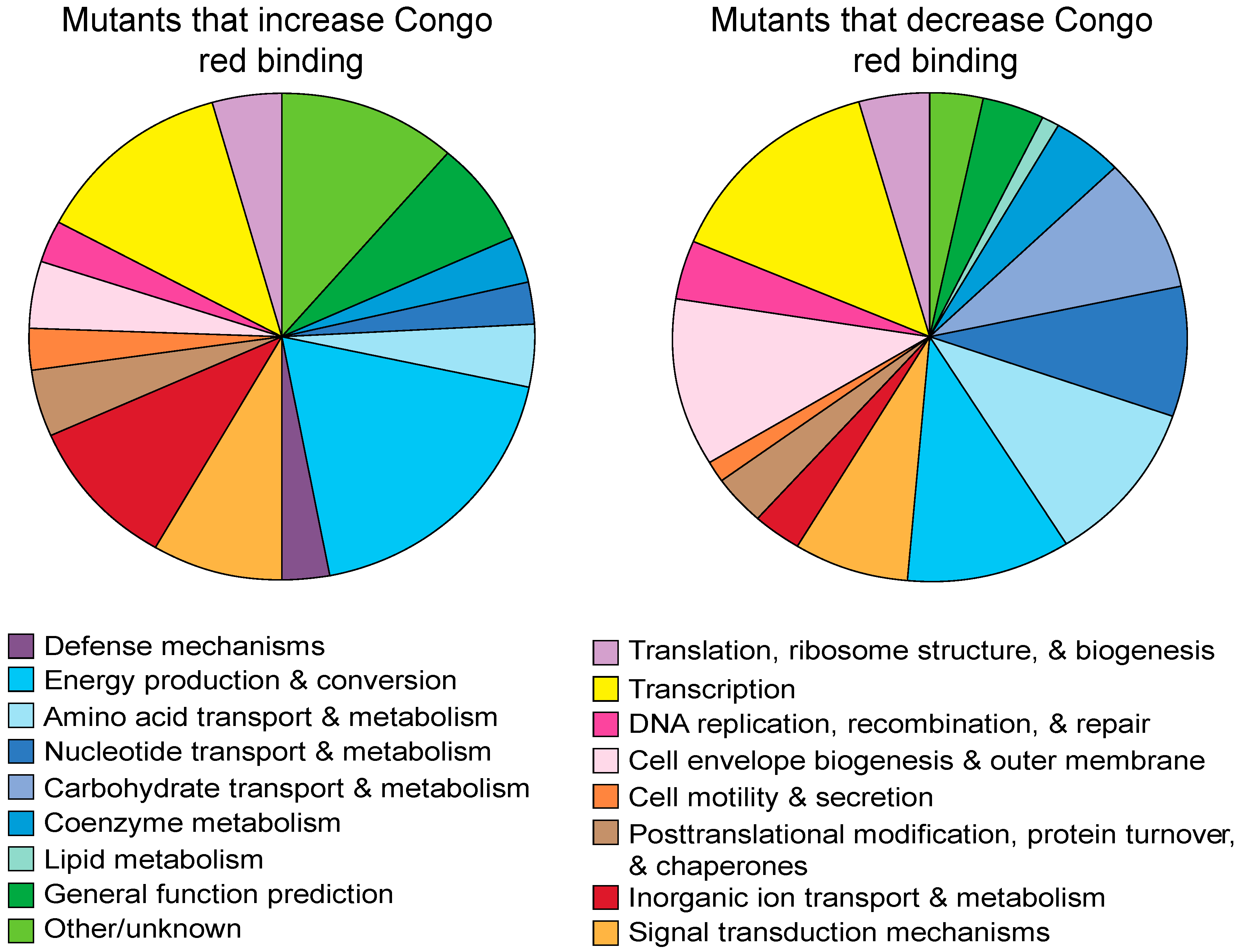
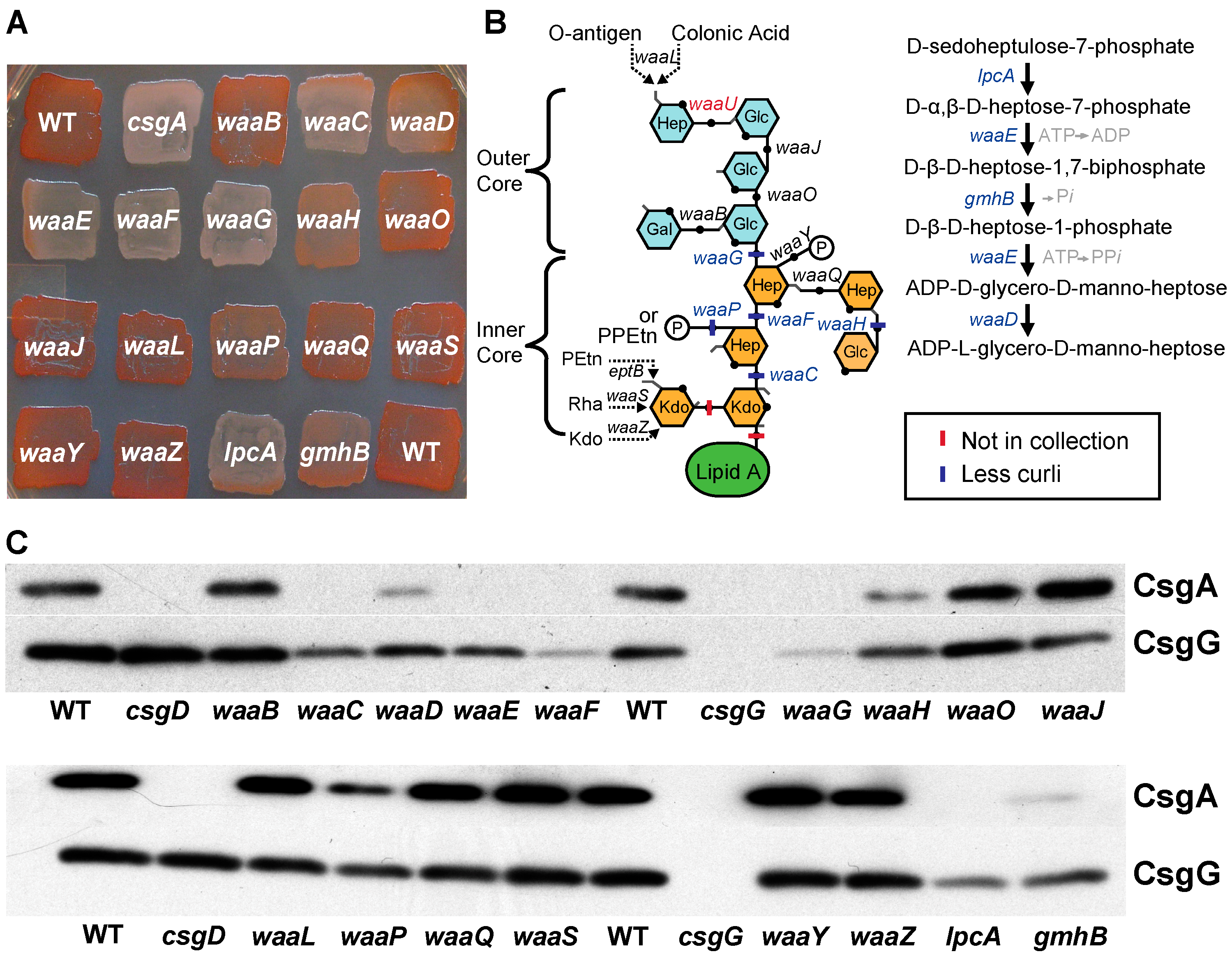
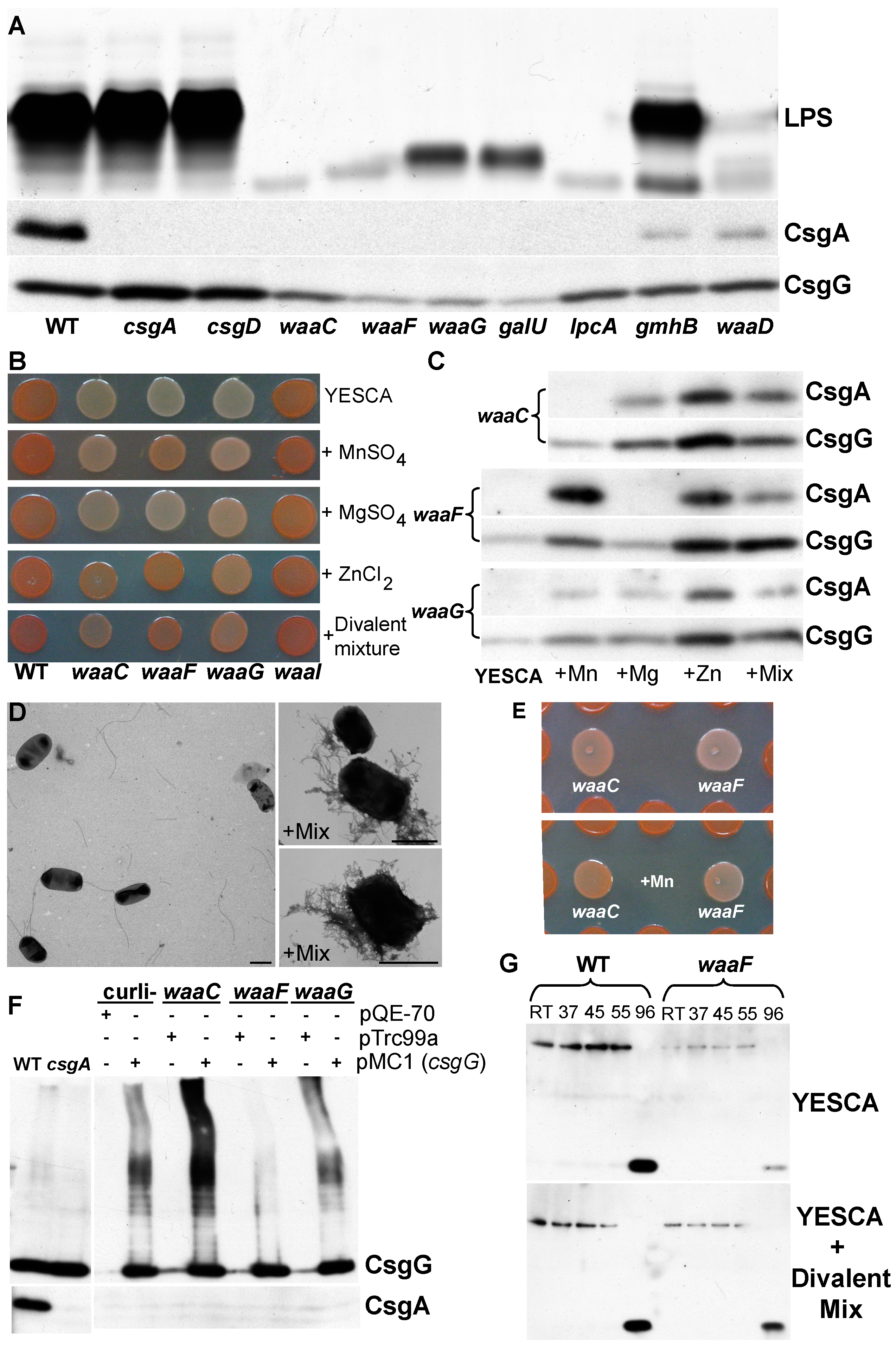
| Function | COG Group | Number (% Total) | Genes (Dark Mutants Are Bold) |
|---|---|---|---|
| Information Storage | |||
| Translation, ribosome structure, and biogenesis | J | 13 (4.2) | efp, miaA, pcnB, poxA, rbfA, rimK, rimM, rplA, rpsF, rpsT, rsgA, srmB, truB |
| Transcription | K | 43 (13.8) | aaeR, arcA, asnC, cpxR, crp, cra, csgD, cusR, cysB, cytR, dksA, fliT, fhlA, flgM, gcvA, greA, hdfR, hfq, ihfA, ihfB, mlrA, mtlR, nagK, nanK, nusB, ompR, perR, purR, puuR, rcsB, rffC, rpoN, rpoS, rpoZ, rstA, sdiA, srlR, treR, waaH, xapR, ydcI, yieP, ynaK |
| DNA replication, recombination, and repair | L | 11 (3.5) | atl, dam, dnaG, dnaT, ihfA, ihfB, nudC, nudL, priA, rnhA, rppH |
| Cellular processes | |||
| Cell envelope biogenesis, outer membrane | M | 29 (9.3) | csgA, csgB, csgE, csgF, csgG, cusB, galU, lpp, mdoH, mltA, mltB , nlpD, ompC, ompF, rcsF, pal, rfe, rffA, rffT, slp, tolc, waaC, waaD, waaE, waaF, waaG, waaP, wzxE, ycgV |
| Cell motility and secretion | N | 5 (1.6) | cpxP, flgM, fliI, tolA, ycbR |
| Posttranslational modification, protein turnover, chaperones | O | 11 (3.5) | ccmA, clpA, clpP, clpX, dnaK, lon, sspA, surA, yfgC, yjjW, yncG |
| Inorganic ion transport and metabolism | P | 14 (4.5) | cpxP, cysC, cysI, ddpD, dps, fepB, fepC, fepD, fepG, fes, mdfA, mdoG, nhaA, yoeE |
| Signal transduction mechanisms | T | 23 (7.4) | arcA, clpX, cpxA, cpxP, cpxR, crp, cusR, dksA, envZ, fhlA, gmr, kdpD, narQ, ompR, qseC, rseA, rstA, rstB, uspE, ydaM, yedV, yeiL, zraR |
| Defense Mechanism | V | 2 (0.6) | cusB, yfeW |
| Metabolism | |||
| Energy production and conversion | C | 37 (11.9) | aceE, aegA, appC, aspA, atpA, atpB, atpC, atpE, atpF, atpG, atpH, dlgD, fadH, hybD, lpd, nuoA, nuoB, nuoC, nuoE, nuoF, nuoG, nuoJ, nuoK, nuoM, nuoN, putA, racC, sdhA, sdhB, sdhC, sdhD, sucA, sucC, ubiF, ubiH, ydiT, yjjW |
| Amino acid transport and metabolism | E | 28 (9.0) | argC, aroA, aroB, aroC, aroD, aroE, aroH, aroK, aroP, aspA, aspC, carA, carB, cysE, ddpD, eamA, glnA, gmhB, mdfA, metC, pepT, proW, speB, trpA, trpB, trpC, trpD, trpE |
| Nucleotide transport and metabolism | F | 22 (7.1) | carA, carB, cmk, cyaA, guaB, ndk, purA, purC, purD, purE, purF, purH, purK, purL, purM, purU, pyrB, pyrC, pyrD, pyrE, pyrF, thyA |
| Carbohydrate transport and metabolism | G | 21 (6.7) | eamA, fbp, glmM, glvG, glxK, gmhA, gnd, lapB, lsrF, mdfA, nagA, nagC, nagK, nanE, nanK, pgm, ptsH, ptsI, rafD, tktA, tpiA |
| Coenzyme metabolism | H | 8 (3.8) | cysG, fepC, lipA, lipB, pdxH, rimK, trpA, trpB, ubiE, ubiF, ubiG, ubiH |
| Lipid metabolism | I | 3 (1.0) | fabF, fabH, fadJ |
| Secondary metabolites | Q | 2 (0.6) | fabF, paaI |
| Poorly characterized | |||
| General function prediction | R | 14 (4.5) | essQ, ilvG, iptB, nudL, php, rapZ, rppH, uidC, ybcI, ybgC, ydeJ, yeiR, ygfZ, yheV, ynjD, yqaB |
| Unknown/Other | S,U | 22 (7.1) | mdoC, rodZ, tolB, tolQ, tolR, tomb, torI, tpr, ybaM, ybaP, yciC, ydaF, ydaT, ydbH, yecH, yedQ, yeeN, yfcL, ygiU, yicC, ykgH, ymgE |
| CR Phenotype | Strain | Average Relative to BW25113 (WT) | Standard Deviation | Outcome of t-Test |
|---|---|---|---|---|
| Red | BW25113 | 1 | N/A | N/A |
| White | csgD | N.D. | N/A | N/A |
| White | nhaA | 0.018 | ±0.0068 | p-value < 0.001 |
| White | php | 0.841 | ±0.130 | Not significant |
| White | purD | 0.036 | ±0.053 | p-value < 0.001 |
| White | lon | 0.392 | ±0.187 | p-value < 0.05 |
| Light Pink | waaC | 1.118 | ±0.363 | Not significant |
| Light Pink | dnaK | 0.221 | ±0.034 | p-value < 0.001 |
| Light Pink | speB | 0.578 | ±0.114 | p-value < 0.05 |
| Light Pink | hfq | 0.06 | ±0.080 | p-value < 0.001 |
| Light Pink | aroA | 0.223 | ±0.057 | p-value < 0.05 |
| Pink | fabH | 1.171 | ±0.315 | Not significant |
| Pink | flgM | 0.86 | ±0.254 | Not significant |
| Pink | ddpD | 0.766 | ±0.337 | Not significant |
| Pink | pyrC | 0.045 | ±0.020 | p-value < 0.001 |
| Pink | nagA * | 3.38 | ±1.946 | p-value < 0.05 |
| Pink | fhlA | 0.695 | ±0.132 | p-value < 0.05 |
| Pink | dksA * | 5.115 | ±1.076 | p-value < 0.001 |
| Pink | hybD | 0.278 | ±0.043 | p-value < 0.05 |
| Pink | rstA | 0.492 | ±0.187 | p-value < 0.05 |
| Light Red | priA | 0.023 | ±0.004 | p-value < 0.001 |
| Light Red | aaeR | 2.324 | ±0.755 | p-value < 0.05 |
| Light Red | glvG | 1.316 | ±0.175 | Not significant |
| Light Red | cmr | 1.586 | ±0.561 | Not significant |
| Light Red | dam | 0.967 | ±0.268 | Not significant |
| Light Red | hdfR | 0.752 | ±0.254 | Not significant |
| Light Red | mltA | 0.874 | ±0.170 | Not significant |
| Dark Red | cysB | 3.06 | ±0.982 | p-value < 0.001 |
| Dark Red | pcnB | 1.602 | ±0.354 | p-value < 0.05 |
| Dark Red | truB | 1.371 | ±0.240 | Not significant |
| Dark Red | rcsB | 3.723 | ±0.970 | p-value < 0.05 |
| Dark Red | sdiA | 2.952 | ±0.684 | p-value < 0.05 |
| Dark Red | fes | 0.999 | ±0.217 | Not significant |
| Dark Red | nuoA | 2.302 | ±0.678 | Not significant |
| Dark Red | qseC | 2.861 | ±2.190 | Not significant |
| Dark Red | arcA | 1.603 | ±0.219 | p-value < 0.05 |
| Dark Red | mdoC | 1.581 | ±0.309 | p-value < 0.05 |
| Dark Red | perR | 1.923 | ±0.537 | p-value < 0.05 |
| Dark Red | cusB | 2.224 | ±0.998 | p-value < 0.05 |
| Outcome of t-Test | Number of Strains |
|---|---|
| Not significant | 15 |
| p-value < 0.05 | 14 |
| p-value < 0.001 | 7 |
| Uncertain | 2 |
| Total | 38 |
| Strain | Aveage csgD Levels | Std Dev | p-Value | Average 16s Levels | Std Dev | p-Value |
|---|---|---|---|---|---|---|
| BW25113 | 3.141 × 10−2 | 1.652 × 10−2 | N/A | 1.38 × 10−2 | 7.56 × 10−3 | N/A |
| dam | 4.605 × 10−2 | 1.173 × 10−2 | 0.107 | 2.05 × 10−2 | 4.31 × 10−3 | 8.93 × 10−2 |
| hfq | 1.742 × 10−4 | 3.279 × 10−5 | 9.36 × 10−4 | 6.46 × 10−3 | 6.48 × 10−3 | 1.02 × 10−1 |
| nagA | 2.407 × 10−2 | 2.143 × 10−3 | 0.306 | 4.79 × 10−3 | 4.23 × 10−3 | 2.94 × 10−2 |
| dksA | 3.777 × 10−2 | 9.779 × 10−3 | 0.436 | 3.14 × 10−3 | 8.50 × 10−4 | 6.51× 10−3 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, D.R.; Price, J.E.; Burby, P.E.; Blanco, L.P.; Chamberlain, J.; Chapman, M.R. The Production of Curli Amyloid Fibers Is Deeply Integrated into the Biology of Escherichia coli. Biomolecules 2017, 7, 75. https://doi.org/10.3390/biom7040075
Smith DR, Price JE, Burby PE, Blanco LP, Chamberlain J, Chapman MR. The Production of Curli Amyloid Fibers Is Deeply Integrated into the Biology of Escherichia coli. Biomolecules. 2017; 7(4):75. https://doi.org/10.3390/biom7040075
Chicago/Turabian StyleSmith, Daniel R., Janet E. Price, Peter E. Burby, Luz P. Blanco, Justin Chamberlain, and Matthew R. Chapman. 2017. "The Production of Curli Amyloid Fibers Is Deeply Integrated into the Biology of Escherichia coli" Biomolecules 7, no. 4: 75. https://doi.org/10.3390/biom7040075
APA StyleSmith, D. R., Price, J. E., Burby, P. E., Blanco, L. P., Chamberlain, J., & Chapman, M. R. (2017). The Production of Curli Amyloid Fibers Is Deeply Integrated into the Biology of Escherichia coli. Biomolecules, 7(4), 75. https://doi.org/10.3390/biom7040075





