Research Progress on the Functional Regulation Mechanisms of ZKSCAN3
Abstract
1. Introduction
2. Gene Structure and Transcript Variants of ZKSCAN3
3. ZKSCAN3 Exerts Multifunctional Regulatory Mechanisms in Normal Physiological Processes
4. The Function and Regulatory Mechanisms of ZKSCAN3 in Tumors
4.1. ZKSCAN3 Drives Colorectal Cancer Progression by Activating Wnt/β-Catenin and ITGβ4/FAK/AKT Pathways
4.2. ZKSCAN3 Promotes Hepatocellular Carcinoma Progression via the FAK/AKT-Autophagy Inhibition Axis
4.3. ZKSCAN3 Facilitates Gastric Cancer Progression Through Ras/MAPK-MST1R-MMP/VEGF Multi-Axis Signaling
4.4. ZKSCAN3 Accelerates Breast Cancer Proliferation and Invasion via AKT/mTOR-Cyclin D1-MMP Axis
4.5. ZKSCAN3 Drives Cervical Cancer Proliferation and Metastasis Through RAS-MAPK/MST1R/VEGF Pathway
4.6. ZKSCAN3 Promotes Prostate Cancer Proliferation and Metastasis via VEGF/ITGβ4-Cyclin D-NFκB-MMP Axis
4.7. ZKSCAN3 Inhibits Malignant Progression of Pancreatic Cancer by Targeting ULK1/LC3-II Autophagy Axis
4.8. ZKSCAN3 Activates Bladder Cancer Invasion and Proliferation via c-Myc/FGFR3-MMP2/9 Signaling Axis
4.9. ZKSCAN3 Synergizes with EGFR to Activate Pro-Survival, Anti-Apoptotic, and Tumor Microenvironment Remodeling Pathways in Ovarian Cancer
4.10. ZKSCAN3 Enhances Cell Cycle Progression and Angiogenesis in Multiple Myeloma by Regulating CCND2 and VEGF
5. The Pathological Regulatory Role and Therapeutic Potential of ZKSCAN3 in Multiple Diseases
6. ZKSCAN3 Bridges Cancer, Neurodegenerative Disorders, and Metabolic Diseases Through Autophagic Regulation
7. Conclusion and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| KRAB | Krüppel-associated box |
| KRAB-ZFPs | KRAB zinc finger proteins |
| H3K9me3 | H3K9 trimethylation |
| TFIIIA | Transcription factor IIIA |
| BRCA1 | Breast cancer susceptibility gene 1 |
| LC3b | Microtubule-associated protein 1A/1B-light chain 3B |
| ITGβ4 | Integrin β4 |
| VEGF | Vascular endothelial growth factor |
| CCND2 | Cyclin D2 |
| hMSCs | Human mesenchymal stem cells |
| LADs | Lamina-associated domains |
| hESCs | Human embryonic stem cells |
| TAC | Transverse aortic constriction |
| REP | Retinal pigment epithelial |
| M1BP | Motif 1-binding protein |
| Wg | Wingless |
| Pol II | Polymerase II |
| Xist | X-inactive-specific transcript |
| AD | Alzheimer’s disease |
| PD | Parkinson’s disease |
| AP | Acute pancreatitis |
| CHIT1 | Chitosanase 1 |
| CGRP | Calcitonin gene-related peptide |
| SIRT1 | Sirtuin 1 |
| TFEB | Transcription factor EB |
| LRRK2 | Leucine repeat kinase 2 |
| T2DM | Type 2 diabetes mellitus |
| AS | Atherosclerosis |
| CEL | Nε-carboxyethyl lysine |
| SBMA | Spinal and bulbar muscular atrophy |
| ALKBH5 | Alkb homolog 5 |
| MEK2 | Mitogen-activated protein kinase kinase 2 |
| RasGRP2 | Ras guanyl releasing protein 2 |
| IGF-2 | Insulin-like growth factor 2 |
| ALI | Acute lung injury |
| LPS | Lipopolysaccharide |
| COPD | Chronic obstructive pulmonary disease |
| LC3-II | Microtubule-associated protein 1A/1B-light chain 3-II |
| LAMP1 | Lysosome-associated membrane protein 1 |
| TFE3 | Transcription factor e3 |
| FAK | Focal adhesion kinase |
| EMT | Epithelial–mesenchymal transition |
| Ct-OATP1B3 | Cancer-type OATP1B3 |
| MMPs | Matrix metalloproteinases |
| FA | Focal adhesion |
| OS | Overall survival |
| shRNA | Short hairpin RNA |
| CCND1 | Cyclin D1 |
| BCL-2 | B-cell lymphoma/leukemia 2 |
| BAX | BCL-2 associated X protein |
| PFS | Progression-free survival |
| MMP26 | Matrix metalloproteinase 26 |
| PRSS3 | Serine protease 3 |
| EGFR | Epidermal growth factor receptor |
| cryo-EM | Cryo-electron microscopy |
References
- Li, X.M.; Wen, J.H.; Feng, Z.S.; Wu, Y.S.; Li, D.Y.; Liang, S.; Wu, D.; Wu, H.L.; Li, S.M.; Ye, Z.N.; et al. Effect of Lacking ZKSCAN3 on Autophagy, Lysosomal Biogenesis and Senescence. Int. J. Mol. Sci. 2023, 24, 7786. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.; McLachlan, A.D.; Klug, A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985, 4, 1609–1614. [Google Scholar] [CrossRef] [PubMed]
- Hoovers, J.M.; Mannens, M.; John, R.; Bliek, J.; van Heyningen, V.; Porteous, D.J.; Leschot, N.J.; Westerveld, A.; Little, P.F. High-resolution localization of 69 potential human zinc finger protein genes: A number are clustered. Genomics 1992, 12, 254–263. [Google Scholar] [CrossRef]
- Li, W.; Huang, C.; Qiu, L.; Tang, Y.; Zhang, X.; Zhang, L.; Zhao, H.; Miyagishi, M.; Kasim, V.; Wu, S. p52-ZER6/IGF1R axis maintains cancer stem cell population to promote cancer progression by enhancing pro-survival mitophagy. Oncogene 2024, 43, 2115–2131. [Google Scholar] [CrossRef]
- Li, W.; Zhang, H.; Xu, J.; Maimaitijiang, A.; Su, Z.; Fan, Z.; Li, J. The Biological Roles of ZKSCAN3 (ZNF306) in the Hallmarks of Cancer: From Mechanisms to Therapeutics. Int. J. Mol. Sci. 2024, 25, 11532. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, S.; Goodwin, J.G.; Chauhan, S.; Manyam, G.; Wang, J.; Kamat, A.M.; Boyd, D.D. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol. Cell 2013, 50, 16–28. [Google Scholar] [CrossRef]
- Sun, M.; Ju, J.; Ding, Y.; Zhao, C.; Tian, C. The signaling pathways regulated by KRAB zinc-finger proteins in cancer. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188731. [Google Scholar] [CrossRef] [PubMed]
- Rosspopoff, O.; Trono, D. Take a walk on the KRAB side. Trends Genet. 2023, 39, 844–857, Erratum in Trends Genet. 2024, 40, 203–205. [Google Scholar] [CrossRef]
- Sobocińska, J.; Molenda, S.; Machnik, M.; Oleksiewicz, U. KRAB-ZFP Transcriptional Regulators Acting as Oncogenes and Tumor Suppressors: An Overview. Int. J. Mol. Sci. 2021, 22, 2212. [Google Scholar] [CrossRef]
- Huang, M.; Chen, Y.; Han, D.; Lei, Z.; Chu, X. Role of the zinc finger and SCAN domain-containing transcription factors in cancer. Am. J. Cancer Res. 2019, 9, 816–836. [Google Scholar]
- Ecco, G.; Imbeault, M.; Trono, D. KRAB zinc finger proteins. Development 2017, 144, 2719–2729. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.L.; Zhang, M.L.; Lin, H.P.; Gao, C.; Song, J.B.; Zheng, Z.; Li, L.; Zhang, Y.; Shen, X.; Zhang, H.; et al. The Zscan4-Tet2 Transcription Nexus Regulates Metabolic Rewiring and Enhances Proteostasis to Promote Reprogramming. Cell Rep. 2020, 32, 107877. [Google Scholar] [CrossRef]
- Liang, Y.; Choo, S.H.; Rossbach, M.; Baburajendran, N.; Palasingam, P.; Kolatkar, P.R. Crystal optimization and preliminary diffraction data analysis of the SCAN domain of Zfp206. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Groner, A.C.; Meylan, S.; Ciuffi, A.; Zangger, N.; Ambrosini, G.; Dénervaud, N.; Bucher, P.; Trono, D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010, 6, e1000869. [Google Scholar] [CrossRef]
- Saftig, P.; Puertollano, R. How Lysosomes Sense, Integrate, and Cope with Stress. Trends Biochem. Sci. 2021, 46, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Klionsky, D.J. Transcriptional regulation of autophagy and its implications in human disease. Cell Death Differ. 2023, 30, 1416–1429. [Google Scholar] [CrossRef]
- Füllgrabe, J.; Klionsky, D.J.; Joseph, B. The return of the nucleus: Transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 2014, 15, 65–74. [Google Scholar] [CrossRef]
- Li, S.; Song, Y.; Quach, C.; Guo, H.; Jang, G.B.; Maazi, H.; Zhao, S.; Sands, N.A.; Liu, Q.; In, G.K.; et al. Transcriptional regulation of autophagy-lysosomal function in BRAF-driven melanoma progression and chemoresistance. Nat. Commun. 2019, 10, 1693. [Google Scholar] [CrossRef]
- Yang, L.; Hamilton, S.R.; Sood, A.; Kuwai, T.; Ellis, L.; Sanguino, A.; Lopez-Berestein, G.; Boyd, D.D. The previously undescribed ZKSCAN3 (ZNF306) is a novel “driver” of colorectal cancer progression. Cancer Res. 2008, 68, 4321–4330. [Google Scholar] [CrossRef]
- Urrutia, R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003, 4, 231. [Google Scholar] [CrossRef]
- Wang, W.; Cai, J.; Wu, Y.; Hu, L.; Chen, Z.; Hu, J.; Chen, Z.; Li, W.; Guo, M.; Huang, Z. Novel activity of KRAB domain that functions to reinforce nuclear localization of KRAB-containing zinc finger proteins by interacting with KAP1. Cell Mol. Life Sci. 2013, 70, 3947–3958. [Google Scholar] [CrossRef] [PubMed]
- Helleboid, P.Y.; Heusel, M.; Duc, J.; Piot, C.; Thorball, C.W.; Coluccio, A.; Pontis, J.; Imbeault, M.; Turelli, P.; Aebersold, R.; et al. The interactome of KRAB zinc finger proteins reveals the evolutionary history of their functional diversification. EMBO J. 2019, 38, e101220. [Google Scholar] [CrossRef]
- Allen, I.; Hassan, H.; Walburga, Y.; Huntley, C.; Loong, L.; Rahman, T.; Allen, S.; Garrett, A.; Torr, B.; Bacon, A.; et al. Second Primary Cancer Risks After Breast Cancer in BRCA1 and BRCA2 Pathogenic Variant Carriers. J. Clin. Oncol. 2025, 43, 651–661. [Google Scholar] [CrossRef]
- Khalizieva, A.; Moser, S.C.; Bouwman, P.; Jonkers, J. BRCA1 and BRCA2: From cancer susceptibility to synthetic lethality. Genes. Dev. 2025, 39, 86–108. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Lemacon, D.S.; Li, S.; Cheruiyot, A.; Kong, L.; Tan, K.; Cheng, C.; Turkay, E.; He, D.; You, Z. Context-dependent pro- and anti-resection roles of ZKSCAN3 in the regulation of fork processing during replication stress. J. Biol. Chem. 2022, 298, 102215. [Google Scholar] [CrossRef]
- Moore, D.; Wong, E.; Arnal, C.; Schoenfelder, S.; Spivakov, M.; Andrews, S.; Christophorou, M.A. The KRAB-Zinc Finger protein ZKSCAN3 represses enhancers via embedded retrotransposons. bioRxiv 2025. [Google Scholar] [CrossRef]
- Huang, C.; Wu, S.; Li, W.; Herkilini, A.; Miyagishi, M.; Zhao, H.; Kasim, V. Zinc-finger protein p52-ZER6 accelerates colorectal cancer cell proliferation and tumour progression through promoting p53 ubiquitination. EBioMedicine 2019, 48, 248–263. [Google Scholar] [CrossRef]
- Qiu, L.; Li, W.; Zhang, L.; Zhang, X.; Zhao, H.; Miyagishi, M.; Wu, S.; Kasim, V. p52-ZER6/DAZAP1 axis promotes ferroptosis resistance and colorectal cancer progression via regulating SLC7A11 mRNA stabilization. Acta Pharm. Sin. B 2025, 15, 2039–2058. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Diao, Z.; Ji, Q.; Wu, Z.; Zhang, W.; Cai, Y.; Wang, Z.; Hu, J.; Liu, Z.; Wang, Q.; Bi, S.; et al. SIRT3 consolidates heterochromatin and counteracts senescence. Nucleic Acids Res. 2021, 49, 4203–4219. [Google Scholar] [CrossRef]
- Hu, H.; Ji, Q.; Song, M.; Ren, J.; Liu, Z.; Wang, Z.; Liu, X.; Yan, K.; Hu, J.; Jing, Y.; et al. ZKSCAN3 counteracts cellular senescence by stabilizing heterochromatin. Nucleic Acids Res. 2020, 48, 6001–6018. [Google Scholar] [CrossRef] [PubMed]
- Hosea, R.; Duan, W.; Meliala, I.T.S.; Li, W.; Wei, M.; Hillary, S.; Zhao, H.; Miyagishi, M.; Wu, S.; Kasim, V. YY2/BUB3 Axis promotes SAC hyperactivation and inhibits colorectal cancer progression via regulating chromosomal instability. Adv. Sci. 2024, 11, 2308690. [Google Scholar] [CrossRef]
- Wu, Z.; Qu, J.; Liu, G.H. Roles of chromatin and genome instability in cellular senescence and their relevance to ageing and related diseases. Nat. Rev. Mol. Cell Biol. 2024, 25, 979–1000. [Google Scholar] [CrossRef] [PubMed]
- Dhankhar, M.; Guo, Z.; Kant, A.; Basir, R.; Joshi, R.; Heo, S.C.; Mauck, R.L.; Lakadamyali, M.; Shenoy, V.B. Revealing the Biophysics of Lamina-Associated Domain Formation by Integrating Theoretical Modeling and High-Resolution Imaging. bioRxiv 2024. [Google Scholar] [CrossRef]
- Li, Z.; Sheng, B.; Zhang, T.; Wang, T.; Chen, D.; An, G.; Wang, X.; Meng, H.; Yang, L. Zkscan3 affects erythroblast development by regulating the transcriptional activity of GATA1 and KLF1 in mice. J. Mol. Histol. 2022, 53, 423–436. [Google Scholar] [CrossRef]
- Gutiérrez, L.; Caballero, N.; Fernández-Calleja, L.; Karkoulia, E.; Strouboulis, J. Regulation of GATA1 levels in erythropoiesis. IUBMB Life 2020, 72, 89–105. [Google Scholar] [CrossRef]
- Mansoor, A.; Mansoor, M.O.; Patel, J.L.; Zhao, S.; Natkunam, Y.; Bieker, J.J. KLF1/EKLF expression in acute leukemia is correlated with chromosomal abnormalities. Blood Cells Mol. Dis. 2020, 83, 102434. [Google Scholar] [CrossRef]
- Zhu, G.; Fan, Z.; Ding, M.; Mu, L.; Liang, J.; Ding, Y.; Fu, Y.; Huang, B.; Wu, W. DNA damage induces the accumulation of Tiam1 by blocking β-TrCP-dependent degradation. J. Biol. Chem. 2014, 289, 15482–15494. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, J.; Long, Y.; Maimaitijiang, A.; Su, Z.; Li, W.; Li, J. Unraveling the Guardian: p53’s Multifaceted Role in the DNA Damage Response and Tumor Treatment Strategies. Int. J. Mol. Sci. 2024, 25, 12928. [Google Scholar] [CrossRef]
- Wu, X.; Liu, Z.; Yu, X.Y.; Xu, S.; Luo, J. Autophagy and cardiac diseases: Therapeutic potential of natural products. Med. Res. Rev. 2021, 41, 314–341. [Google Scholar] [CrossRef]
- Bosch, L.; de Haan, J.J.; Bastemeijer, M.; van der Burg, J.; van der Worp, E.; Wesseling, M.; Viola, M.; Odille, C.; El Azzouzi, H.; Pasterkamp, G.; et al. The transverse aortic constriction heart failure animal model: A systematic review and meta-analysis. Heart Fail. Rev. 2021, 26, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, X.; Bakshi, S.; Benavides, G.A.; Sun, Z.; Hernandez-Moreno, G.; Collins, H.E.; Kane, M.S.; Litovsky, S.; Young, M.E.; Chatham, J.C.; et al. Cardiomyocyte ZKSCAN3 regulates remodeling following pressure-overload. Physiol. Rep. 2023, 11, e15686. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.Y.; Valapala, M. Role of the Transcriptional Repressor Zinc Finger with KRAB and SCAN Domains 3 (ZKSCAN3) in Retinal Pigment Epithelial Cells. Cells 2021, 10, 2504. [Google Scholar] [CrossRef]
- Poliacikova, G.; Barthez, M.; Rival, T.; Aouane, A.; Luis, N.M.; Richard, F.; Daian, F.; Brouilly, N.; Schnorrer, F.; Maurel-Zaffran, C.; et al. M1BP is an essential transcriptional activator of oxidative metabolism during Drosophila development. Nat. Commun. 2023, 14, 3187. [Google Scholar] [CrossRef] [PubMed]
- Chimata, A.V.; Darnell, H.; Raj, A.; Kango-Singh, M.; Singh, A. Transcriptional pausing factor M1BP regulates cellular homeostasis by suppressing autophagy and apoptosis in Drosophila eye. Autophagy Rep. 2023, 2, 2252307. [Google Scholar] [CrossRef]
- Barthez, M.; Poplineau, M.; Elrefaey, M.; Caruso, N.; Graba, Y.; Saurin, A.J. Human ZKSCAN3 and Drosophila M1BP are functionally homologous transcription factors in autophagy regulation. Sci. Rep. 2020, 10, 9653. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, W.; Li, X.; Lue, Z.; Liu, Y.; Wu, J.; Zhang, X. The autophagic regulation of rosiglitazone-promoted adipocyte browning. Front. Pharmacol. 2024, 15, 1412520. [Google Scholar] [CrossRef]
- Alharbi, A.B.; Schmitz, U.; Bailey, C.G.; Rasko, J.E.J. CTCF as a regulator of alternative splicing: New tricks for an old player. Nucleic Acids Res. 2021, 49, 7825–7838. [Google Scholar] [CrossRef]
- Charrier, A.; Ockunzzi, J.; Main, L.; Ghanta, S.V.; Buchner, D.A. Molecular regulation of PPARγ/RXRα signaling by the novel cofactor ZFP407. PLoS ONE 2024, 19, e0294003. [Google Scholar] [CrossRef]
- Li, Z.; Wen, C.; Li, J.; Meng, H.; Ji, C.; Han, Z.; An, G.; Yang, L. Zkscan3 gene is a potential negative regulator of plasma cell differentiation. Eur. J. Inflamm. 2019, 17, 2058739219850008. [Google Scholar] [CrossRef]
- Jen, J.; Wang, Y.C. Zinc finger proteins in cancer progression. J. Biomed. Sci. 2016, 23, 53. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, L.; Wu, Q.; Boyd, D.D. Unbiased Screening for Transcriptional Targets of ZKSCAN3 Identifies Integrin β4 and Vascular Endothelial Growth Factor as Downstream Targets. J. Biol. Chem. 2008, 283, 35295–35304. [Google Scholar] [CrossRef]
- Qian, Z.; Chang, T.; Zhang, T.; Wang, J.; Gu, H. Genomic analyses identify biological processes in ZKSCAN3-deficient colorectal cancer cells. bioRxiv 2022. [Google Scholar] [CrossRef]
- Cho, Y.E.; Kim, J.H.; Che, Y.H.; Kim, Y.J.; Sung, J.Y.; Kim, Y.W.; Choe, B.G.; Lee, S.; Park, J.H. Role of the WNT/β-catenin/ZKSCAN3 Pathway in Regulating Chromosomal Instability in Colon Cancer Cell lines and Tissues. Int. J. Mol. Sci. 2022, 23, 9302. [Google Scholar] [CrossRef]
- Tang, W.; Hu, Y.; Tu, K.; Gong, Z.; Zhu, M.; Yang, T.; Sarwar, A.; Dai, B.; Zhang, D.; Zhan, Y.; et al. Targeting Trop2 by Bruceine D suppresses breast cancer metastasis by blocking Trop2/β-catenin positive feedback loop. J. Adv. Res. 2024, 58, 193–210. [Google Scholar] [CrossRef]
- Roh, S.A.; Kim, C.W.; Tak, K.H.; Kim, J.C. Zkscan3 facilitates invasion of colorectal cancer associated with ceacam5. Cancer Res. 2014, 74, 4992. [Google Scholar] [CrossRef]
- Kim, C.W.; Roh, S.A.; Tak, K.H.; Koh, B.M.; Ha, Y.J.; Cho, D.H.; Kim, S.Y.; Kim, Y.S.; Kim, J.C. ZKSCAN3 Facilitates Liver Metastasis of Colorectal Cancer Associated with CEA-expressing Tumor. Anticancer. Res. 2016, 36, 2397–2406. [Google Scholar]
- Li, J.; Hao, N.; Han, J.; Zhang, M.; Li, X.; Yang, N. ZKSCAN3 drives tumor metastasis via integrin β4/FAK/AKT mediated epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Cell Int. 2020, 20, 216. [Google Scholar] [CrossRef]
- Sun, Y.; Piñón Hofbauer, J.; Harada, M.; Wöss, K.; Koller, U.; Morio, H.; Stierschneider, A.; Kitamura, K.; Hashimoto, M.; Chiba, K.; et al. Cancer-type organic anion transporting polypeptide 1B3 is a target for cancer suicide gene therapy using RNA trans-splicing technology. Cancer Lett. 2018, 433, 107–116. [Google Scholar] [CrossRef]
- Haberkorn, B.; Löwen, D.; Meier, L.; Fromm, M.F.; König, J. Transcriptional Regulation of Liver-Type OATP1B3 (Lt-OATP1B3) and Cancer-Type OATP1B3 (Ct-OATP1B3) Studied in Hepatocyte-Derived and Colon Cancer-Derived Cell Lines. Pharmaceutics 2023, 15, 738. [Google Scholar] [CrossRef] [PubMed]
- Li, W.F.; Alfason, L.; Huang, C.; Tang, Y.; Qiu, L.; Miyagishi, M.; Wu, S.R.; Kasim, V. p52-ZER6: A determinant of tumor cell sensitivity to MDM2-p53 binding inhibitors. Acta Pharmacol. Sin. 2023, 44, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Nonoyama, K.; Matsuo, Y.; Sugita, S.; Eguchi, Y.; Denda, Y.; Murase, H.; Kato, T.; Imafuji, H.; Saito, K.; Morimoto, M.; et al. Expression of ZKSCAN3 protein suppresses proliferation, migration, and invasion of pancreatic cancer through autophagy. Cancer Sci. 2024, 115, 1964–1978. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, H.; Kornblau, S.M.; Graber, D.A.; Zhang, N.; Matthews, J.A.; Wang, M.; Weber, D.M.; Thomas, S.K.; Shah, J.J.; et al. Evidence of a role for the novel zinc-finger transcription factor ZKSCAN3 in modulating Cyclin D2 expression in multiple myeloma. Oncogene 2011, 30, 1329–1340. [Google Scholar] [CrossRef]
- Braga, E.A.; Fridman, M.V.; Burdennyy, A.M.; Filippova, E.A.; Loginov, V.I.; Pronina, I.V.; Dmitriev, A.A.; Kushlinskii, N.E. Regulation of the Key Epithelial Cancer Suppressor miR-124 Function by Competing Endogenous RNAs. Int. J. Mol. Sci. 2022, 23, 13620. [Google Scholar] [CrossRef]
- Liu, S.; Yao, S.; Yang, H.; Liu, S.; Wang, Y. Autophagy: Regulator of cell death. Cell Death Dis. 2023, 14, 648. [Google Scholar] [CrossRef]
- Adhikari, M.; Adhikari, B.; Ghimire, B.; Baboota, S.; Choi, E.H. Cold Atmospheric Plasma and Silymarin Nanoemulsion Activate Autophagy in Human Melanoma Cells. Int. J. Mol. Sci. 2020, 21, 1939. [Google Scholar] [CrossRef]
- Zhang, X.; Bai, Y.; Huang, L.; Liu, S.; Mo, Y.; Cheng, W.; Wang, G.; Cao, Z.; Chen, X.; Cui, H.; et al. CHD1L augments autophagy-mediated migration of hepatocellular carcinoma through targeting ZKSCAN3. Cell Death Dis. 2021, 12, 950. [Google Scholar] [CrossRef]
- Zhang, T.; Zhu, S.; Huang, G.W. ALKBH5 suppresses autophagic flux via N6-methyladenosine demethylation of ZKSCAN3 mRNA in acute pancreatitis. World J. Gastroenterol. 2024, 30, 1764–1776. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, Y.; Jiang, M.; Tang, Y.; Wang, Q.; Bai, L.; Yu, C.; Yang, X.; Ding, K.; Wang, W.; et al. The demethylase ALKBH5 mediates ZKSCAN3 expression through the m(6)A modification to activate VEGFA transcription and thus participates in MNNG-induced gastric cancer progression. J. Hazard. Mater. 2024, 473, 134690. [Google Scholar] [CrossRef]
- Zhang, L.; Li, C.; Song, X.; Guo, R.; Zhao, W.; Liu, C.; Chen, X.; Song, Q.; Wu, B.; Deng, N. Targeting ONECUT2 inhibits tumor angiogenesis via down-regulating ZKSCAN3/VEGFA. Biochem. Pharmacol. 2024, 225, 116315. [Google Scholar] [CrossRef] [PubMed]
- Takano, Y.; Shida, A.; Fujisaki, M.; Mitsumori, N.; Yanaga, K. Prognostic Significance of ZKSCAN3 (ZNF306) Expression in Gastric Carcinoma. Anticancer. Res. 2020, 40, 81–86. [Google Scholar] [CrossRef]
- Chi, Y.; Xu, H.; Wang, F.; Chen, X.; Shan, Z.; Sun, Y.; Fan, Q. ZKSCAN3 promotes breast cancer cell proliferation, migration and invasion. Biochem. Biophys. Res. Commun. 2018, 503, 2583–2589. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martinez, L.; Zhang, Y.; Nakata, Y.; Chan, H.L.; Morey, L. Epigenetic mechanisms in breast cancer therapy and resistance. Nat. Commun. 2021, 12, 1786. [Google Scholar] [CrossRef]
- Zhang, X.; Jing, Y.; Qin, Y.; Hunsucker, S.; Meng, H.; Sui, J.; Jiang, Y.; Gao, L.; An, G.; Yang, N.; et al. The zinc finger transcription factor ZKSCAN3 promotes prostate cancer cell migration. Int. J. Biochem. Cell Biol. 2012, 44, 1166–1173. [Google Scholar] [CrossRef] [PubMed]
- Glaviano, A.; Foo, A.S.C.; Lam, H.Y.; Yap, K.C.H.; Jacot, W.; Jones, R.H.; Eng, H.; Nair, M.G.; Makvandi, P.; Geoerger, B.; et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol. Cancer 2023, 22, 138. [Google Scholar] [CrossRef]
- Lee, S.; Cho, Y.E.; Kim, J.Y.; Park, J.H. ZKSCAN3 Upregulation and Its Poor Clinical Outcome in Uterine Cervical Cancer. Int. J. Mol. Sci. 2018, 19, 2859. [Google Scholar] [CrossRef]
- Mao, Y.; Jiang, X.; Guo, P.; Ouyang, Y.; Chen, X.; Xia, M.; Wu, L.; Tang, Z.; Liang, T.; Li, Y.; et al. ZXDC enhances cervical cancer metastasis through IGF2BP3-mediated activation of RhoA/ROCK signaling. iScience 2023, 26, 107447. [Google Scholar] [CrossRef]
- Kawahara, T.; Teramoto, Y.; Li, Y.; Ishiguro, H.; Gordetsky, J.; Yang, Z.; Miyamoto, H. Impact of Vasectomy on the Development and Progression of Prostate Cancer: Preclinical Evidence. Cancers 2020, 12, 2295. [Google Scholar] [CrossRef]
- Husby, A.; Wohlfahrt, J.; Melbye, M. Vasectomy and Prostate Cancer Risk: A 38-Year Nationwide Cohort Study. J. Natl. Cancer Inst. 2020, 112, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, E.J.; Anderson, R.L.; Stevens, V.L.; Newton, C.C.; Gansler, T.; Gapstur, S.M. Vasectomy and Prostate Cancer Incidence and Mortality in a Large US Cohort. J. Clin. Oncol. 2016, 34, 3880–3885. [Google Scholar] [CrossRef] [PubMed]
- Mucci, L.A.; Wilson, K.M.; Preston, M.A.; Giovannucci, E.L. Is Vasectomy a Cause of Prostate Cancer? J. Natl. Cancer Inst. 2020, 112, 5–6. [Google Scholar] [CrossRef]
- Kakavas, S.; Kotsiou, O.S.; Perlikos, F.; Mermiri, M.; Mavrovounis, G.; Gourgoulianis, K.; Pantazopoulos, I. Pulmonary function testing in COPD: Looking beyond the curtain of FEV1. NPJ Prim. Care Respir. Med. 2021, 31, 23. [Google Scholar] [CrossRef] [PubMed]
- Soler Artigas, M.; Loth, D.W.; Wain, L.V.; Gharib, S.A.; Obeidat, M.; Tang, W.; Zhai, G.; Zhao, J.H.; Smith, A.V.; Huffman, J.E.; et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat. Genet. 2011, 43, 1082–1090. [Google Scholar] [CrossRef]
- Cho, Y.E.; Kim, Y.J.; Lee, S.; Park, J.H. NOP53 Suppresses Autophagy through ZKSCAN3-Dependent and -Independent Pathways. Int. J. Mol. Sci. 2021, 22, 9318. [Google Scholar] [CrossRef]
- Du, Q.; Zhang, M.; Gao, A.; He, T.; Guo, M. Epigenetic silencing ZSCAN23 promotes pancreatic cancer growth by activating Wnt signaling. Cancer Biol. Ther. 2024, 25, 2302924. [Google Scholar] [CrossRef]
- Menon, S.S.; Guruvayoorappan, C.; Sakthivel, K.M.; Rasmi, R.R. Ki-67 protein as a tumour proliferation marker. Clin. Chim. Acta 2019, 491, 39–45. [Google Scholar] [CrossRef]
- Kawahara, T.; Inoue, S.; Ide, H.; Kashiwagi, E.; Ohtake, S.; Mizushima, T.; Li, P.; Li, Y.; Zheng, Y.; Uemura, H.; et al. ZKSCAN3 promotes bladder cancer cell proliferation, migration, and invasion. Oncotarget 2016, 7, 53599–53610. [Google Scholar] [CrossRef]
- Alfason, L.; Li, W.; Altaf, F.; Wu, S.; Kasim, V. Resuscitating the Guardian: Current Progress in p53-Based Anti-Tumor Therapy. Onco Ther. 2021, 8, 51–92. [Google Scholar] [CrossRef]
- Li, W.F.; Herkilini, A.; Tang, Y.; Huang, P.; Song, G.B.; Miyagishi, M.; Kasim, V.; Wu, S.R. The transcription factor PBX3 promotes tumor cell growth through transcriptional suppression of the tumor suppressor p53. Acta Pharmacol. Sin. 2021, 42, 1888–1899. [Google Scholar] [CrossRef]
- Rajaram, P.; Chandra, P.; Ticku, S.; Pallavi, B.K.; Rudresh, K.B.; Mansabdar, P. Epidermal growth factor receptor: Role in human cancer. Indian. J. Dent. Res. 2017, 28, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Voldborg, B.R.; Damstrup, L.; Spang-Thomsen, M.; Poulsen, H.S. Epidermal growth factor receptor (EGFR) and EGFR mutations, function and possible role in clinical trials. Ann. Oncol. 1997, 8, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- De Luca, A.; Carotenuto, A.; Rachiglio, A.; Gallo, M.; Maiello, M.R.; Aldinucci, D.; Pinto, A.; Normanno, N. The role of the EGFR signaling in tumor microenvironment. J. Cell Physiol. 2008, 214, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Corona, R.I.; Seo, J.-H.; Lin, X.; Hazelett, D.J.; Reddy, J.; Fonseca, M.A.; Abassi, F.; Lin, Y.G.; Mhawech-Fauceglia, P.Y.; Shah, S.P. Non-coding somatic mutations converge on the PAX8 pathway in ovarian cancer. Nat. Commun. 2020, 11, 2020. [Google Scholar] [CrossRef]
- Yang, L.; Orlowski, R.Z. ZKSCAN3, a Novel Zinc Finger Transcription Factor, Regulates Cyclin D2 Expression in Multiple Myeloma (MM) Cells. Blood 2008, 112, 749. [Google Scholar] [CrossRef]
- Egan, J.B.; Shi, C.X.; Tembe, W.; Christoforides, A.; Kurdoglu, A.; Sinari, S.; Middha, S.; Asmann, Y.; Schmidt, J.; Braggio, E.; et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood 2012, 120, 1060–1066. [Google Scholar] [CrossRef]
- Montero-Vergara, J.; Plachetta, K.; Kinch, L.; Bernhardt, S.; Kashyap, K.; Levine, B.; Thukral, L.; Vetter, M.; Thomssen, C.; Wiemann, S.; et al. GRB2 is a BECN1 interacting protein that regulates autophagy. Cell Death Dis. 2024, 15, 14. [Google Scholar] [CrossRef]
- Zhang, J.; Zeng, W.; Han, Y.; Lee, W.R.; Liou, J.; Jiang, Y. Lysosomal LAMP proteins regulate lysosomal pH by direct inhibition of the TMEM175 channel. Mol. Cell 2023, 83, 2524–2539.e2527. [Google Scholar] [CrossRef]
- Bu, S.; Lv, Y.; Liu, Y.; Qiao, S.; Wang, H. Zinc Finger Proteins in Neuro-Related Diseases Progression. Front. Neurosci. 2021, 15, 760567. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yu, W.; Lv, Y. Neuroprotective effects of chitinase-1 and calcitonin gene-related peptide on Alzheimer’s disease by promoting lysosomal function. J. Alzheimers Dis. 2025, 103, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Lafferty, T.K.; Sehrawat, A.; Chen, Y.; Ferreira, P.C.L.; Bellaver, B.; Povala, G.; Kamboh, M.I.; Klunk, W.E.; Cohen, A.D.; et al. Multi-analyte proteomic analysis identifies blood-based neuroinflammation, cerebrovascular and synaptic biomarkers in preclinical Alzheimer’s disease. Mol. Neurodegener. 2024, 19, 68. [Google Scholar] [CrossRef] [PubMed]
- de Vries, T.; Villalón, C.M.; MaassenVanDenBrink, A. Pharmacological treatment of migraine: CGRP and 5-HT beyond the triptans. Pharmacol. Ther. 2020, 211, 107528. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; Wang, J.; Xia, Y.; Zhang, J.; Chen, L. Recent advances in Alzheimer’s disease: Mechanisms, clinical trials and new drug development strategies. Signal Transduct. Target. Ther. 2024, 9, 211. [Google Scholar] [CrossRef]
- Chang, N.; Li, J.; Lin, S.; Zhang, J.; Zeng, W.; Ma, G.; Wang, Y. Emerging roles of SIRT1 activator, SRT2104, in disease treatment. Sci. Rep. 2024, 14, 5521. [Google Scholar] [CrossRef]
- Wu, X.; Ren, Y.; Wen, Y.; Lu, S.; Li, H.; Yu, H.; Li, W.; Zou, F. Deacetylation of ZKSCAN3 by SIRT1 induces autophagy and protects SN4741 cells against MPP(+)-induced oxidative stress. Free Radic. Biol. Med. 2022, 181, 82–97. [Google Scholar] [CrossRef]
- Baeken, M.W. Sirtuins and their influence on autophagy. J. Cell. Biochem. 2024, 125, e30377. [Google Scholar] [CrossRef]
- Mueller, T.; Jeffrey, P.; He, Y.; Ouyang, X.; Westbrook, D.; Darley-Usmar, V.; Goldberg, M.S.; Volpicelli-Daley, L.; Zhang, J. Alpha-synuclein preformed fibril-induced aggregation and dopaminergic cell death in cathepsin D overexpression and ZKSCAN3 knockout mice. bioRxiv 2024. [Google Scholar] [CrossRef]
- Zhao, Q.; Gao, S.M.; Wang, M.C. Molecular Mechanisms of Lysosome and Nucleus Communication. Trends Biochem. Sci. 2020, 45, 978–991. [Google Scholar] [CrossRef]
- Rusmini, P.; Cortese, K.; Crippa, V.; Cristofani, R.; Cicardi, M.E.; Ferrari, V.; Vezzoli, G.; Tedesco, B.; Meroni, M.; Messi, E.; et al. Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration. Autophagy 2019, 15, 631–651. [Google Scholar] [CrossRef] [PubMed]
- Chua, J.P.; Reddy, S.L.; Merry, D.E.; Adachi, H.; Katsuno, M.; Sobue, G.; Robins, D.M.; Lieberman, A.P. Transcriptional activation of TFEB/ZKSCAN3 target genes underlies enhanced autophagy in spinobulbar muscular atrophy. Hum. Mol. Genet. 2014, 23, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Lin, S.; Sun, B.; Chen, W.; Liu, J.; Chen, M. ZKSCAN3 affects the autophagy-lysosome pathway through TFEB in Parkinson’s disease. Biomed. Rep. 2025, 22, 74. [Google Scholar] [CrossRef]
- Sosero, Y.L.; Gan-Or, Z. LRRK2 and Parkinson’s disease: From genetics to targeted therapy. Ann. Clin. Transl. Neurol. 2023, 10, 850–864. [Google Scholar] [CrossRef]
- Nabar, N.R.; Heijjer, C.N.; Shi, C.S.; Hwang, I.Y.; Ganesan, S.; Karlsson, M.C.I.; Kehrl, J.H. LRRK2 is required for CD38-mediated NAADP-Ca(2+) signaling and the downstream activation of TFEB (transcription factor EB) in immune cells. Autophagy 2022, 18, 204–222. [Google Scholar] [CrossRef]
- Zhang, K.; Zhu, S.; Li, J.; Jiang, T.; Feng, L.; Pei, J.; Wang, G.; Ouyang, L.; Liu, B. Targeting autophagy using small-molecule compounds to improve potential therapy of Parkinson’s disease. Acta Pharm. Sin. B 2021, 11, 3015–3034. [Google Scholar] [CrossRef]
- Paquette, M.; El-Houjeiri, L.; Zirden, L.C.; Puustinen, P.; Blanchette, P.; Jeong, H.; Dejgaard, K.; Siegel, P.M.; Pause, A. AMPK-dependent phosphorylation is required for transcriptional activation of TFEB and TFE3. Autophagy 2021, 17, 3957–3975. [Google Scholar] [CrossRef]
- Xue, W.; Li, Y. Enhanced lysosome biogenesis ameliorates neurodegenerative diseases. Aging 2022, 14, 8582–8584. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Cao, G.; Wei, G. A30P mutant α-synuclein impairs autophagic flux by inactivating JNK signaling to enhance ZKSCAN3 activity in midbrain dopaminergic neurons. Cell Death Dis. 2019, 10, 133. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, J.; Wang, X.; Zhang, Y.; Ma, Y.; Guan, G.; Yuwen, Y.; He, N.; Liu, H.; Yu, X.; et al. N(ε)-carboxyethyl-lysin influences atherosclerotic plaque stability through ZKSCAN3 acetylation-regulated macrophage autophagy via the RAGE/LKB1/AMPK1/SIRT1 pathway. Cardiovasc. Diabetol. 2025, 24, 36. [Google Scholar] [CrossRef]
- Yin, L.; Zhou, J.; Li, T.; Wang, X.; Xue, W.; Zhang, J.; Lin, L.; Wang, N.; Kang, X.; Zhou, Y.; et al. Inhibition of the dopamine transporter promotes lysosome biogenesis and ameliorates Alzheimer’s disease-like symptoms in mice. Alzheimers Dement. 2023, 19, 1343–1357. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, M.; Ding, X.; Yan, C.; Song, Z.; Chen, L.; Huang, X.; Wang, X.; Jian, Y.; Tang, G.; et al. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat. Cell Biol. 2016, 18, 1065–1077. [Google Scholar] [CrossRef]
- Zhang, W.; Li, X.; Wang, S.; Chen, Y.; Liu, H. Regulation of TFEB activity and its potential as a therapeutic target against kidney diseases. Cell Death Discov. 2020, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Gukovskaya, A.S.; Gukovsky, I.; Algül, H.; Habtezion, A. Autophagy, Inflammation, and Immune Dysfunction in the Pathogenesis of Pancreatitis. Gastroenterology 2017, 153, 1212–1226. [Google Scholar] [CrossRef]
- Ning, L.; Shishi, Z.; Bo, W.; Huiqing, L. Targeting immunometabolism against acute lung injury. Clin. Immunol. 2023, 249, 109289. [Google Scholar] [CrossRef]
- Zhang, L.; Xiao, X.; Ding, H. Silencing ZKSCAN3 gene expression aggravates lung injury induced by LPS in mice. Chin. J. Clin. Pharmacol. Ther. 2023, 28, 164. [Google Scholar]
- Zhang, H.; Hu, D.; Xu, Y.; Wu, L.; Lou, L. Effect of pulmonary rehabilitation in patients with chronic obstructive pulmonary disease: A systematic review and meta-analysis of randomized controlled trials. Ann. Med. 2022, 54, 262–273. [Google Scholar] [CrossRef]
- Cheng, X.T.; Xie, Y.X.; Zhou, B.; Huang, N.; Farfel-Becker, T.; Sheng, Z.H. Revisiting LAMP1 as a marker for degradative autophagy-lysosomal organelles in the nervous system. Autophagy 2018, 14, 1472–1474. [Google Scholar] [CrossRef] [PubMed]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 and Autophagy. Methods Mol. Biol. 2008, 445, 77–88. [Google Scholar] [CrossRef]
- Ouyang, X.; Becker, E., Jr.; Bone, N.B.; Johnson, M.S.; Craver, J.; Zong, W.X.; Darley-Usmar, V.M.; Zmijewski, J.W.; Zhang, J. ZKSCAN3 in severe bacterial lung infection and sepsis-induced immunosuppression. Lab. Invest. 2021, 101, 1467–1474. [Google Scholar] [CrossRef]
- New-Aaron, M.; Thomes, P.G.; Ganesan, M.; Dagur, R.S.; Donohue, T.M., Jr.; Kusum, K.K.; Poluektova, L.Y.; Osna, N.A. Alcohol-Induced Lysosomal Damage and Suppression of Lysosome Biogenesis Contribute to Hepatotoxicity in HIV-Exposed Liver Cells. Biomolecules 2021, 11, 1497. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Yan, Y.; Liu, C.; Finkel, T. The role of ZKSCAN3 in the transcriptional regulation of autophagy. Autophagy 2017, 13, 1235–1238. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, Z.; Li, X.; Li, X.; Li, Z.; Chen, H.; Gong, S.; Zhang, M.; Zhang, Y.; Li, Z.; Yang, L.; et al. The kidney-expressed transcription factor ZKSCAN3 is dispensable for autophagy transcriptional regulation and AKI progression in mouse. Mutat. Res. 2022, 825, 111790. [Google Scholar] [CrossRef]
- Yu, B.; Qi, Y.; Li, R.; Shi, Q.; Satpathy, A.T.; Chang, H.Y. B cell-specific XIST complex enforces X-inactivation and restrains atypical B cells. Cell 2021, 184, 1790–1803.e1717. [Google Scholar] [CrossRef]



| Tumor | Cell Lines | Phenotypes | Mechanisms | Refs |
|---|---|---|---|---|
| CRC | HCT116, LoVo, LS174T, SW480, RKO, CT26, MCA38 | ZKSCAN3 promotes proliferation, invasion, and metastasis | ZKSCAN3 enhances anchorage-independent growth and orthotopic tumor growth; regulates VEGF, ITGβ4, CEA, and AKT expression. | [52,54,57,84] |
| HCC | Huh7, QGY-7703 | ZKSCAN3 inhibits autophagy and promotes migration | ZKSCAN3 binds to the ITGβ4 promoter to activate the FAK/AKT axis; suppresses ULK1 and LC3b expression. | [58] |
| GC | BGC823, HGC-27 | ZKSCAN3 promotes proliferation, invasion, metastasis, and tumor angiogenesis | ZKSCAN3 up-regulates MEK2, RasGRP2, IGF-2, and ITGβ4; activates MST1R to induce MMP-2/9, cathepsin D, and VEGF expression. | [69,71] |
| BCA | MCF-7, MDA-MB-231 | ZKSCAN3 regulates cell viability, migration, and invasion | ZKSCAN3 modulates CCND1, BCL-2, MMP-2/9, Bax, and Akt/mTOR signaling pathways. | [63,72,75] |
| CC | HeLa, C33a, Caski, HeLa, SiHa | ZKSCAN3 promotes cell proliferation | ZKSCAN3 fails to activate autophagy and lysosomal gene expression. | [1,76] |
| PCa | PC3, LNCaP, VcaP, PC3, DU145, C4-s2 | ZKSCAN3 promotes tumor proliferation, angiogenesis, and metastasis | ZKSCAN3 activates VEGF, ITGβ4, cyclin D1/D2, NF-κB, and MMPs. | [74,78] |
| PaCa | MIA PaCa-2 | ZKSCAN3 suppresses cancer cell proliferation, migration, and invasion | ZKSCAN3 activates ULK1 and LC3-II to promote autophagosome formation and lysosomal degradation. | [62,85] |
| BLCA | UC13, UMUC3, 647V, 5637 | ZKSCAN3 regulates autophagy, proliferation, migration, and invasion | Silencing ZKSCAN3 promotes vacuolization, inhibits cell growth, and induces senescence/autophagy Via MMP-2, MMP-9, c-myc/FGFR3, and p53/PTEN. | [6,35,87] |
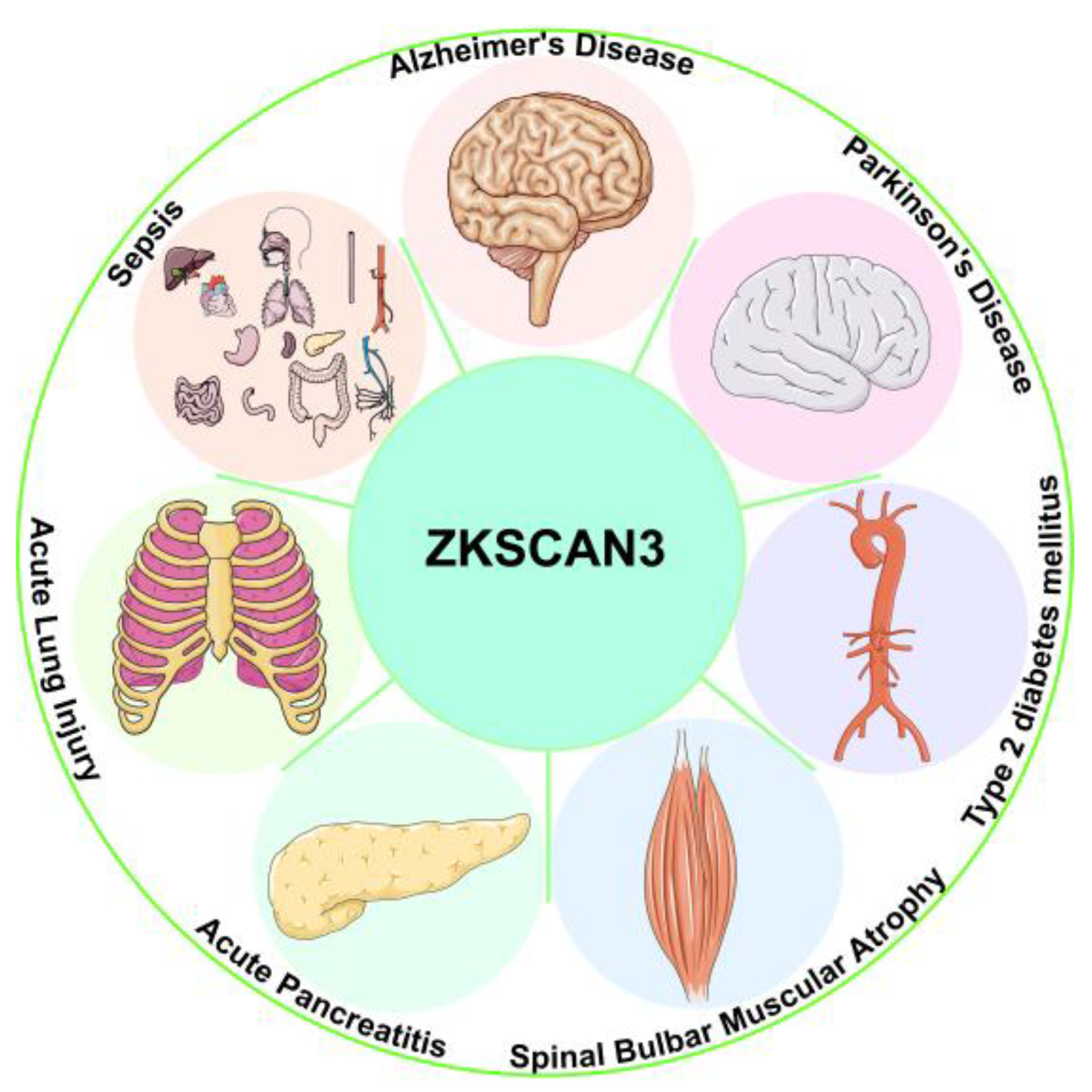
| Disease | Mechanisms | Phenotypes | Refs |
|---|---|---|---|
| AD | Down-regulation of ZKSCAN3 leads to up-regulation of GRB2 and Lamp2. Combined intervention of CHIT1 and CGRP inhibits ZKSCAN3. | Enhanced lysosomal activity, reduced neuronal damage, improved synaptic structure. Reduces Aβ toxicity-induced neuronal apoptosis. | [99,100,101,103] |
| PD | SIRT1 deacetylates ZKSCAN3, thereby promoting its nuclear export. ZKSCAN3 suppresses TFEB nuclear translocation and inhibits LAMP1 expression. LRRK2 mutations activate TFEB independently of the mTOR-ZKSCAN3 pathway. | Restored autophagy flux without alleviating α-synuclein aggregation. Targeting SIRT1-ZKSCAN3 axis or TFEB activation may mitigate neuronal damage. | [103,104,106,110,115,116] |
| T2DM-AS | CEL regulates ZKSCAN3 acetylation through the RAGE/LKB1/AMPK1/SIRT1 pathway. | Impaired macrophage autophagy and reduced plaque stability. | [117] |
| SBMA | HEP14/HEP15 induces TFEB nuclear translocation and suppresses ZKSCAN3 activity Via JNK/p38 MAPK signaling. | Enhanced lysosomal biogenesis and improved autophagic function. | [15,118,119] |
| AP | ALKBH5-mediated ZKSCAN3 mRNA demethylation causes its up-regulation. | Reduced autophagic activity and accelerated AP progression. | [68] |
| ALI | ZKSCAN3 inhibition leads to decreased antioxidant defense and exacerbated tissue necrosis. Negative correlation between ZKSCAN3 expression and lung function in COPD patients. | Aggravated LPS-induced lung injury. | [123] |
| Sepsis | ZKSCAN3 up-regulation suppresses LC3-II and Lamp1 expression. Acetaldehyde blocks lysosomal repair Via ZKSCAN3. | Reduced autophagic-lysosomal activity and impaired pathogen clearance. Immune dysregulation and multi-organ damage. | [125,127,128] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Li, X.; Xia, J.; Li, W.; Su, Z. Research Progress on the Functional Regulation Mechanisms of ZKSCAN3. Biomolecules 2025, 15, 1016. https://doi.org/10.3390/biom15071016
Xu J, Li X, Xia J, Li W, Su Z. Research Progress on the Functional Regulation Mechanisms of ZKSCAN3. Biomolecules. 2025; 15(7):1016. https://doi.org/10.3390/biom15071016
Chicago/Turabian StyleXu, Jianxiong, Xinzhe Li, Jingjing Xia, Wenfang Li, and Zhengding Su. 2025. "Research Progress on the Functional Regulation Mechanisms of ZKSCAN3" Biomolecules 15, no. 7: 1016. https://doi.org/10.3390/biom15071016
APA StyleXu, J., Li, X., Xia, J., Li, W., & Su, Z. (2025). Research Progress on the Functional Regulation Mechanisms of ZKSCAN3. Biomolecules, 15(7), 1016. https://doi.org/10.3390/biom15071016







