The Role of BRCT Domain from LmjPES in Leishmania major Pathogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Cell Culture Conditions
2.3. Generation of the Transgenic L. major Cell Lines
2.4. RNA-Sequencing Analysis
2.5. Quantitative Real Time-PCR (qPCR)
2.6. In Vivo Infection and Evaluation
2.7. Hematoxylin and Eosin Staining
2.8. Statistical Analysis
3. Results
3.1. Generation of L. major Parasites Exhibiting a Constitutive Overexpression of BRCT Domain from LmjPES (LmjPES BRCT) or an Expression of LmjPES Lacking BRCT (LmjPES ΔBRCT)
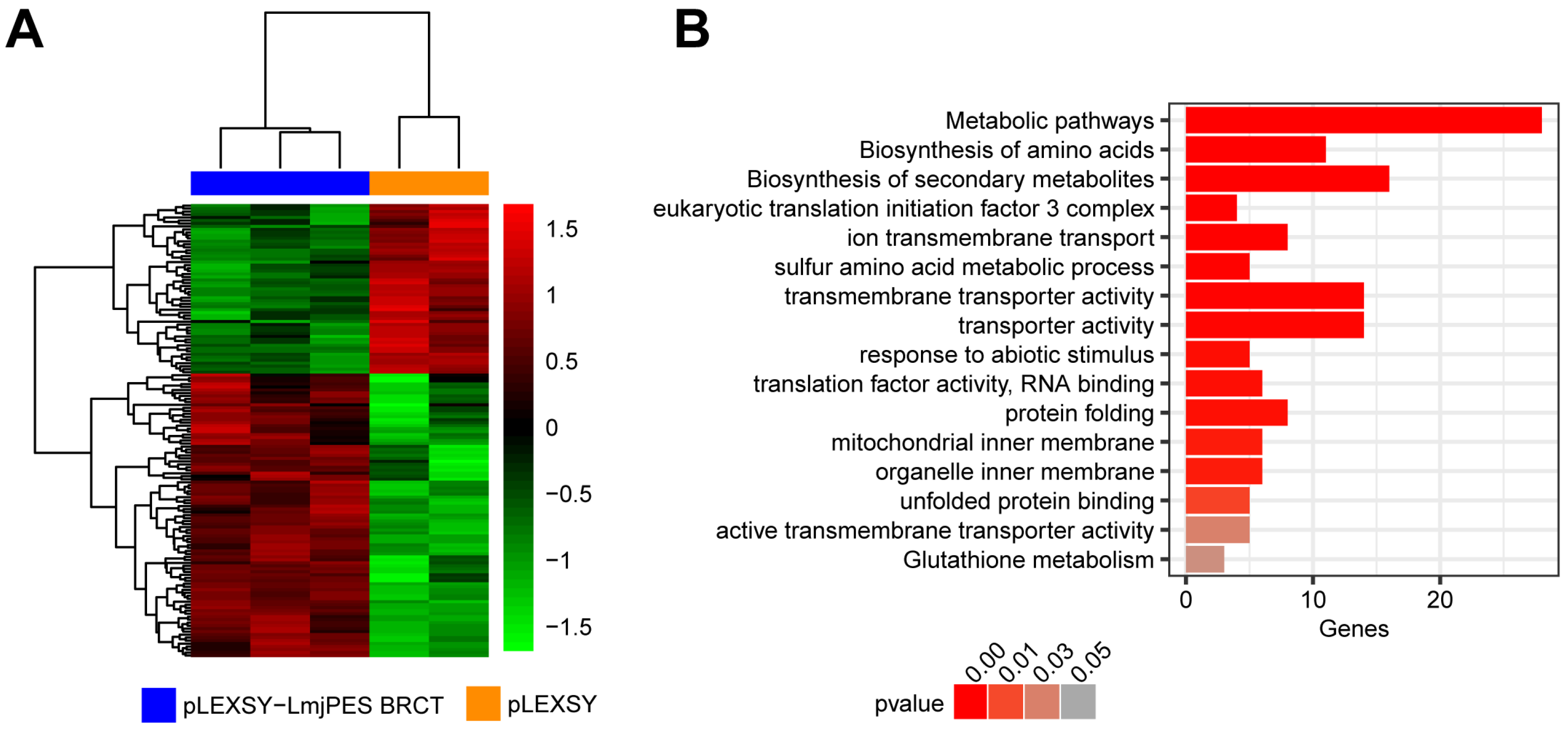
3.2. BRCT Domain from LmjPES May Regulate Genes Involved in Metabolic Pathways, Transporter Activity, and Protein Folding
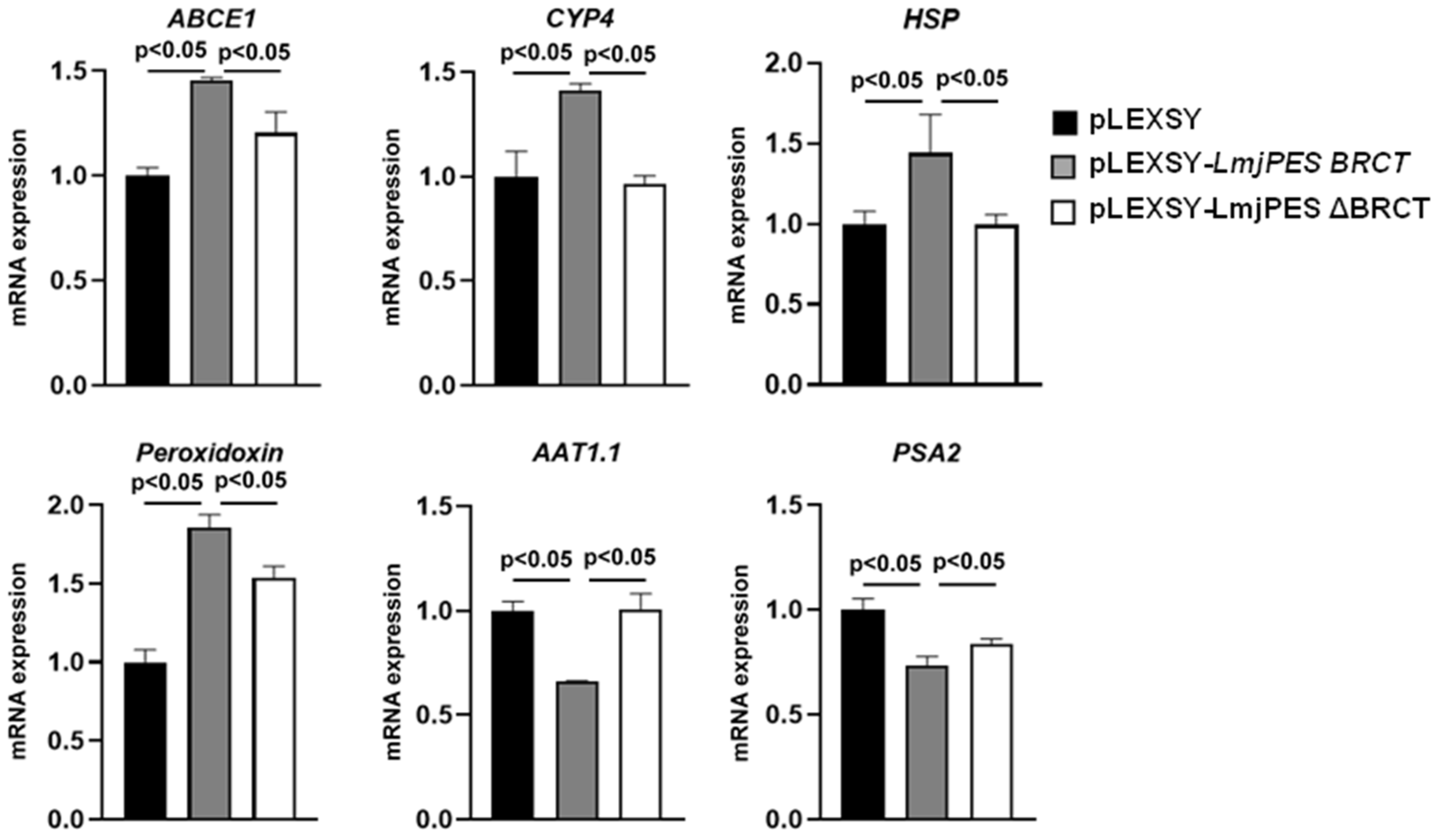
3.3. BRCT Domain from LmjPES May Be Involved in L. major Pathogenesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BRCT | Breast cancer associated 1 C-terminal domain, |
| CL | Cutaneous Leishmaniasis |
| PES1 | Pescadillo ribosomal biogenesis factor 1 |
| VL | Visceral Leishmaniasis |
| WHO | World Health Organization |
References
- Leishmaniasis. Available online: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis (accessed on 24 February 2025).
- Burguete-Mikeo, A.; Fernández-Rubio, C.; Peña-Guerrero, J.; El-Dirany, R.; Gainza, L.; Carasa Buj, B.; Nguewa, P.A. Characterization of Leishmania parasites isolated from naturally infected mammals. Animals 2023, 13, 2153. [Google Scholar] [CrossRef]
- Serafim, T.D.; Coutinho-Abreu, I.V.; Dey, R.; Kissinger, R.; Valenzuela, J.G.; Oliveira, F.; Kamhawi, S. Leishmaniasis: The act of transmission. Trends Parasitol. 2021, 76, 976–987. [Google Scholar] [CrossRef] [PubMed]
- Ranatunga, M.; Deacon, A.; Harbige, L.S.; Dyer, P.; Boateng, J.; Getti, G.T.M. Ex vivo analysis of the association of GFP-expressing L. aethiopica and L. mexicana with human peripheral blood-derived (PBD) leukocytes over 24 hours. Microorganisms 2024, 12, 1909. [Google Scholar] [CrossRef] [PubMed]
- dos Santos Meira, C.; Gedamu, L. Protective or detrimental? understanding the role of host immunity in leishmaniasis. Microorganisms 2019, 7, 695. [Google Scholar] [CrossRef]
- Belkaid, Y.; Mendez, S.; Lira, R.; Kadambi, N.; Milon, G.; Sacks, D. A natural model of Leishmania major infection reveals a prolonged “Silent” phase of parasite amplification in the skin before the onset of lesion formation and immunity. J. Immunol. 2000, 165, 969–977. [Google Scholar] [CrossRef]
- Kumar, R.; Bumb, R.A.; Salotra, P. Correlation of parasitic load with interleukin-4 response in patients with cutaneous leishmaniasis due to Leishmania tropica. FEMS Immunol. Med. Microbiol. 2009, 57, 239–246. [Google Scholar] [CrossRef]
- Hanau, S.; Maritati, M.; Contini, C.; Trentini, A.; Manfrinato, M.C.; Almugadam, S.H. Commentary on the issue of Leishmania infection: Focus on some pathogenetic, clinical, and epidemiological aspects. Vet. Sci. 2025, 12, 536. [Google Scholar] [CrossRef]
- Kumari, D.; Singh, K. Exploring the paradox of defense between host and Leishmania parasite. Int. Immunopharmacol. 2022, 102, 108400. [Google Scholar] [CrossRef]
- Karamysheva, Z.N.; Gutierrez Guarnizo, S.A.; Karamyshev, A.L. Regulation of translation in the protozoan parasite Leishmania. Int. J. Mol. Sci. 2020, 21, 2981. [Google Scholar] [CrossRef]
- Domagalska, M.A.; Barrett, M.P.; Dujardin, J.C. Drug resistance in Leishmania: Does it really matter? Trends Parasitol. 2023, 39, 251–259. [Google Scholar] [CrossRef]
- Cosma, C.; Maia, C.; Khan, N.; Infantino, M.; Del Riccio, M. Leishmaniasis in humans and animals: A one health approach for surveillance, prevention and control in a changing world. Trop. Med. Infect. Dis. 2024, 9, 258. [Google Scholar] [CrossRef]
- Ivens, A.C.; Peacock, C.S.; Worthey, E.A.; Murphy, L.; Aggarwal, G.; Berriman, M.; Sisk, E.; Rajandream, M.A.; Adlem, E.; Aert, R.; et al. The genome of the kinetoplastid parasite, Leishmania major. Science 2005, 309, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Algarabel, M.; Fernández-rubio, C.; Musilova, K.; Peña-guerrero, J.; Vacas, A.; Larrea, E.; Nguewa, P.A. In Leishmania major, the homolog of the oncogene Pes1 may play a critical role in parasite infectivity. Int. J. Mol. Sci. 2021, 22, 12592. [Google Scholar] [CrossRef]
- Yuan, S.; Xu, N.; Yang, J.; Yuan, B. Emerging role of PES1 in disease: A promising therapeutic target? Gene 2025, 932, 48896. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Wang, B.; Meng, Z.; Zhao, J.; Jin, X. PES1 Is Transcriptionally regulated by BRD4 and promotes cell proliferation and glycolysis in hepatocellular carcinoma. Int. J. Biochem. Cell Biol. 2018, 104, 1–8. [Google Scholar] [CrossRef]
- Li, Y.Z.; Zhang, C.; Pei, J.P.; Zhang, W.C.; Zhang, C.D.; Dai, D.Q. The functional role of Pescadillo ribosomal biogenesis factor 1 in cancer. J. Cancer 2022, 13, 268–277. [Google Scholar] [CrossRef]
- Kinoshita, Y.; Jarell, A.D.; Flaman, J.M.; Foltz, G.; Schuster, J.; Sopher, B.L.; Irvin, D.K.; Kanning, K.; Kornblum, H.I.; Nelson, P.S.; et al. Pescadillo, a novel cell cycle regulatory protein abnormally expressed in malignant cells. J. Biol. Chem. 2001, 276, 6656–6665. [Google Scholar] [CrossRef]
- Dai, L.; Dai, Y.; Han, J.; Huang, Y.; Wang, L.; Huang, J.; Zhou, Z. Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin. Mol. Cell 2021, 81, 2765–2777. [Google Scholar] [CrossRef]
- Glover, J.N.M. Insights into the molecular basis of human hereditary breast cancer from studies of the BRCA1 BRCT domain. Fam. Cancer 2006, 5, 89–93. [Google Scholar] [CrossRef]
- Masi, A.; Antoccia, A. NBS1 heterozygosity and cancer Risk. Curr. Genom. 2008, 9, 275–281. [Google Scholar] [CrossRef]
- Hölzel, M.; Grimm, T.; Rohrmoser, M.; Malamoussi, A.; Harasim, T.; Gruber-Eber, A.; Kremmer, E.; Eick, D. The BRCT domain of mammalian Pes1 is crucial for nucleolar localization and rRNA processing. Nucleic Acids Res. 2007, 35, 789–800. [Google Scholar] [CrossRef]
- Peña-Guerrero, J.; Fernández-Rubio, C.; Burguete-Mikeo, A.; El-Dirany, R.; García-Sosa, A.T.; Nguewa, P. Discovery and validation of Lmj_04_BRCT domain, a novel therapeutic target: Identification of candidate drugs for leishmaniasis. Int. J. Mol. Sci. 2021, 22, 10493. [Google Scholar] [CrossRef]
- Larrea, E.; Fernández-Rubio, C.; Peña-Guerrero, J.; Guruceaga, E.; Nguewa, P.A. The BRCT domain from the homologue of the oncogene PES1 in Leishmania major (LmjPES) promotes malignancy and drug resistance in mammalian cells. Int. J. Mol. Sci. 2022, 23, 13203. [Google Scholar] [CrossRef]
- Medina-Acosta, E.; Cross, G.A.M. Rapid Isolation of DNA from Trypanosomatid protozoa using a simple “mini-Prep” procedure. Mol. Biochem. Parasitol. 1993, 59, 327–329. [Google Scholar] [CrossRef] [PubMed]
- Sacks, D.L.; Hieny, S.; Sher, A. Identification of cell surface carbohydrate and antigenic changes between noninfective and infective developmental stages of Leishmania major promastigotes. J. Immunol. 1985, 135, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.A.; Beverley, S.M. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 2003, 128, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-Seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Yates, A.D.; Allen, J.; Amode, R.M.; Azov, A.G.; Barba, M.; Becerra, A.; Bhai, J.; Campbell, L.I.; Carbajo Martinez, M.; Chakiachvili, M.; et al. Ensembl Genomes 2022: An expanding genome resource for non-vertebrates. Nucleic Acids Res. 2022, 50, D996–D1003. [Google Scholar] [CrossRef]
- Heinemann, J. Cluster analysis of untargeted metabolomic experiments. Methods Mol. Biol. 2019, 1859, 275–285. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2022, 51, D587–D592. [Google Scholar] [CrossRef]
- Vacas, A.; Fernández-Rubio, C.; Larrea, E.; Peña-Guerrero, J.; Nguewa, P.A. LmjF.22.0810 from Leishmania major modulates the Th2-type immune response and is involved in leishmaniasis outcome. Biomedicines 2020, 8, 452. [Google Scholar] [CrossRef]
- Amiri-Dashatan, N.; Rezaei-Tavirani, M.; Zali, H.; Koushki, M.; Ahmadi, N. Quantitative proteomic analysis reveals differentially expressed proteins in Leishmania major metacyclogenesis. Microb. Pathog. 2020, 149, 104557. [Google Scholar] [CrossRef]
- Pramanik, P.K.; Alam, M.N.; Roy Chowdhury, D.; Chakraborti, T. Drug resistance in protozoan parasites: An incessant wrestle for survival. J. Glob. Antimicrob. Resist. 2019, 18, 1–11. [Google Scholar] [CrossRef]
- Bolhassani, A.; Taheri, T.; Taslimi, Y.; Zamanilui, S.; Zahedifard, F.; Seyed, N.; Torkashvand, F.; Vaziri, B.; Rafati, S. Fluorescent Leishmania species: Development of stable GFP expression and its application for in vitro and in vivo studies. Exp. Parasitol. 2011, 127, 637–645. [Google Scholar] [CrossRef]
- Mißlitz, A.; Mottram, J.C.; Overath, P.; Aebischer, T. Targeted integration into a rRNA locus results in uniform and high level expression of transgenes in Leishmania amastigotes. Mol. Biochem. Parasitol. 2000, 107, 251–261. [Google Scholar] [CrossRef]
- Gadelha, F.R.; Gonçalves, C.C.; Mattos, E.C.; Alves, M.J.M.; Piñeyro, M.D.; Robello, C.; Peloso, E.F. Release of the cytosolic tryparedoxin peroxidase into the incubation medium and a different profile of cytosolic and mitochondrial peroxiredoxin expression in H2O2-treated Trypanosoma cruzi tissue culture-derived trypomastigotes. Exp. Parasitol. 2013, 133, 287–293. [Google Scholar] [CrossRef]
- Da Fonseca Pires, S.; Fialho, L.C.; Silva, S.O.; Melo, M.N.; De Souza, C.C.; Tafuri, W.L.; Bruna Romero, O.; De Andrade, H.M. Identification of virulence factors in Leishmania infantum strains by a proteomic approach. J. Proteome Res. 2014, 13, 1860–1872. [Google Scholar] [CrossRef] [PubMed]
- Alcolea, P.J.; Alonso, A.; García-Tabares, F.; Larraga, J.; Martins, L.T.C.; Loayza, F.J.; Ruiz-García, S.; Larraga, V. An insight into differential protein abundance throughout Leishmania donovani promastigote growth and differentiation. Int. Microbiol. 2023, 26, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Clos, J.; Grünebast, J.; Holm, M. Promastigote-to-amastigote conversion in Leishmania spp.-A molecular view. Pathogens 2022, 11, 1052. [Google Scholar] [CrossRef] [PubMed]
- Boozhmehrani, M.J.; Eslami, G.; Khamesipour, A.; Jafari, A.A.; Vakili, M.; Hosseini, S.S.; Askari, V. The role of ATP-binding cassette transporter genes expression in treatment failure cutaneous leishmaniasis. AMB Express 2022, 12, 78. [Google Scholar] [CrossRef]
- Roy, G.; Bhattacharya, A.; Leprohon, P.; Ouellette, M. Decreased glutamate transport in acivicin resistant Leishmania tarentolae. PLoS Negl. Trop. Dis. 2021, 15, e0010046. [Google Scholar] [CrossRef]
- Leprohon, P.; Légaré, D.; Girard, I.; Papadopoulou, B.; Ouellette, M. Modulation of Leishmania ABC protein gene expression through life stages and among drug-resistant parasites. Eukaryot. Cell 2006, 5, 1713–1725. [Google Scholar] [CrossRef]
- Kemp, M.; Handman, E.; Kemp, K.; Ismail, A.; Mustafa, M.D.; Kordofani, A.Y.; Bendtzen, K.; Kharazmi, A.; Theander, T.G. The Leishmania promastigote surface antigen-2 (PSA-2) is specifically recognised by Th1 cells in humans with naturally acquired immunity to L. major. FEMS Immunol. Med. Microbiol. 1998, 20, 209–218. [Google Scholar] [CrossRef]
- Reiling, L.; Chrobak, M.; Schmetz, C.; Clos, J. Overexpression of a single Leishmania major gene enhances parasite infectivity in vivo and in vitro. Mol. Microbiol. 2010, 76, 1175–1190. [Google Scholar] [CrossRef]
- Smirlis, D.; Bisti, S.N.; Xingi, E.; Konidou, G.; Thiakaki, M.; Soteriadou, K.P. Leishmania histone H1 overexpression delays parasite cell-cycle progression, parasite differentiation and reduces Leishmania infectivity in vivo. Mol. Microbiol. 2006, 60, 1457–1473. [Google Scholar] [CrossRef]
- Mckean, P.G.; Denny, P.W.; Knuepfer, E.; Keen, J.K.; Smith, D.F. Phenotypic changes associated with deletion and overexpression of a stage-regulated gene family in Leishmania. Cell. Microbiol. 2001, 3, 511–523. [Google Scholar] [CrossRef]


| Oligo Name | Oligo Sequence (5′-3′) |
|---|---|
| BRCTF1 | ACCAGATCTGCCATGGACACCATGCGCGGGCTAACCTTCTTCATATCG |
| BRCTR2 | TGGTGATGGTGGTGGGTACCGCGGTAGCCCGTCACCGG |
| PES∆BRCTF1 | CCATGGGAAATGGTCCATAAGAAGCAGGCA |
| PES∆BRCTR2 | GCGGAACAGCTCGCGCA |
| PES∆BRCTF3 | TGCGCGAGCTGTTCCGCAACGCGCGGCTGGTG |
| PES∆BRCTR4 | GGTACCCTGCACCCACTTGGGCAGTTT |
| Target Gene | Sense Primer (5′-3′) | Antisense Primer (5′-3′) | Amplicon Length (pb) |
|---|---|---|---|
| AAT1.1 | GCAGGTGATTATGCCGTATG | GCACAAAGGAGTAAATCGCC | 170 |
| HSP | CCTTTAAAGTGACGGAGTGC | TCGACAGTGTTTACCTTGCC | 235 |
| ABCE1 | TTCGTATCATCAACCTCCCC | CCCAGGCTCATTCATGTATC | 209 |
| CYP4 | TTCACTGAAAGTGTCCCTCC | TTGAAGAGCTCCATCTCGAC | 162 |
| PSA2 | GCACTCGATGACATCTTTGG | TTAAGAGAGACGGAAGCCAG | 254 |
| Peroxidoxin | ACATGAACGACTACAAGGGC | GATTCTTCGATCAGCACACC | 279 |
| GAPDH | CATCAAGTGCGTGAAGGCGC | CGTCGGCGAGTACTCGTGCTG | 216 |
| LmjPES BRCT | TCTTCATATCGCGTGAGGTG | CATGCTTTTTCATCCCTGGC | 147 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larrea, E.; Peña-Guerrero, J.; Fernández-Rubio, C.; Burguete-Mikeo, A.; Guruceaga, E.; Nguewa, P. The Role of BRCT Domain from LmjPES in Leishmania major Pathogenesis. Biomolecules 2025, 15, 1191. https://doi.org/10.3390/biom15081191
Larrea E, Peña-Guerrero J, Fernández-Rubio C, Burguete-Mikeo A, Guruceaga E, Nguewa P. The Role of BRCT Domain from LmjPES in Leishmania major Pathogenesis. Biomolecules. 2025; 15(8):1191. https://doi.org/10.3390/biom15081191
Chicago/Turabian StyleLarrea, Esther, José Peña-Guerrero, Celia Fernández-Rubio, Aroia Burguete-Mikeo, Elizabeth Guruceaga, and Paul Nguewa. 2025. "The Role of BRCT Domain from LmjPES in Leishmania major Pathogenesis" Biomolecules 15, no. 8: 1191. https://doi.org/10.3390/biom15081191
APA StyleLarrea, E., Peña-Guerrero, J., Fernández-Rubio, C., Burguete-Mikeo, A., Guruceaga, E., & Nguewa, P. (2025). The Role of BRCT Domain from LmjPES in Leishmania major Pathogenesis. Biomolecules, 15(8), 1191. https://doi.org/10.3390/biom15081191







