Characterization of Natural Products as Inhibitors of Shikimate Dehydrogenase from Methicillin-Resistant Staphylococcus aureus: Kinetic and Molecular Dynamics Simulations, and Biological Activity Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Purification of the Recombinant SaSDH
2.2. Enzymatic Activity Assays
2.3. Inhibition Screening Assays
2.4. Evaluation of IC50 Values
2.5. Inhibition Mechanism Characterization
2.6. Evaluation of Biological Activity
2.7. Molecular Docking
2.8. Molecular Dynamics Simulations
2.9. Linear Interaction Energy Calculation
2.10. Drug-like and ADME-Tox Properties
3. Results
3.1. Natural Products Derivatives Screening
3.2. SaSDH Inhibitor Characterization
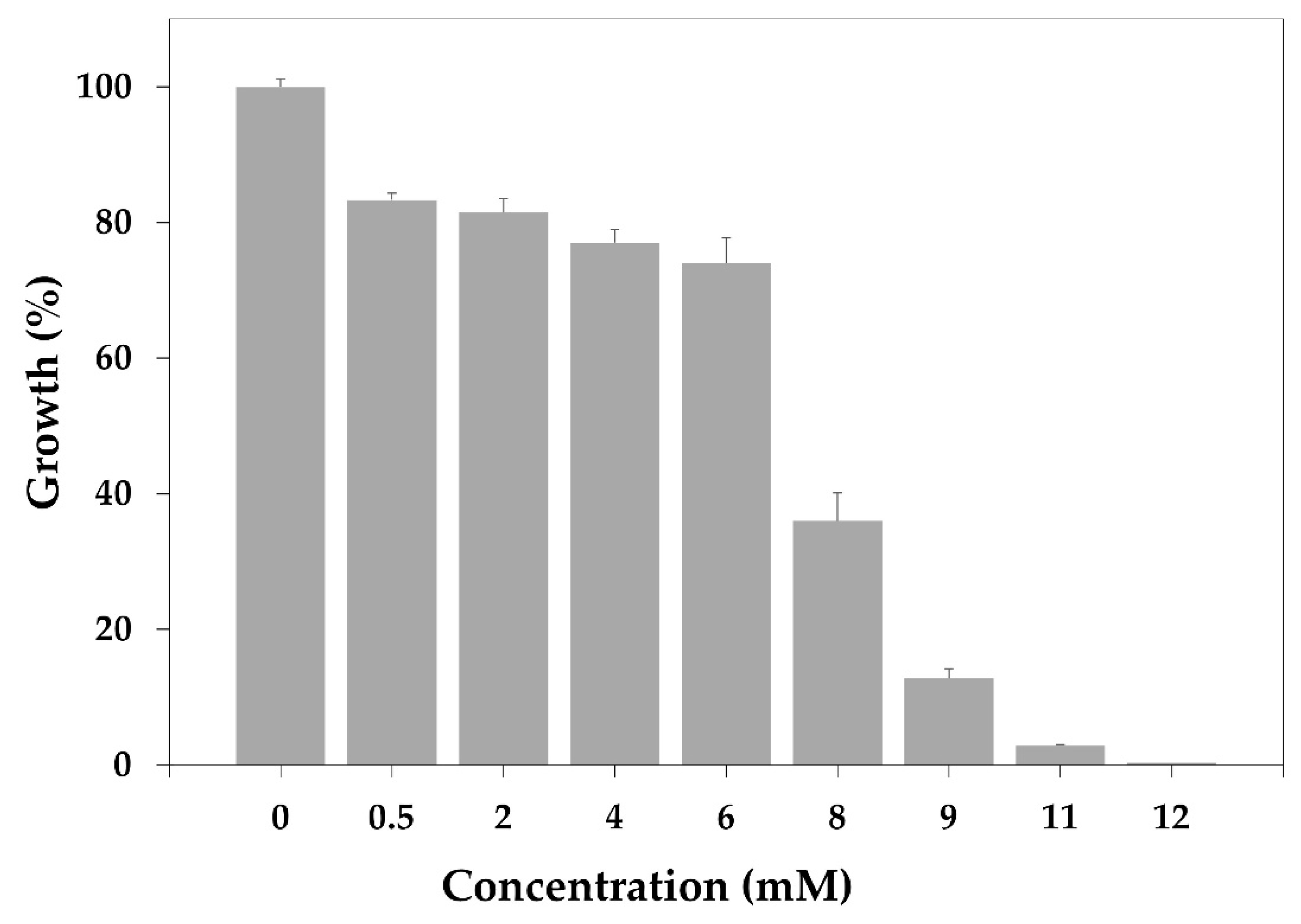
3.3. Minimum Inhibitory Concentration (MIC) in MRSA
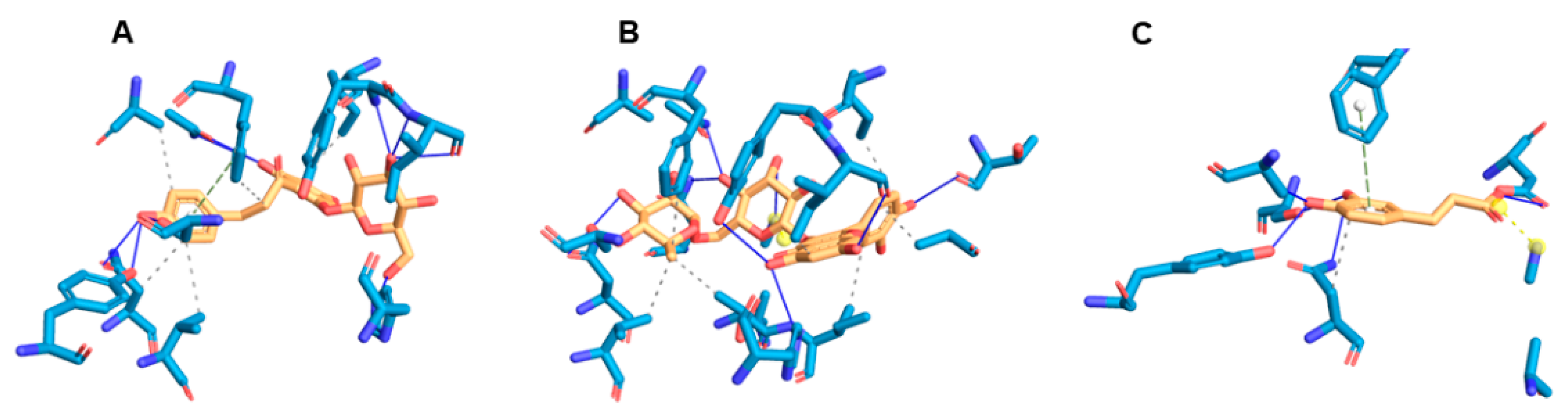
3.4. Molecular Docking Analysis
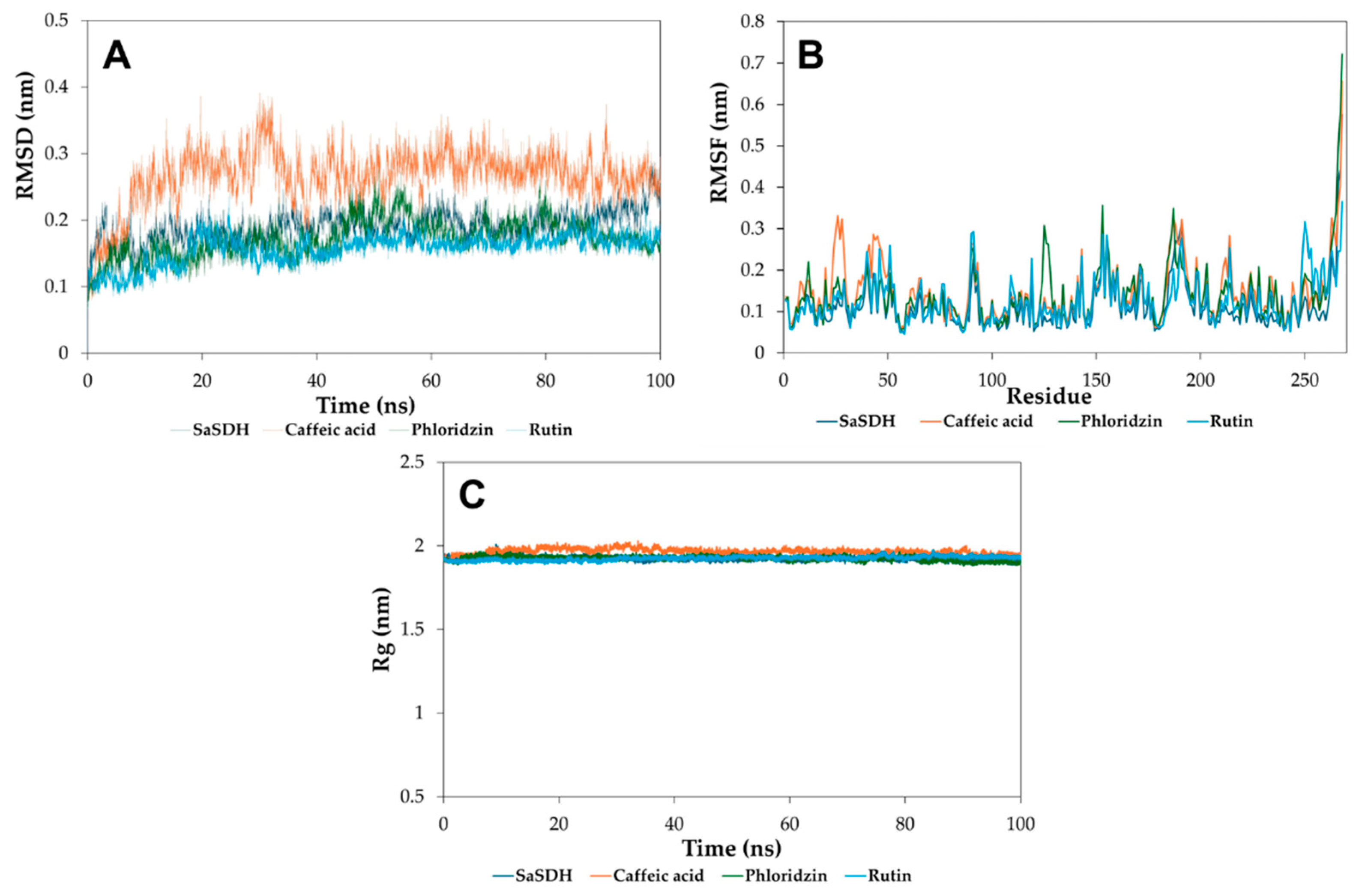
3.5. Molecular Dynamics Simulation Studies
3.5.1. Root Mean Square Deviation, Root Mean Square Fluctuation, and Radius of Gyrate
3.5.2. Linear Interaction Energy
3.6. Physicochemical and Toxicological Properties
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Antimicrobial Resistance: Accelerating National and Global Responses. Available online: https://apps.who.int/gb/ebwha/pdf_files/EB154/B154_13-en.pdf (accessed on 1 February 2025).
- Okeke, I.N.; de Kraker, M.E.A.; Van Boeckel, T.P.; Kumar, C.K.; Schmitt, H.; Gales, A.C.; Bertagnolio, S.; Sharland, M.; Laxminarayan, R. The Scope of the Antimicrobial Resistance Challenge. Lancet 2024, 403, 2426–2438. [Google Scholar] [CrossRef]
- Malik, S.S.; Mundra, S. Increasing Consumption of Antibiotics during the COVID-19 Pandemic: Implications for Patient Health and Emerging Anti-Microbial Resistance. Antibiotics 2023, 12, 45. [Google Scholar] [CrossRef]
- Gajdács, M.; Urbán, E.; Stájer, A.; Baráth, Z. Antimicrobial Resistance in the Context of the Sustainable Development Goals: A Brief Review. Eur. J. Investig. Health Psychol. Educ. 2021, 11, 71–82. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Impalli, I.; Rangarajan, R.; Cohn, J.; Ramjeet, K.; Trainor, B.W.; Strathdee, S.; Sumpradit, N.; Berman, D.; Wertheim, H.; et al. Expanding Antibiotic, Vaccine, and Diagnostics Development and Access to Tackle Antimicrobial Resistance. Lancet 2024, 403, 2534–2550. [Google Scholar] [CrossRef] [PubMed]
- Lade, H.; Kim, J.-S. Molecular Determinants of β-Lactam Resistance in Methicillin-Resistant Staphylococcus aureus (MRSA): An Updated Review. Antibiotics 2023, 12, 1362. [Google Scholar] [CrossRef]
- Almeida, A.M.; Marchiosi, R.; Abrahão, J.; Constantin, R.P.; dos Santos, W.D.; Ferrarese-Filho, O. Revisiting the Shikimate Pathway and Highlighting Their Enzyme Inhibitors. Phytochem. Rev. 2024, 23, 421–457. [Google Scholar] [CrossRef]
- Dev, A.; Tapas, S.; Pratap, S.; Kumar, P. Structure and Function of Enzymes of Shikimate Pathway. Curr. Bioinform. 2012, 7, 374–391. [Google Scholar] [CrossRef]
- Mir, R.; Jallu, S.; Singh, T.P. The Shikimate Pathway: Review of Amino Acid Sequence, Function and Three-Dimensional Structures of the Enzymes. Crit. Rev. Microbiol. 2015, 41, 172–189. [Google Scholar] [CrossRef] [PubMed]
- Kapnick, S.M.; Zhang, Y. New Tuberculosis Drug Development: Targeting the Shikimate Pathway. Expert Opin. Drug Discov. 2008, 3, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Michel, G.; Roszak, A.W.; Sauvé, V.; Maclean, J.; Matte, A.; Coggins, J.R.; Cygler, M.; Lapthorn, A.J. Structures of Shikimate Dehydrogenase AroE and Its Paralog YdiB. J. Biol. Chem. 2003, 278, 19463–19472. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Qu, J.; Deng, Q. Identification of Potential Inhibitors against Staphylococcus aureus Shikimate Dehydrogenase through Virtual Screening and Susceptibility Test. J. Enzym. Inhib. Med. Chem. 2024, 39, 2301768. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Quiroz, D.C.; Cardona-Félix, C.S.; Viveros-Ceballos, J.L.; Reyes-González, M.A.; Bolívar, F.; Ordoñez, M.; Escalante, A. Synthesis, Biological Activity and Molecular Modelling Studies of Shikimic Acid Derivatives as Inhibitors of the Shikimate Dehydrogenase Enzyme of Escherichia coli. J. Enzym. Inhib. Med. Chem. 2018, 33, 397–404. [Google Scholar] [CrossRef]
- Peek, J.; Shi, T.; Christendat, D. Identification of Novel Polyphenolic Inhibitors of Shikimate Dehydrogenase (AroE). J. Biomol. Screen. 2014, 19, 1090–1098. [Google Scholar] [CrossRef]
- Han, C.; Wang, L.; Yu, K.; Chen, L.; Hu, L.; Chen, K.; Jiang, H.; Shen, X. Biochemical Characterization and Inhibitor Discovery of Shikimate Dehydrogenase from Helicobacter pylori. FEBS J. 2006, 273, 4682–4692. [Google Scholar] [CrossRef]
- Lima, M.C.; Paiva de Sousa, C.; Fernandez-Prada, C.; Harel, J.; Dubreuil, J.D.; de Souza, E.L. A Review of the Current Evidence of Fruit Phenolic Compounds as Potential Antimicrobials against Pathogenic Bacteria. Microb. Pathog. 2019, 130, 259–270. [Google Scholar] [CrossRef]
- Mandal, S.M.; Dias, R.O.; Franco, O.L. Phenolic Compounds in Antimicrobial Therapy. J. Med. Food 2017, 20, 1031–1038. [Google Scholar] [CrossRef]
- Miklasińska-Majdanik, M.; Kępa, M.; Wojtyczka, R.D.; Idzik, D.; Wąsik, T.J. Phenolic Compounds Diminish Antibiotic Resistance of Staphylococcus aureus Clinical Strains. Int. J. Env. Res. Public Health 2018, 15, 2321. [Google Scholar] [CrossRef]
- Moloney, M.G. Natural Products as a Source for Novel Antibiotics. Trends Pharmacol. Sci. 2016, 37, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Martínez, F.J.; Barrajón-Catalán, E.; Micol, V. Tackling Antibiotic Resistance with Compounds of Natural Origin: A Comprehensive Review. Biomedicines 2020, 8, 405. [Google Scholar] [CrossRef] [PubMed]
- Jadimurthy, R.; Mayegowda, S.B.; Nayak, S.C.; Mohan, C.D.; Rangappa, K.S. Escaping Mechanisms of ESKAPE Pathogens from Antibiotics and Their Targeting by Natural Compounds. Biotechnol. Rep. 2022, 34, e00728. [Google Scholar] [CrossRef]
- Wright, G.D. Opportunities for Natural Products in 21 st Century Antibiotic Discovery. Nat. Prod. Rep. 2017, 34, 694–701. [Google Scholar] [CrossRef]
- Punkvang, A.; Kamsri, P.; Mulholland, A.; Spencer, J.; Hannongbua, S.; Pungpo, P. Simulations of Shikimate Dehydrogenase from Mycobacterium tuberculosis in Complex with 3-Dehydroshikimate and NADPH Suggest Strategies for MtbSDH Inhibition. J. Chem. Inf. Model. 2019, 59, 1422–1433. [Google Scholar] [CrossRef]
- Isa, M.A.; Majumdar, R.S.; Haider, S. In Silico Identification of Potential Inhibitors against Shikimate Dehydrogenase through Virtual Screening and Toxicity Studies for the Treatment of Tuberculosis. Int. Microbiol. 2019, 22, 7–17. [Google Scholar] [CrossRef]
- Enríquez-Mendiola, D.; Téllez-Valencia, A.; Sierra-Campos, E.; Campos-Almazán, M.; Valdez-Solana, M.; Gómez Palacio-Gastélum, M.; Avitia-Domínguez, C. Kinetic and Molecular Dynamic Studies of Inhibitors of Shikimate Dehydrogenase from Methicillin-Resistant Staphylococcus aureus. Chem. Biol. Drug Des. 2019, 94, 1504–1517. [Google Scholar] [CrossRef]
- Avitia-Domínguez, C.; Sierra-Campos, E.; Salas-Pacheco, J.M.; Nájera, H.; Rojo-Domínguez, A.; Cisneros-Martínez, J.; Téllez-Valencia, A. Inhibition and Biochemical Characterization of Methicillin-Resistant Staphylococcus aureus Shikimate Dehydrogenase: An in Silico and Kinetic Study. Molecules 2014, 19, 4491–4509. [Google Scholar] [CrossRef]
- dos Santos Nascimento, I.J.; de Aquino, T.M.; da Silva-Júnior, E.F. The New Era of Drug Discovery: The Power of Computer-Aided Drug Design (CADD). Lett. Drug Des. Discov. 2022, 19, 951–955. [Google Scholar] [CrossRef]
- Sabe, V.T.; Ntombela, T.; Jhamba, L.A.; Maguire, G.E.M.; Govender, T.; Naicker, T.; Kruger, H.G. Current Trends in Computer Aided Drug Design and a Highlight of Drugs Discovered via Computational Techniques: A Review. Eur. J. Med. Chem. 2021, 224, 113705. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Fu, A.; Zhang, L. Progress in Molecular Docking. Quant. Biol. 2019, 7, 83–89. [Google Scholar] [CrossRef]
- Paggi, J.M.; Pandit, A.; Dror, R.O. The Art and Science of Molecular Docking. Annu. Rev. Biochem. 2025, 55, 389–410. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Gülçin, I. Antioxidant Activity of Caffeic Acid (3,4-Dihydroxycinnamic Acid). Toxicology 2006, 217, 213–220. [Google Scholar] [CrossRef]
- Téllez-Valencia, A.; Ávila-Rı, S.; Pérez-Montfort, R.; Rodrı, A.; de Gómez-Puyou, M.T.; López-Calahorra, F.; Gómez-Puyou, A. Highly Specific Inactivation of Triosephosphate Isomerase from Trypanosoma cruzi. Biochem. Biophys. Res. Commun. 2002, 295, 958–963. [Google Scholar] [CrossRef] [PubMed]
- Goličnik, M.; Bavec, A. Evaluation of the Paraoxonase-1 Kinetic Parameters of the Lactonase Activity by Nonlinear Fit of Progress Curves. J. Enzyme. Inhib. Med. Chem. 2020, 35, 261–264. [Google Scholar] [CrossRef]
- Sierra-Campos, E.; Valdez-Solana, M.A.; Ruiz-Baca, E.; Ventura-García, E.K.; Avitia-Domínguez, C.I.; Aguilera-Ortiz, M.; Téllez-Valencia, A. Anti-Sporotrichotic Activity, Lambert-W Inhibition Kinetics and 3D Structural Characterization of Sporothrix schenckii Catalase as Target of Glucosinolates from Moringa Oleifera. Sci. Pharm. 2022, 90, 70. [Google Scholar] [CrossRef]
- Cantón, R.; García Sánchez, J.E.; Gómez-Luis, M.L.; Martínez Martínez, L.; Rodríguez-Avial, C.; Villa, J. Métodos Básicos Para El Estudio de La Sensibilidad a Los Microbianos. In Procedimientos en Microbiología Clínica; Picazo, J.J., Ed.; Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica (SEIMC): Madrid, España, 2000; Volume 11, pp. 1–54. [Google Scholar]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational Protein–Ligand Docking and Virtual Drug Screening with the AutoDock Suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, J.; Marsili, M. Iterative Partial Equalization of Orbital Electronegativity—A Rapid Access to Atomic Charges. Tetrahedrom 1980, 36, 3219–3288. [Google Scholar] [CrossRef]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the Scope of the Protein-Ligand Interaction Profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, 530–534. [Google Scholar] [CrossRef] [PubMed]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully Automated Protein-Ligand Interaction Profiler. Nucleic Acids Res. 2015, 43, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Kagami, L.; Wilter, A.; Diaz, A.; Vranken, W. The ACPYPE Web Server for Small-Molecule MD Topology Generation. Bioinformatics 2023, 39, btad350. [Google Scholar] [CrossRef]
- Hansen, N.; Öehlknecht, C.; de Ruiter, A.; Lier, B.; van Gunsteren, W.F.; Oostenbrink, C.; Gebhardt, J. A Suite of Advanced Tutorials for the GROMOS Biomolecular Simulation Software [Article v1.0]. Living J. Comput. Mol. Sci. 2020, 2, 18552. [Google Scholar] [CrossRef]
- Schmid, N.; Christ, C.D.; Christen, M.; Eichenberger, A.P.; van Gunsteren, W.F. Architecture, Implementation and Parallelisation of the GROMOS Software for Biomolecular Simulation. Comput. Phys. Commun. 2012, 183, 890–903. [Google Scholar] [CrossRef]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. Ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory. Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef]
- Pastor, R.W.; Brooks, B.R.; Szabo, A. An Analysis of the Accuracy of Langevin and Molecular Dynamics Algorithms. Mol. Phys. 1988, 65, 1409–1419. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Brandsdal, B.O.; Österberg, F.; Almlöf, M.; Feierberg, I.; Luzhkov, V.B.; Åqvist, J. Free Energy Calculations and Ligand Binding. J. Adv. Protein. Chem. 2003, 66, 123–158. [Google Scholar] [CrossRef]
- Åqvist, J.; Medina, C.; Samuelsson, J.-E. A New Method for Predicting Binding Affinity in Computer-Aided Drug Design. Protein Eng. Des. Sel. 1994, 7, 385–391. [Google Scholar] [CrossRef]
- Aqvist, J.; Marelius, J. The Linear Interaction Energy Method for Predicting Ligand Binding Free Energies. Comb. Chem. High Throughput Screen 2001, 4, 613–626. [Google Scholar] [CrossRef]
- Hansson, T.; Marelius, J.; Åqvist, J. Ligand Binding Affinity Prediction by Linear Interaction Energy Methods. J. Comput.-Aided Mol. Des. 1998, 12, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Chang, G.S.; Chung, J.E.; Sung, K.Y.; No, K.T. The PreADME: PC-Based Program for Batch Prediction of ADME Properties. EuroQSAR 2004, 9, 5–10. [Google Scholar]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A Web Server for Target Prediction of Bioactive Small Molecules. Nucleic Acids Res. 2014, 42, 32–38. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; Von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program for Chemistry Aware Data Visualization and Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Valdés, L.; Cuervo, A.; Salazar, N.; Ruas-Madiedo, P.; Gueimonde, M.; González, S. The Relationship between Phenolic Compounds from Diet and Microbiota: Impact on Human Health. Food Funct. 2015, 6, 2424–2439. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Shahrajabian, M.H. Therapeutic Potential of Phenolic Compounds in Medicinal Plants—Natural Health Products for Human Health. Molecules 2023, 28, 1845. [Google Scholar] [CrossRef]
- Sanhueza, L.; Melo, R.; Montero, R.; Maisey, K.; Mendoza, L.; Wilkens, M. Synergistic Interactions between Phenolic Compounds Identified in Grape Pomace Extract with Antibiotics of Different Classes against Staphylococcus aureus and Escherichia coli. PLoS ONE 2017, 12, e0172273. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Burgos, E.C.; Burgos-Hernández, A.; Noguera-Artiaga, L.; Kačániová, M.; Hernández-García, F.; Cárdenas-López, J.L.; Carbonell-Barrachina, Á.A. Antimicrobial Activity of Pomegranate Peel Extracts as Affected by Cultivar. J. Sci. Food Agric. 2017, 97, 802–810. [Google Scholar] [CrossRef]
- Shende, V.V.; Bauman, K.D.; Moore, B.S. The Shikimate Pathway: Gateway to Metabolic Diversity. Nat. Prod. Rep. 2024, 41, 604–648. [Google Scholar] [CrossRef]
- Fonseca, I.O.; Silva, R.G.; Fernandes, C.L.; de Souza, O.N.; Basso, L.A.; Santos, D.S. Kinetic and Chemical Mechanisms of Shikimate Dehydrogenase from Mycobacterium tuberculosis. Arch. Biochem. Biophys. 2007, 457, 123–133. [Google Scholar] [CrossRef]
- Barreca, D.; Bellocco, E.; Laganà, G.; Ginestra, G.; Bisignano, C. Biochemical and Antimicrobial Activity of Phloretin and Its Glycosilated Derivatives Present in Apple and Kumquat. Food Chem. 2014, 160, 292–297. [Google Scholar] [CrossRef]
- Salaheen, S.; Peng, M.; Joo, J.; Teramoto, H.; Biswas, D. Eradication and Sensitization of Methicillin Resistant Staphylococcus aureus to Methicillin with Bioactive Extracts of Berry Pomace. Front. Microbiol. 2017, 8, 253. [Google Scholar] [CrossRef]
- Tiwari, R.; Siddiqui, M.H.; Mahmood, T.; Farooqui, A.; Bagga, P.; Ahsan, F.; Shamim, A. An Exploratory Analysis on the Toxicity & Safety Profile of Polyherbal Combination of Curcumin, Quercetin and Rutin. Clin. Phytosci. 2020, 6, 82. [Google Scholar] [CrossRef]
- Yazar, M.; Sevindik, M.; Uysal, I.; Polat, A.O. Effects of Caffeic Acid on Human Health: Pharmacological and Therapeutic Effects, Biological Activity and Toxicity. Pharm. Chem. J. 2025, 59, 29–55. [Google Scholar] [CrossRef]
- Zeiger, E.; Tice, R. Chlorogenic Acid and Caffeic Acid: Review of Toxicological Literature; Integrated Laboratory Systems, Research Triangle Park: Piedmont, NC, USA, 1998. [Google Scholar]
- Aijaz, M.; Keserwani, N.; Yusuf, M.; Ansari, N.H.; Ushal, R.; Kalia, P. Chemical, Biological, and Pharmacological Prospects of Caffeic Acid. Biointerface Res. Appl. Chem. 2023, 13, 324. [Google Scholar] [CrossRef]

| Compound | Structure | % Inhibition of SaSDH at 500 µM |
|---|---|---|
| Phloridzin | 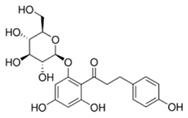 | 93 |
| Rutin |  | 87 |
| Caffeic acid | 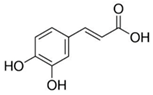 | 70 |
| 2,3-Diaminonaphthalene | 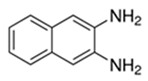 | 45 |
| 6-Bromo-2-hydroxy-3-methoxybenzaldehyde | 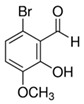 | 45 |
| 1H-indole-2-carbaldehyde | 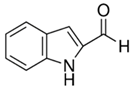 | 34 |
| 1-(3-aminopropyl)-2-methyl-1H-imidazole | 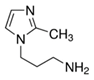 | 34 |
| Limonene | 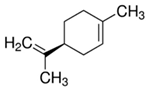 | 32 |
| Compound | Vmax (U/mg) | Km (µM) | Inhibition Mechanism |
|---|---|---|---|
| SaSDH Control | 0.52 | 4 | - |
| Phloridzin | 0.25 | 7 | Mixed with a predominant competitive component |
| Rutin | 0.39 | 4 | Non-competitive |
| Caffeic acid | 0.32 | 4 | Non-competitive |
| Compound | Type of Interactions | ||||
|---|---|---|---|---|---|
| Hydropobic | Hydrogen Bonds | Salt Bridges | Π-Stacking | Binding Score Kcal/mol | |
| Phloridzin | Phe236 (3.64) Asn58 (3.71) Val5 (3.89) Gln239 (3.98) Ile209 (3.99) | Tyr211 (2.85) | Phe236 (4.9) | ||
| Ser15 (3.0) | |||||
| His12 (3.02) | |||||
| Gln239 (3.04) | −8.7 | ||||
| Asn58 (3.08) | |||||
| Tyr32 (3.19) | |||||
| Ile212 (3.26) | |||||
| Gln239 (3.93) | |||||
| Rutin | Phe236 (3.38) Ile212 (3.48) Ile209 (3.61) Ile65 (3.64) Val5 (3.70) Ala185 (3.72) Thr60 (3.84) | Ser15 (2.79) | Lys64 (4.08) | ||
| Ile212 (3.06) | |||||
| Gln239 (3.06) | |||||
| Thr183 (3.10) | |||||
| Asn85 (3.49) | −9.6 | ||||
| His12 (3.73) | |||||
| Lys64 (3.81) | |||||
| Asn58 (3.99) | |||||
| Tyr211 (4.04) | |||||
| Caffeic acid | Asn58 (3.0) | Ser15 (1.99) | Phe236 (5.38) | Lys64 (3.10) | |
| Tyr32 (2.26) | |||||
| Tyr32 (2.71) | −6.3 | ||||
| Asn58 (2.82) | |||||
| Asp100 (2.89) | |||||
| Ser243 (3.54) | |||||
| Complex | (VLJ)bound | (VLJ)free | (VCL)bound | (VCL)free | ∆Gbind |
|---|---|---|---|---|---|
| SaSDH-Phloridzin | −40.8 ± 2.60 | −13.9 ± 1.27 | −8.6 ± 0.74 | −12.8 ± 1.20 | −2.75 |
| SaSDH-Rutin | −55.7 ± 0.36 | −15.2 ± 0.31 | −9.1 ± 0.45 | −14.2 ± 0.36 | −4.4 |
| SaSDH-Caffeic acid | −26.3 ± 0.36 | −3.1 ± 0.13 | −3.4 ± 0.26 | −3.0 ± 0.31 | −4.8 |
| Parameters | Phloridzin | Rutin | Caffeic Acid |
|---|---|---|---|
| Physicochemical | |||
| Molecular weight ^ | 436.41 | 610.52 | 180.16 |
| Log P * | 0.055 | −1.25 | 0.78 |
| Log S * | −2.40 | −2.39 | −1.40 |
| H-bridge donors * | 7 | 10 | 3 |
| H-bridge acceptors * | 10 | 16 | 4 |
| Rotatable bonds ^ | 7 | 6 | 2 |
| ADME | |||
| Blood–brain barrier penetration ^ | No | No | No |
| Gastrointestinal absorption ^ | Low | Low | High |
| Plasma protein binding ° | Weak | Weak | Weak |
| Caco2 cell permeability ° | Moderate | Moderate | Moderate |
| Toxicological | |||
| Irritant * | No | No | No |
| Effects on reproduction * | Low | No | High |
| Tumorigenic * | No | No | High |
| Mutagenic * | No | No | High |
| CYP450 inhibition ° | 2C19, 2C9 and 3A4 | 2C19, 2C9 and 3A4 | 2C9 and 3A4 |
| Drug-Like | |||
| Drug-likeness Score * | −4.87 | 1.93 | 0.1675 |
| CMC-like rule ° | Compliant | Non-compliant | Compliant |
| Lead-like rule ° | Non-compliant | Non-compliant | Compliant |
| Ro5 ° | Adequate | Non-compliant | Adequate |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corral-Rodríguez, N.F.; Moreno-Contreras, V.I.; Sierra-Campos, E.; Valdez-Solana, M.; Cisneros-Martínez, J.; Téllez-Valencia, A.; Avitia-Domínguez, C. Characterization of Natural Products as Inhibitors of Shikimate Dehydrogenase from Methicillin-Resistant Staphylococcus aureus: Kinetic and Molecular Dynamics Simulations, and Biological Activity Studies. Biomolecules 2025, 15, 1137. https://doi.org/10.3390/biom15081137
Corral-Rodríguez NF, Moreno-Contreras VI, Sierra-Campos E, Valdez-Solana M, Cisneros-Martínez J, Téllez-Valencia A, Avitia-Domínguez C. Characterization of Natural Products as Inhibitors of Shikimate Dehydrogenase from Methicillin-Resistant Staphylococcus aureus: Kinetic and Molecular Dynamics Simulations, and Biological Activity Studies. Biomolecules. 2025; 15(8):1137. https://doi.org/10.3390/biom15081137
Chicago/Turabian StyleCorral-Rodríguez, Noé Fabián, Valeria Itzel Moreno-Contreras, Erick Sierra-Campos, Mónica Valdez-Solana, Jorge Cisneros-Martínez, Alfredo Téllez-Valencia, and Claudia Avitia-Domínguez. 2025. "Characterization of Natural Products as Inhibitors of Shikimate Dehydrogenase from Methicillin-Resistant Staphylococcus aureus: Kinetic and Molecular Dynamics Simulations, and Biological Activity Studies" Biomolecules 15, no. 8: 1137. https://doi.org/10.3390/biom15081137
APA StyleCorral-Rodríguez, N. F., Moreno-Contreras, V. I., Sierra-Campos, E., Valdez-Solana, M., Cisneros-Martínez, J., Téllez-Valencia, A., & Avitia-Domínguez, C. (2025). Characterization of Natural Products as Inhibitors of Shikimate Dehydrogenase from Methicillin-Resistant Staphylococcus aureus: Kinetic and Molecular Dynamics Simulations, and Biological Activity Studies. Biomolecules, 15(8), 1137. https://doi.org/10.3390/biom15081137







