Parental Histone Recycling During Chromatin Replication
Abstract
1. Introduction
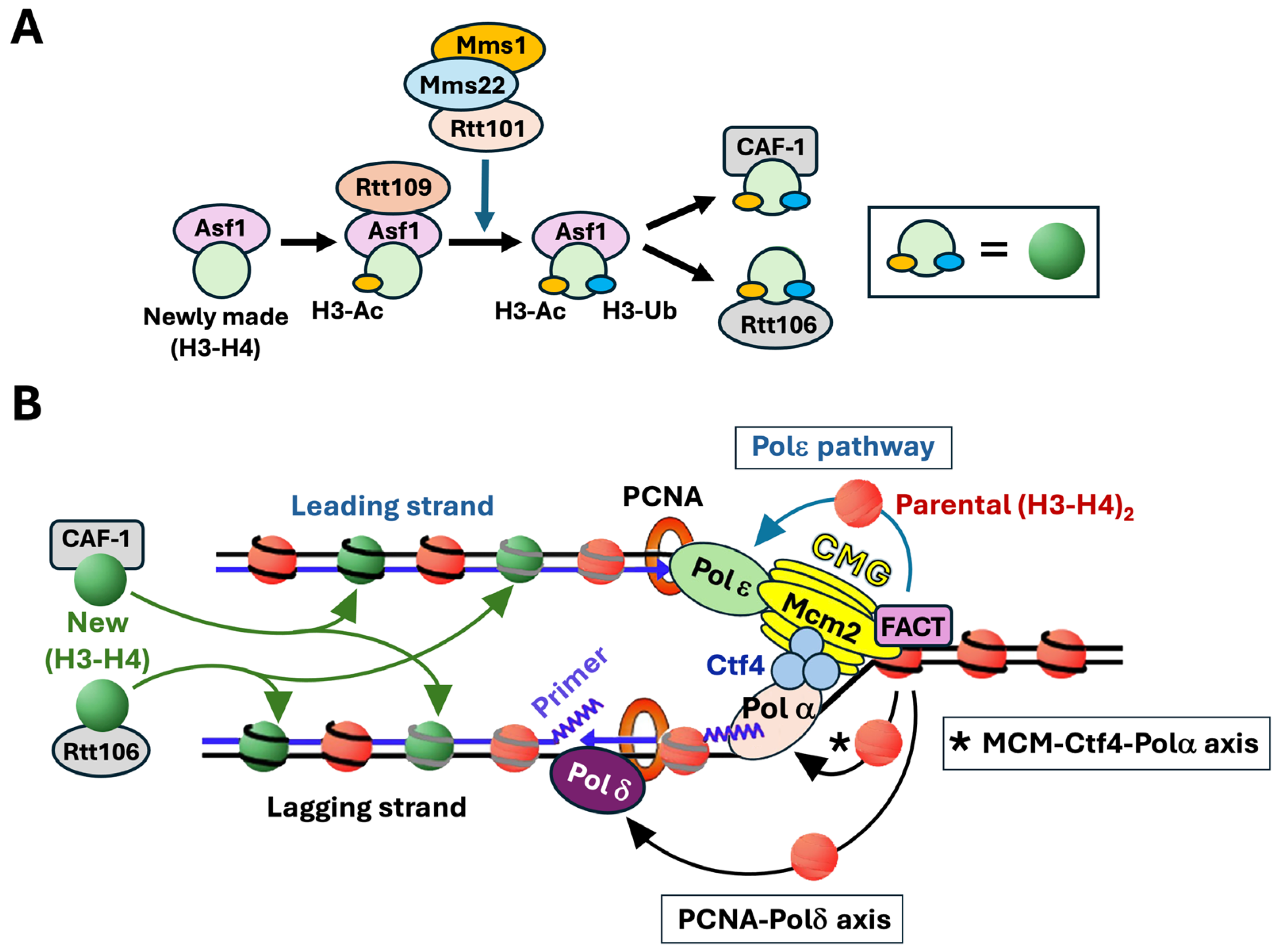
2. Discovery of Strand-Specific Pathways for the Recycling of Parental Histones H3-H4 During DNA Replication
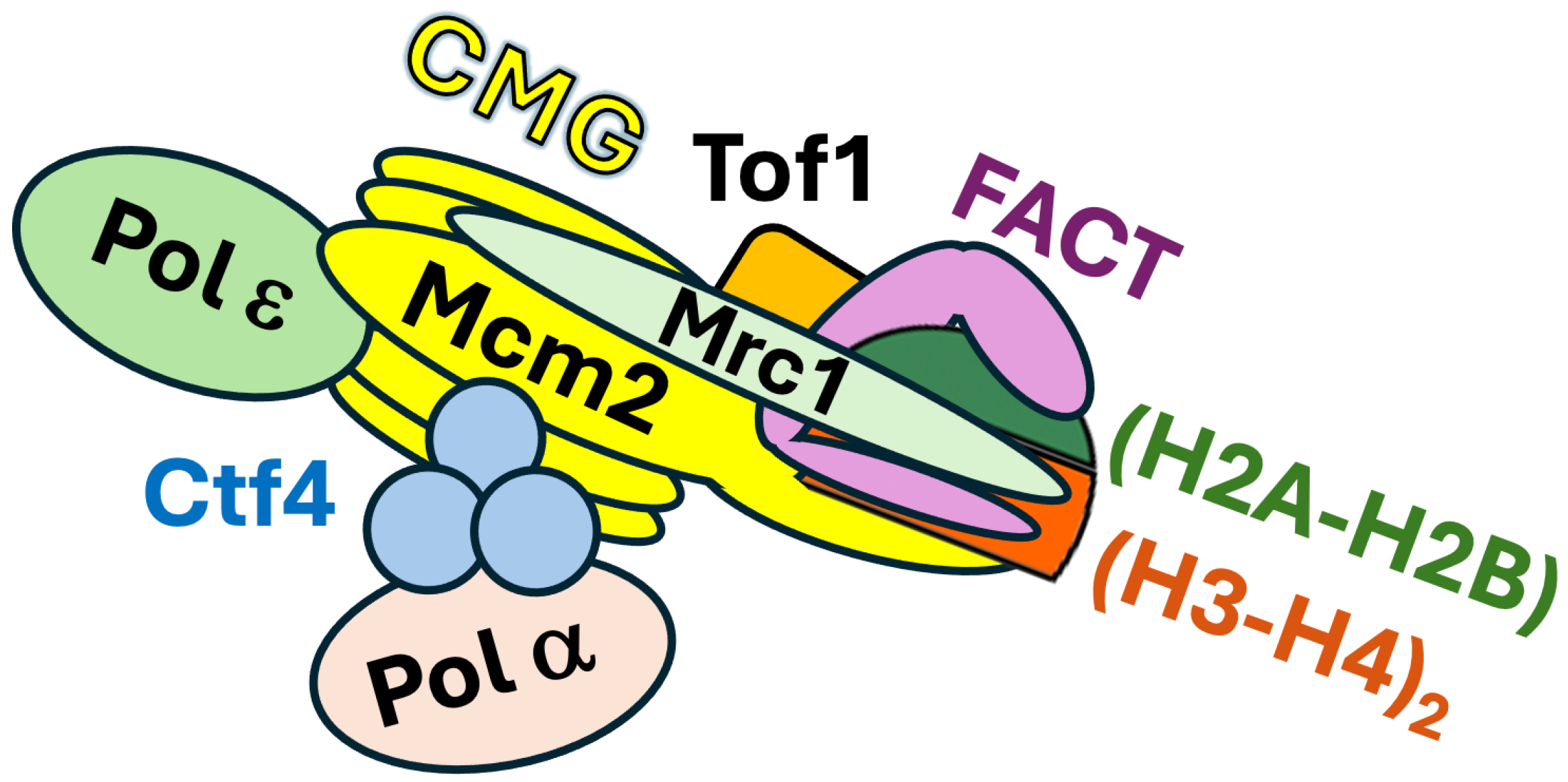
3. Role of Mrc1 in Coordinating Symmetric Parental Histone Transfer During DNA Replication
4. FACT Participates in Parental Histone Transfer to Both Leading and Lagging Strands
5. Molecular Mechanisms of Parental Histone Recycling Mediated by the Replisome
6. Conclusions and Outlook
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, D.; O’Donnell, M. The Eukaryotic Replication Machine. Enzymes 2016, 39, 191–229. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.X.; Lujan, S.A.; Burkholder, A.B.; Garbacz, M.A.; Kunkel, T.A. Roles for DNA polymerase δ in initiating and terminating leading strand DNA replication. Nat. Commun. 2019, 10, 3992. [Google Scholar] [CrossRef] [PubMed]
- Formosa, T.; Winston, F. The role of FACT in managing chromatin: Disruption, assembly, or repair? Nucleic Acids Res. 2020, 48, 11929–11941. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.L.; Aria, V.; Baris, Y.; Yeeles, J.T.P. How Pol α-primase is targeted to replisomes to prime eukaryotic DNA replication. Mol. Cell 2023, 83, 2911–2924.e16. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, G.L.; Pfander, B.; Jentsch, S. PCNA, the maestro of the replication fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Grabarczyk, D.B. The Fork Protection Complex: A Regulatory Hub at the Head of the Replisome. Subcell Biochem. 2022, 99, 83–107. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, W.; Li, G. Structures and Functions of Chromatin Fibers. Annu. Rev. Biophys. 2021, 50, 95–116. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Gaullier, G.; Luger, K. Nucleosome structure and dynamics are coming of age. Nat. Struct. Mol. Biol. 2019, 26, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Millán-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone post-translational modifications—Cause and consequence of genome function. Nat. Rev. Genet. 2022, 23, 563–580. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Long, C.; Chen, X.; Huang, C.; Chen, S.; Zhu, B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 2010, 328, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Serra-Cardona, A.; Zhang, Z. Replication-Coupled Nucleosome Assembly in the Passage of Epigenetic Information and Cell Identity. Trends Biochem. Sci. 2018, 43, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Almouzni, G.; Cedar, H. Maintenance of Epigenetic Information. Cold Spring Harb. Perspect. Biol. 2016, 8, a019372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Cooper, S.; Brockdorff, N. The interplay of histone modifications—Writers that read. Embo Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Ransom, M.; Dennehey, B.K.; Tyler, J.K. Chaperoning histones during DNA replication and repair. Cell 2010, 140, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Zhang, H.; Zhang, H.; Wang, Z.; Zhou, H.; Zhang, Z. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell 2013, 155, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Hammond-Martel, I.; Verreault, A.; Wurtele, H. Chromatin dynamics and DNA replication roadblocks. DNA Repair 2021, 104, 103140. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, X.; Feng, J.; Leng, H.; Li, S.; Xiao, J.; Liu, S.; Xu, Z.; Xu, J.; Li, D.; et al. The histone chaperone FACT contributes to DNA replication-coupled nucleosome assembly. Cell Rep. 2016, 14, 1128–1141. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.P.; Xu, R.M. Structure and function of histone chaperones in replication-coupled chromatin assembly. Curr. Opin. Struct. Biol. 2025, 92, 103059. [Google Scholar] [CrossRef] [PubMed]
- Kurat, C.F.; Yeeles, J.T.P.; Patel, H.; Early, A.; Diffley, J.F.X. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol. Cell 2017, 65, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Devbhandari, S.; Jiang, J.; Kumar, C.; Whitehouse, I.; Remus, D. Chromatin Constrains the Initiation and Elongation of DNA Replication. Mol. Cell 2017, 65, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Ishimi, Y.; Ichinose, S.; Omori, A.; Sato, K.; Kimura, H. Binding of human minichromosome maintenance proteins with histone H3. J. Biol. Chem. 1996, 271, 24115–24122. [Google Scholar] [CrossRef] [PubMed]
- Ishimi, Y.; Komamura, Y.; You, Z.; Kimura, H. Biochemical function of mouse minichromosome maintenance 2 protein. J. Biol. Chem. 1998, 273, 8369–8375. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Strømme, C.B.; Saredi, G.; Hödl, M.; Strandsby, A.; González-Aguilera, C.; Chen, S.; Groth, A.; Patel, D.J. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat. Struct. Mol. Biol. 2015, 22, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Foltman, M.; Evrin, C.; De Piccoli, G.; Jones, R.C.; Edmondson, R.D.; Katou, Y.; Nakato, R.; Shirahige, K.; Labib, K. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep. 2013, 3, 892–904. [Google Scholar] [CrossRef] [PubMed]
- Gan, H.; Serra-Cardona, A.; Hua, X.; Zhou, H.; Labib, K.; Yu, C.; Zhang, Z. The Mcm2-Ctf4-Polα Axis Facilitates Parental Histone H3-H4 Transfer to Lagging Strands. Mol. Cell 2018, 72, 140–151.e3. [Google Scholar] [CrossRef] [PubMed]
- Petryk, N.; Dalby, M.; Wenger, A.; Stromme, C.B.; Strandsby, A.; Andersson, R.; Groth, A. MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 2018, 361, 1389–1392. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Gan, H.; Serra-Cardona, A.; Zhang, L.; Gan, S.; Sharma, S.; Johansson, E.; Chabes, A.; Xu, R.M.; Zhang, Z. A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 2018, 361, 1386–1389. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Gan, H.; Han, J.; Zhou, Z.X.; Jia, S.; Chabes, A.; Farrugia, G.; Ordog, T.; Zhang, Z. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol. Cell 2014, 56, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Ukomadu, C.; Jha, S.; Senga, T.; Dhar, S.K.; Wohlschlegel, J.A.; Nutt, L.K.; Kornbluth, S.; Dutta, A. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 2007, 21, 2288–2299. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Simon, A.C.; Ortiz Bazan, M.A.; Kilkenny, M.L.; Wirthensohn, D.; Wightman, M.; Matak-Vinkovíc, D.; Pellegrini, L.; Labib, K. Ctf4 Is a Hub in the Eukaryotic Replisome that Links Multiple CIP-Box Proteins to the CMG Helicase. Mol. Cell 2016, 63, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hua, X.; Serra-Cardona, A.; Xu, X.; Gan, S.; Zhou, H.; Yang, W.S.; Chen, C.L.; Xu, R.M.; Zhang, Z. DNA polymerase α interacts with H3-H4 and facilitates the transfer of parental histones to lagging strands. Sci. Adv. 2020, 6, eabb5820. [Google Scholar] [CrossRef] [PubMed]
- Bellelli, R.; Belan, O.; Pye, V.E.; Clement, C.; Maslen, S.L.; Skehel, J.M.; Cherepanov, P.; Almouzni, G.; Boulton, S.J. POLE3-POLE4 Is a Histone H3-H4 Chaperone that Maintains Chromatin Integrity during DNA Replication. Mol. Cell 2018, 72, 112–126.e5. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Zhang, Q.; Jia, J.; Zhou, J.; Zhang, Z.; Karri, S.; Jiang, J.; Dickinson, Q.; Yao, Y.; Tang, X.; et al. DNA polymerase delta governs parental histone transfer to DNA replication lagging strand. Proc. Natl. Acad. Sci. USA 2024, 121, e2400610121. [Google Scholar] [CrossRef] [PubMed]
- Serra-Cardona, A.; Hua, X.; McNutt, S.W.; Zhou, H.; Toda, T.; Jia, S.; Chu, F.; Zhang, Z. The PCNA-Pol δ complex couples lagging strand DNA synthesis to parental histone transfer for epigenetic inheritance. Sci. Adv. 2024, 10, eadn5175. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Yang, C.; Wu, J.; Lei, Y.; Hu, J.; Feng, J.; Li, Q. DNA polymerase δ subunit Pol32 binds histone H3-H4 and couples nucleosome assembly with Okazaki fragment processing. Sci. Adv. 2024, 10, eado1739. [Google Scholar] [CrossRef] [PubMed]
- Johansson, E.; Garg, P.; Burgers, P.M. The Pol32 subunit of DNA polymerase delta contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 2004, 279, 1907–1915. [Google Scholar] [CrossRef] [PubMed]
- Madhani, H.D. Deep learning meets histones at the replication fork. Cell 2024, 187, 4824–4826. [Google Scholar] [CrossRef] [PubMed]
- Pardo, B.; Crabbé, L.; Pasero, P. Signaling pathways of replication stress in yeast. FEMS Yeast Res. 2016, 17, fow101. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Alcasabas, A.A.; Elledge, S.J. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 2001, 15, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, Y.; Fang, Y.; Paulo, J.A.; Yaghoubi, D.; Hua, X.; Shipkovenska, G.; Toda, T.; Zhang, Z.; Gygi, S.P.; et al. A replisome-associated histone H3-H4 chaperone required for epigenetic inheritance. Cell 2024, 187, 5010–5028.e24. [Google Scholar] [CrossRef] [PubMed]
- Charlton, S.J.; Flury, V.; Kanoh, Y.; Genzor, A.V.; Kollenstart, L.; Ao, W.; Brøgger, P.; Weisser, M.B.; Adamus, M.; Alcaraz, N.; et al. The fork protection complex promotes parental histone recycling and epigenetic memory. Cell 2024, 187, 5029–5047.e21. [Google Scholar] [CrossRef] [PubMed]
- Toda, T.; Fang, Y.; Shan, C.M.; Hua, X.; Kim, J.K.; Tang, L.C.; Jovanovic, M.; Tong, L.; Qiao, F.; Zhang, Z.; et al. Mrc1 regulates parental histone segregation and heterochromatin inheritance. Mol. Cell 2024, 84, 3223–3236.e4. [Google Scholar] [CrossRef] [PubMed]
- Orphanides, G.; LeRoy, G.; Chang, C.H.; Luse, D.S.; Reinberg, D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 1998, 92, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Belotserkovskaya, R.; Oh, S.; Bondarenko, V.A.; Orphanides, G.; Studitsky, V.M.; Reinberg, D. FACT facilitates transcription-dependent nucleosome alteration. Science 2003, 301, 1090–1093. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, F.K.; Kulaeva, O.I.; Patel, S.S.; Dyer, P.N.; Luger, K.; Reinberg, D.; Studitsky, V.M. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. USA 2013, 110, 7654–7659. [Google Scholar] [CrossRef] [PubMed]
- Kemble, D.J.; McCullough, L.L.; Whitby, F.G.; Formosa, T.; Hill, C.P. FACT Disrupts Nucleosome Structure by Binding H2A-H2B with Conserved Peptide Motifs. Mol. Cell 2015, 60, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Winkler, D.D.; Luger, K. The histone chaperone FACT: Structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 2011, 286, 18369–18374. [Google Scholar] [CrossRef] [PubMed]
- Hondele, M.; Stuwe, T.; Hassler, M.; Halbach, F.; Bowman, A.; Zhang, E.T.; Nijmeijer, B.; Kotthoff, C.; Rybin, V.; Amlacher, S.; et al. Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature 2013, 499, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Stuwe, T.; Hothorn, M.; Lejeune, E.; Rybin, V.; Bortfeld, M.; Scheffzek, K.; Ladurner, A.G. The FACT Spt16 “peptidase” domain is a histone H3-H4 binding module. Proc. Natl. Acad. Sci. USA 2008, 105, 8884–8889. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Liu, Y.; Luger, K. Histone chaperone FACT FAcilitates Chromatin Transcription: Mechanistic and structural insights. Curr. Opin. Struct. Biol. 2020, 65, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Chien, C.T.; Hirose, S.; Lee, S.C. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 2006, 25, 3975–3985. [Google Scholar] [CrossRef] [PubMed]
- Safaric, B.; Chacin, E.; Scherr, M.J.; Rajappa, L.; Gebhardt, C.; Kurat, C.F.; Cordes, T.; Duderstadt, K.E. The fork protection complex recruits FACT to reorganize nucleosomes during replication. Nucleic Acids Res. 2022, 50, 1317–1334. [Google Scholar] [CrossRef] [PubMed]
- Formosa, T. The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta 2013, 1819, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tang, Y.; Xu, J.; Leng, H.; Shi, G.; Hu, Z.; Wu, J.; Xiu, Y.; Feng, J.; Li, Q. The N-terminus of Spt16 anchors FACT to MCM2-7 for parental histone recycling. Nucleic Acids Res. 2023, 51, 11549–11567. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Hua, X.; Shan, C.M.; Toda, T.; Qiao, F.; Zhang, Z.; Jia, S. Coordination of histone chaperones for parental histone segregation and epigenetic inheritance. Genes Dev. 2024, 38, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Gao, Y.; Zhang, Y.; Yu, D.; Lin, J.; Feng, J.; Li, J.; Xu, Z.; Zhang, Y.; Dang, S.; et al. Parental histone transfer caught at the replication fork. Nature 2024, 627, 890–897. [Google Scholar] [CrossRef] [PubMed]
- Baretić, D.; Jenkyn-Bedford, M.; Aria, V.; Cannone, G.; Skehel, M.; Yeeles, J.T.P. Cryo-EM structure of the fork protection complex bound to CMG at a replication fork. Mol. Cell 2020, 78, 926–940.e13. [Google Scholar] [CrossRef] [PubMed]
- Koç, A.; Wheeler, L.J.; Mathews, C.K.; Merrill, G.F. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 2004, 279, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.E.; Mihelich, M.N.; Whitted, J.E.; Reitman, H.J.; Timmerman, A.J.; Tehseen, M.; Hamdan, S.M.; Schauer, G.D. Revised mechanism of hydroxyurea-induced cell cycle arrest and an improved alternative. Proc. Natl. Acad. Sci. USA 2024, 121, e2404470121. [Google Scholar] [CrossRef] [PubMed]
- Calzada, A.; Hodgson, B.; Kanemaki, M.; Bueno, A.; Labib, K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005, 19, 1905–1919. [Google Scholar] [CrossRef] [PubMed]
- Byun, T.S.; Pacek, M.; Yee, M.C.; Walter, J.C.; Cimprich, K.A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005, 19, 1040–1052. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.L.; Baris, Y.; Taylor, M.R.G.; Yeeles, J.T.P. Structure of a human replisome shows the organisation and interactions of a DNA replication machine. EMBO J. 2021, 40, e108819. [Google Scholar] [CrossRef] [PubMed]
- Rzechorzek, N.J.; Hardwick, S.W.; Jatikusumo, V.A.; Chirgadze, D.Y.; Pellegrini, L. CryoEM structures of human CMG-ATPγS-DNA and CMG-AND-1 complexes. Nucleic Acids Res. 2020, 48, 6980–6995. [Google Scholar] [CrossRef] [PubMed]
- Jenkyn-Bedford, M.; Jones, M.L.; Baris, Y.; Labib, K.P.M.; Cannone, G.; Yeeles, J.T.P.; Deegan, T.D. A conserved mechanism for regulating replisome disassembly. Nature 2021, 600, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Komata, M.; Katou, Y.; Guan, Z.; Reis, C.C.; Budd, M.; Shirahige, K.; Campbell, J.L. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol. Cell 2008, 32, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Komata, M.; Bando, M.; Araki, H.; Shirahige, K. The direct binding of Mrc1, a checkpoint mediator, to Mcm6, a replication helicase, is essential for the replication checkpoint against methyl methanesulfonate-induced stress. Mol. Cell. Biol. 2009, 29, 5008–5019. [Google Scholar] [CrossRef] [PubMed]
- Bando, M.; Katou, Y.; Komata, M.; Tanaka, H.; Itoh, T.; Sutani, T.; Shirahige, K. Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J. Biol. Chem. 2009, 284, 34355–34365. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.S.; Spenkelink, L.M.; Schauer, G.D.; Hill, F.R.; Georgescu, R.E.; O’Donnell, M.E.; van Oijen, A.M. Single-molecule visualization of Saccharomyces cerevisiae leading-strand synthesis reveals dynamic interaction between MTC and the replisome. Proc. Natl. Acad. Sci. USA 2017, 114, 10630–10635. [Google Scholar] [CrossRef] [PubMed]
- Goswami, P.; Abid Ali, F.; Douglas, M.E.; Locke, J.; Purkiss, A.; Janska, A.; Eickhoff, P.; Early, A.; Nans, A.; Cheung, A.M.C.; et al. Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome. Nat. Commun. 2018, 9, 5061. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Shi, Y.; Georgescu, R.E.; Yuan, Z.; Chait, B.T.; Li, H.; O’Donnell, M.E. The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol. 2015, 22, 976–982. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.S.; Spenkelink, L.M.; Schauer, G.D.; Yurieva, O.; Mueller, S.H.; Natarajan, V.; Kaur, G.; Maher, C.; Kay, C.; O’Donnell, M.E.; et al. Tunability of DNA Polymerase Stability during Eukaryotic DNA Replication. Mol. Cell 2020, 77, 17–25.e5. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.E.; Le Douarin, B.; Henry, C.; Galibert, F. The Saccharomyces cerevisiae protein YJR043C (Pol32) interacts with the catalytic subunit of DNA polymerase alpha and is required for cell cycle progression in G2/M. Mol. Gen. Genet. 1999, 260, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Stewart-Morgan, K.R.; Petryk, N.; Groth, A. Chromatin replication and epigenetic cell memory. Nat. Cell Biol. 2020, 22, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Evrin, C.; Maman, J.D.; Diamante, A.; Pellegrini, L.; Labib, K. Histone H2A-H2B binding by Pol α in the eukaryotic replisome contributes to the maintenance of repressive chromatin. EMBO J. 2018, 37, e99021. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Bi, X. Parental Histone Recycling During Chromatin Replication. Biomolecules 2026, 16, 13. https://doi.org/10.3390/biom16010013
Bi X. Parental Histone Recycling During Chromatin Replication. Biomolecules. 2026; 16(1):13. https://doi.org/10.3390/biom16010013
Chicago/Turabian StyleBi, Xin. 2026. "Parental Histone Recycling During Chromatin Replication" Biomolecules 16, no. 1: 13. https://doi.org/10.3390/biom16010013
APA StyleBi, X. (2026). Parental Histone Recycling During Chromatin Replication. Biomolecules, 16(1), 13. https://doi.org/10.3390/biom16010013





