The α5-α6-α7-Pba3-Pba4 Complex: A Starting Unit in Proteasome Core Particle Assembly
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains, Cultures, and Media
2.2. Protein Extraction, Gel Filtration, SDS- and Native PAGE, and Immunoblots
2.3. Immunoaffinity Purification of the Pba4-FLAG Complex
2.4. Expression of 8His-SUMO1-Fused Proteins in E. coli and Their Purification
2.5. Generation, Purification, and Validation of Anti-α5 and Anti-α6 Antibodies
2.6. In Vitro Binding Assay on HA-Resin
2.7. In Vitro Assembly of Pba3-Pba4 Complexes
3. Results
3.1. Pba3-Pba4 Is Detected in Several Proteasome Precursor Complexes
3.2. Pba4-Containing Complexes Are Dependent on α5, α6, and α7 Subunits
3.3. Deletion of PBA1 Gene Affects Incorporation of Pba3-Pba4 into High MW Complexes
3.4. Blm10 Deletion Abolishes Pba4-High Molecular Complexes Detected in Absence of PBA1
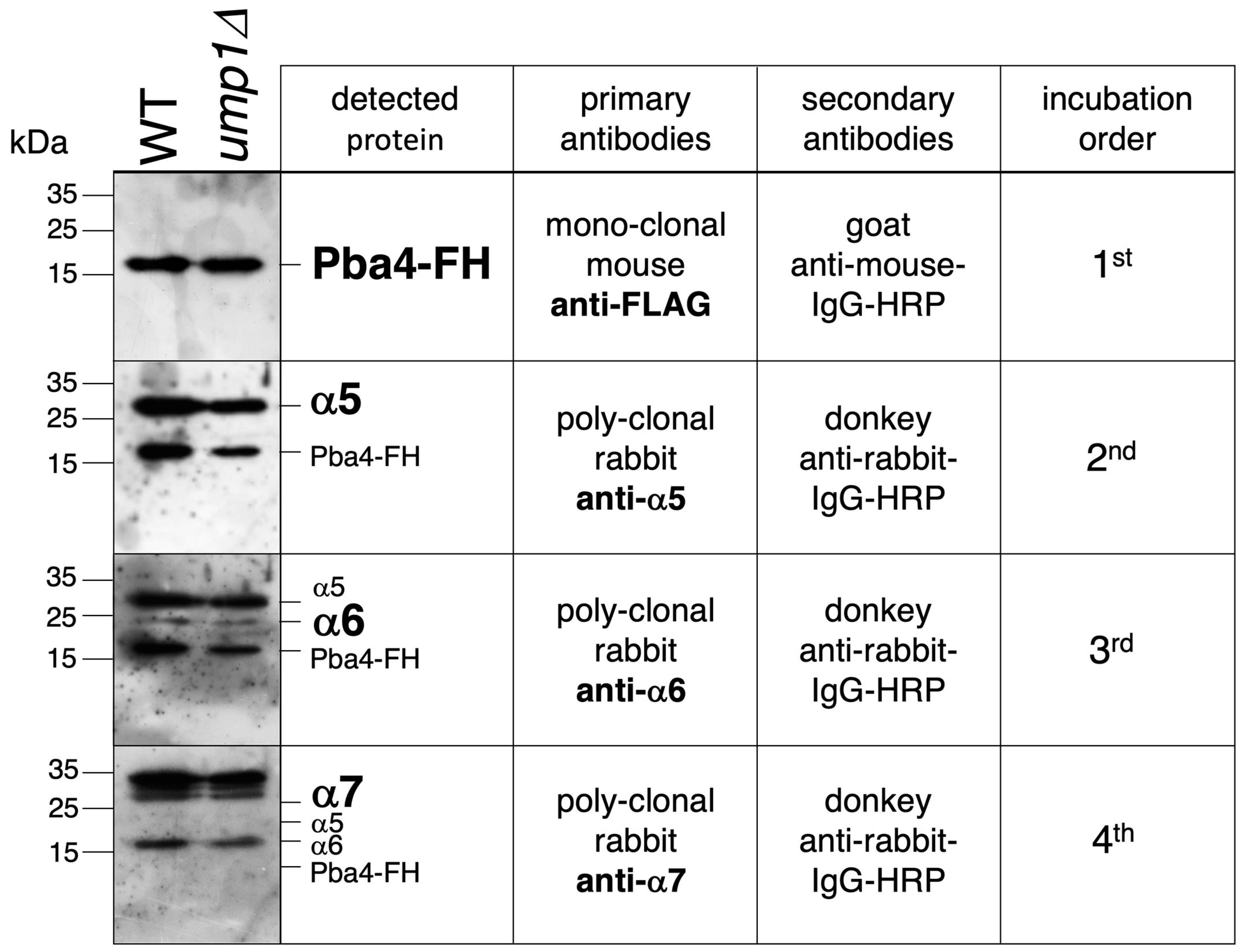
3.5. Detection of α5-α6-α7-Pba3-Pba4 Complexes in S. cerevisiae
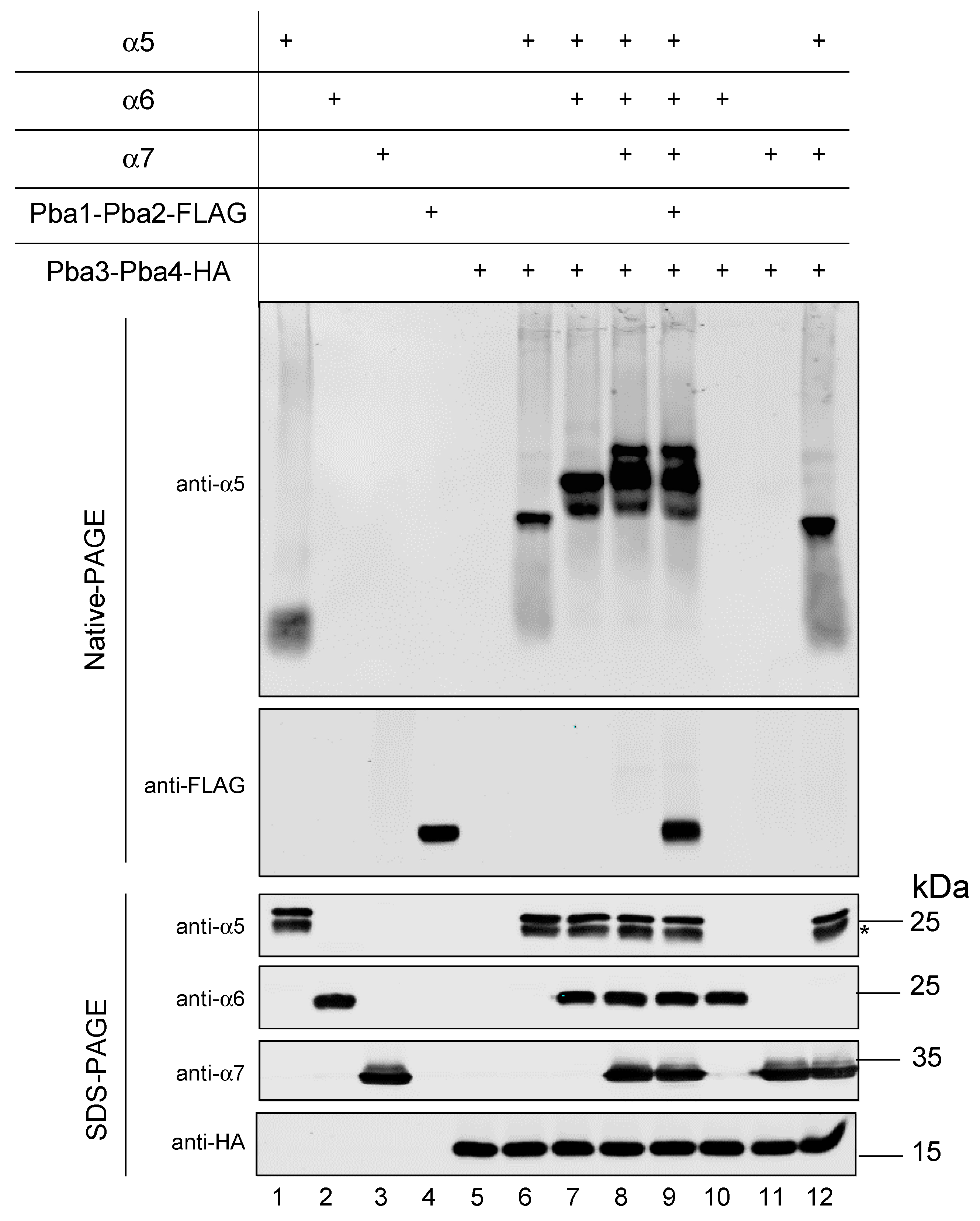
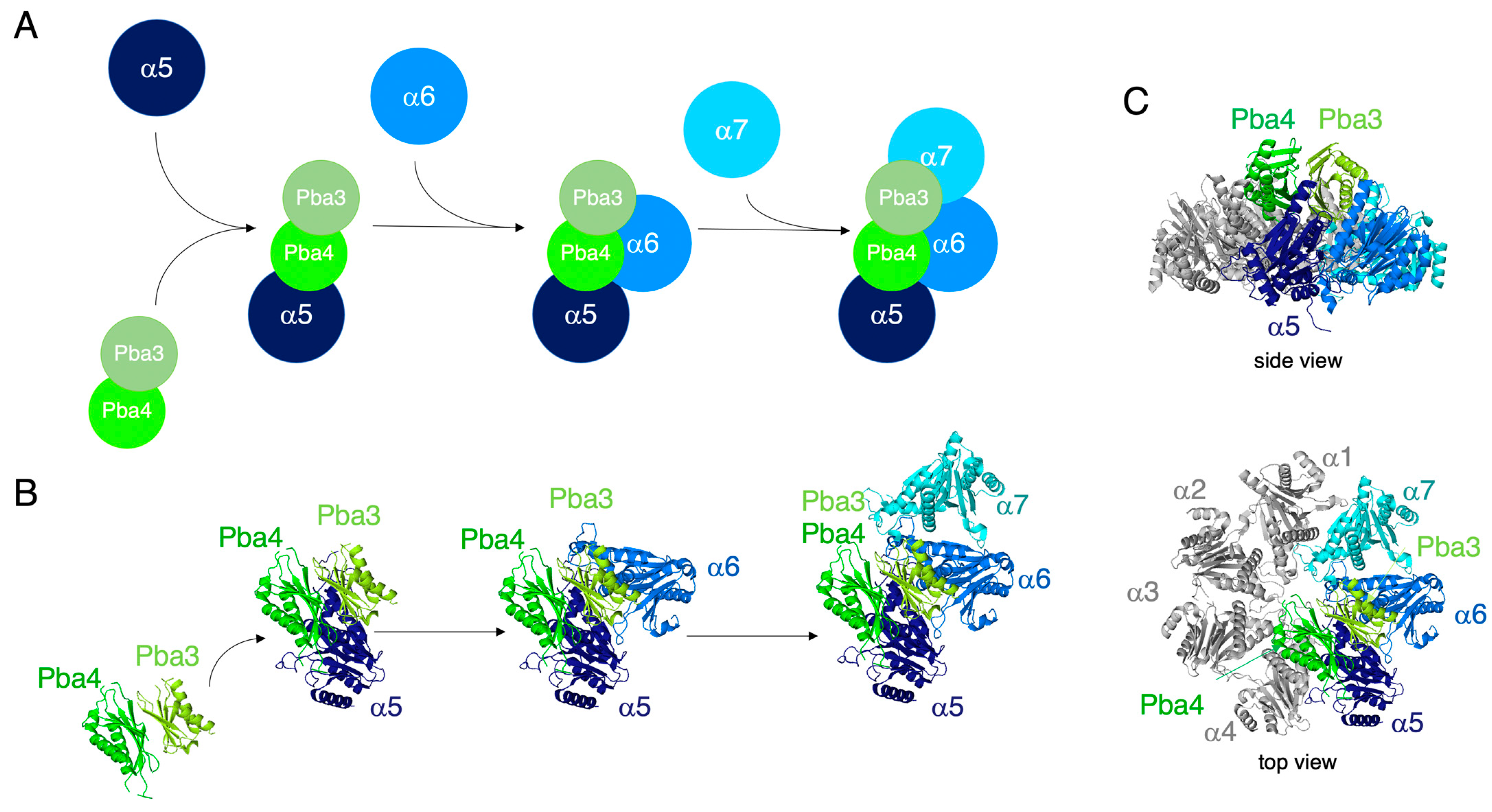
3.6. In Vitro Reconstitution of α5-α6-α7-Pba3-Pba4 Precursor Complex Assembly
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marques, A.J.; Palanimurugan, R.; Matias, A.C.; Ramos, P.C.; Dohmen, R.J. Catalytic mechanism and assembly of the proteasome. Chem. Rev. 2009, 109, 1509–1536. [Google Scholar] [CrossRef] [PubMed]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef] [PubMed]
- Budenholzer, L.; Cheng, C.L.; Li, Y.; Hochstrasser, M. Proteasome structure and assembly. J. Mol. Biol. 2017, 429, 3500–3524. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Ditzel, L.; Löwe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 1997, 386, 463–471. [Google Scholar] [CrossRef]
- Matias, A.C.; Matos, J.; Dohmen, R.J.; Ramos, P.C. Hsp70 and Hsp110 Chaperones Promote Early Steps of Proteasome Assembly. Biomolecules 2022, 13, 11. [Google Scholar] [CrossRef]
- Murata, S.; Yashiroda, H.; Tanaka, K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 2009, 10, 104–115. [Google Scholar] [CrossRef]
- Hirano, Y.; Hendil, K.B.; Yashiroda, H.; Iemura, S.; Nagane, R.; Hioki, Y.; Natsume, T.; Tanaka, K.; Murata, S. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature 2005, 437, 1381–1385. [Google Scholar] [CrossRef]
- Hirano, Y.; Hayashi, H.; Iemura, S.; Hendil, K.B.; Niwa, S.; Kishimoto, T.; Kasahara, M.; Natsume, T.; Tanaka, K.; Murata, S. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol. Cell 2006, 24, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Yashiroda, H.; Mizushima, T.; Okamoto, K.; Kameyama, T.; Hayashi, H.; Kishimoto, T.; Niwa, S.; Kasahara, M.; Kurimoto, E.; Sakata, E.; et al. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat. Struct. Mol. Biol. 2008, 15, 228–236. [Google Scholar] [CrossRef]
- Kusmierczyk, A.R.; Hochstrasser, M. Some assembly required: Dedicated chaperones in eukaryotic proteasome biogenesis. Biol. Chem. 2008, 389, 1143–1151. [Google Scholar] [CrossRef]
- Takagi, K.; Saeki, Y.; Yashiroda, H.; Yagi, H.; Kaiho, A.; Murata, S.; Yamane, T.; Tanaka, K.; Mizushima, T.; Kato, K. Pba3-Pba4 heterodimer acts as a molecular matchmaker in proteasome alpha-ring formation. Biochem. Biophys. Res. Commun. 2014, 450, 1110–1114. [Google Scholar] [CrossRef] [PubMed]
- Ramos, P.C.; Marques, A.J.; London, M.K.; Dohmen, R.J. Role of C-terminal extensions of subunits beta2 and beta7 in assembly and activity of eukaryotic proteasomes. J. Biol. Chem. 2004, 279, 14323–14330. [Google Scholar] [CrossRef] [PubMed]
- Ramos, P.C.; Höckendorff, J.; Johnson, E.S.; Varshavsky, A.; Dohmen, R.J. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell 1998, 92, 489–499. [Google Scholar] [CrossRef]
- Longtine, M.S.; McKenzie, A., 3rd; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Mumberg, D.; Müller, R.; Funk, M. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994, 22, 5767–5768. [Google Scholar] [CrossRef] [PubMed]
- Janke, C.; Magiera, M.M.; Rathfelder, N.; Taxis, C.; Reber, S.; Maekawa, H.; Moreno-Borchart, A.; Doenges, G.; Schwob, E. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and pro- moter substitution cassettes. Yeast 2004, 21, 947–962. [Google Scholar] [CrossRef]
- Kötter, P.; Weigand, J.E.; Meyer, B.; Entian, K.-D.; Suess, B. A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res. 2009, 37, e120. [Google Scholar] [CrossRef]
- Mark, E.; Ramos, P.C.; Kayser, F.; Höckendorff, J.; Dohmen, R.J.; Wendler, P. Structural roles of Ump1 and β-subunit propeptides in proteasome biogenesis. Life Sci. Alliance 2024, 7, e202402865. [Google Scholar] [CrossRef]
- Stadtmueller, B.M.; Kish-Trier, E.; Ferrell, K.; Petersen, C.N.; Robinson, H.; Myszka, D.G.; Eckert, D.M.; Formosa, T.; Hill, C.P. Structure of a proteasome pba1- pba2 complex: Implications for proteasome assembly, activation, and biological function. J. Biol. Chem. 2012, 287, 37371–37382. [Google Scholar] [CrossRef]
- Walsh, R.M.; Rawson, S.; Schnell, H.M.; Velez, B.; Rajakumar, T.; Hanna, J. Structure of the preholoproteasome reveals late steps in proteasome core particle biogenesis. Nat. Struct. Mol. Biol. 2023, 30, 1516–1524. [Google Scholar] [CrossRef]
- Kusmierczyk, A.R.; Kunjappu, M.J.; Kim, R.Y.; Hochstrasser, M. A conserved 20S proteasome assembly factor requires a C-terminal HbYX motif for proteasomal precursor binding. Nat. Struct. Mol. Biol. 2011, 18, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Sadre-Bazzaz, K.; Whitby, F.G.; Robinson, H.; Formosa, T.; Hill, C.P. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol. Cell 2010, 37, 728–735. [Google Scholar] [CrossRef]
- Tian, G.; Park, S.; Lee, M.J.; Huck, B.; McAllister, F.; Hill, C.P.; Gygi, S.P.; Finley, D. An asymmetric interface between the regulatory and core particles of the proteasome. Nat. Struct. Mol. Biol. 2011, 18, 1259–1267. [Google Scholar] [CrossRef]
- Dange, T.; Smith, D.; Noy, T.; Rommel, P.C.; Jurzitza, L.; Cordero, R.J.; Legendre, A.; Finley, D.; Goldberg, A.L.; Schmidt, M. Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism. J. Biol. Chem. 2011, 286, 42830–42839. [Google Scholar] [CrossRef] [PubMed]
- Barthelme, D.; Chen, L.Z.; Grabenstatter, J.; Baker, T.A.; Sauer, R.T. Architecture and assembly of the archaeal Cdc48·20S proteasome. Proc. Natl. Acad. Sci. USA 2014, 111, E1687–E1694. [Google Scholar] [CrossRef]
- Fehlker, M.; Wendler, P.; Lehmann, A.; Enenkel, C. Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep. 2003, 4, 959–963. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kusmierczyk, A.R.; Wong, P.; Emili, A.; Hochstrasser, M. beta-Subunit appendages promote 20S proteasome assembly byovercoming an Ump1-dependent checkpoint. EMBO J. 2007, 26, 2339–2349. [Google Scholar] [CrossRef]
- Kaur, M.; Chen, X.; Lee, S.Y.; Weaver, T.M.; Freudenthal, B.D.; Walters, K.J.; Roelofs, J. Structure of Blm10:13S proteasome intermediate reveals parallel assembly pathways for the proteasome core particle. bioRxiv 2024. [Google Scholar] [CrossRef]
- Lehmann, A.; Jechow, K.; Enenkel, C. Blm10 binds to pre-activated proteasome core particles with open gate conformation. EMBO Rep. 2008, 9, 1237–1243. [Google Scholar] [CrossRef]
- Schmidt, M.; Haas, W.; Crosas, B.; Santamaria, P.G.; Gygi, S.P.; Walz, T.; Finley, D. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat. Struct. Mol. Biol. 2005, 12, 294–303. [Google Scholar] [CrossRef]
- Burris, A.; Waite, K.A.; Reuter, Z.; Ockerhausen, S.; Roelofs, J. Proteasome activator Blm10 levels and autophagic degradation directly impact the proteasome landscape. J. Biol. Chem. 2021, 296, 100468. [Google Scholar] [CrossRef]
- Hermanns, T.; Kolek, S.; Uthoff, M.; de Heiden, R.A.; Mulder, M.P.C.; Baumann, U.; Hofmann, K. A family of bacterial Josephin-like deubiquitinases with an irreversible cleavage mode. Mol. Cell 2025, 85, 1202–1215.e5. [Google Scholar] [CrossRef]
- Le Tallec, B.; Barrault, M.B.; Courbeyrette, R.; Guérois, R.; Marsolier-Kergoat, M.C.; Peyroche, A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol. Cell 2007, 27, 660–674. [Google Scholar] [CrossRef]
- Velichutina, I.; Connerly, P.L.; Arendt, C.S.; Li, X.; Hochstrasser, M. Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. EMBO J. 2004, 23, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.M.; Chang, S.-C.; Park, S.; Finley, D.; Yifan Cheng, Y.; Goldberg, A.L. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell 2007, 27, 731–744. [Google Scholar] [CrossRef]
- Wani, P.S.; Rowland, M.A.; Ondracek, A.; Deeds, E.J.; Roelofs, J. Maturation of the proteasome core particle induces an affinity switch that controls regulatory particle association. Nat. Commun. 2015, 6, 6384. [Google Scholar] [CrossRef] [PubMed]
- Schnell, H.M.; Walsh, R.M.; Rawson, S.; Kaur, M.; Bhanu, M.K.; Tian, G.; Prado, M.A.; Guerra-Moreno, A.; Paulo, J.A.; Gygi, S.P.; et al. Structures of chaperone-associated assembly intermediates reveal coordinated mechanisms of proteasome biogenesis. Nat. Struct. Mol. Biol. 2021, 28, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Weberruss, M.H.; Savulescu, A.F.; Jando, J.; Bissinger, T.; Harel, A.; Glickman, M.H.; Enenkel, C. Blm10 facilitates nuclear import of proteasome core particles. EMBO J. 2013, 32, 2697–2707. [Google Scholar] [CrossRef]
- Enenkel, C.; Ernst, O.P. Proteasome dynamics in response to metabolic changes. Front. Cell. Dev. Biol. 2025, 3, 1523382. [Google Scholar] [CrossRef]



| Name | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 metΔ0 ura3Δ0 | EuroScarf |
| CM2 | MATa PRE6-HA::YIplac211 | [5] |
| CM134 | MATa PGALS-SCL1 (α1)::kanMX PRE6 (α4)-HA::YIplac211 | [5] |
| CM144 | MATa PGALS-PRE6 (α4)::KanMX PRE6 (α4)-HA::YIplac211 | this study |
| CM192 | MATa pba1Δ::HIS3 PBA4-HA::YIplac211 | this study |
| CM223 | MATa pre9Δ::HIS3 PBA4-HA::YIplac211 | this study |
| CM233 | MATa PGALS-PRE10 (α7)::KanMXPBA4-HA::YIplac211 | this study |
| CM241 | MATa PGALS-PRE6 (α4)::KanMXPBA4-HA::YIplac211 | this study |
| CM257 | MATa PRE9 (α3)-HA::YIplac211 | this study |
| CM310 | MATa pba1Δ::NAT pba3Δ::HIS3 PUP2 (α5)-HA::YIplac211 | this study |
| CM342 | MATa PGAL1-PUP1 (β2)::KanMX PBA4-HA::YIplac211 | this study |
| FP8 | MATa PRE6 (α4)-HA::YIplac211 pre9Δ::HIS3MX6 | [5] |
| FP16 | MATa PBA4-HA::YIplac211 | [5] |
| JD47-13C | MATa his3-Δ200 leu2-3,112lys2-801 trp1-Δ63 ura3-52 | [13] |
| JD129 | MATa UMP1-HA::YIplac128 | [13] |
| MN6 | MATa ump1Δ::HIS3MX PBA4-HA::YIplac211 | this study |
| MN48 | MATa PGAL1-PRE1 (β4)::KanMXPBA4-HA::YIplac211 | this study |
| MN49 | MATa PGAL1-PRE2 (β5)::KanMXPBA4-HA::YIplac211 | this study |
| MN50 | MATa PGAL1-PRE7 (β6)::KanMXPBA4-HA::YIplac211 | this study |
| MN55 | MATa blm10Δ::KanMX PBA4-HA::YIplac211 (BY4741) | this study |
| MO47 | MATa PGAL1-PBA1::TRP1 PGAL1 PBA2::HIS3 PBA4-HA::YIplac211 | this study |
| PR20 | MATa PGALS-SCL1 (α1)::KanMXPBA4-HA::YIplac211 | this study |
| PR21 | MATa PGALS-PRE8 (α2)::KanMXSCL1 (α1)-HA::YIplac211 | this study |
| PR22 | MATa PGALS-PRE8 (α2)::KanMXPRE8 (α2)-HA::YIplac128 | this study |
| PR23 | MATa PGALS-PRE8 (α2)::KanMXPRE9 (α3)-HA::YIplac211 | this study |
| PR24 | MATa PGALS-PRE8 (α2)::KanMXPRE6 (α4)-HA::YIplac211 | this study |
| PR25 | MATa PGALS-PRE8 (α2)::KanMXPUP2 (α5)-HA::YIplac211 | this study |
| PR26 | MATa PGALS-PRE8 (α2)::KanMXPRE5(α6)-HA::YIplac211 | this study |
| PR27 | MATa PGALS-PRE8 (α2)::KanMXPRE10 (α7)-HA::YIplac211 | this study |
| PR28 | MATa PGALS-PRE8 (α2)::KanMXUMP1-HA::YIplac128 | this study |
| PR30 | MATa PGALS-PRE8 (α2)::KanMXPBA4-HA::YIplac211 | this study |
| PR60 | MATa PGALS-PUP2(α5)::KanMXPBA4-HA::YIplac211 | this study |
| PR70 | MATa PGALS-PRE5(α6)::KanMXPBA4-HA::YIplac211 | this study |
| PR118 | MATa pba1Δ::HIS3 | this study |
| PR119 | MATa pba3Δ::HIS3 | this study |
| PR130 | MATa pba1Δ::HIS3 PUP2 (α5)-HA::YIplac211 | this study |
| PR132 | MATa pba3Δ::HIS3 PRE6 (α4)-HA::YIplac211 | this study |
| PR193 | MATa pba3Δ::HIS3 SCL1 (α1)-HA::YIplac211 | this study |
| PR194 | MATa pba3Δ::HIS3 PRE8 (α2)-HA::YIplac211 | this study |
| PR207 | MATa pba1Δ::HIS3 blm10Δ::KanMX | this study |
| PR211 | MATaPBA4-FLAG6His ::YIplac204 | this study |
| PR213 | MATablm10Δ::KanMX pba1Δ::HIS3 PBA4-HA::YIplac211 (BY4741) | this study |
| PR236 | MATalpha pba1Δ::NAT1 | this study |
| PR238 | MATalpha pba1Δ::NAT1 pba3Δ::HIS3 | this study |
| PR241 | MATalpha ump1Δ::LEU2 pba3Δ::HIS3 | this study |
| PR244 | MATa pba1Δ::NAT1 pba3Δ::HIS3 | this study |
| PR245 | MATa ump1Δ::LEU2 | this study |
| PR247 | MATa pba1Δ::NAT1 ump1Δ::LEU2 pba3Δ::HIS3 | this study |
| PR265 | MATa pba2Δ::KanMX PBA4-HA::YIplac211(BY4741) | this study |
| PR344 | MATaump1Δ::HIS3 PBA4-FLAG-6His::YIplac204 | this study |
| PR357 | JD47-13C PGAL1-tc3-PBA4-2HA::URA | this study |
| PR368 | MATapba1Δ::TRP1 ump1Δ::HIS3 PBA4-HA::YIplac211 | this study |
| blm10Δ | MATa blm10Δ:: KanMX (BY4741) | EuroScarf |
| pba1Δ | MATa pba1Δ:: KanMX (BY4741) | EuroScarf |
| Name | Design | Source |
|---|---|---|
| pJD791 | pUC21-PGAL1-tc3-PBA4-2HA | this study |
| pJZ20 | pET11a-8His-SUMO1-PUP2(α5) | this study |
| pJZ21 | pET11a-8His-SUMO1-PRE5(α6) | this study |
| pJZ22 | pET11a-8His-SUMO1-PRE10(α7) | this study |
| pJZ24 | pET11a-8His-SUMO1-PBA1-PBA2-FLAG | this study |
| pJZ39 | pET11a-8His-SUMO1-PBA3-PBA4-HA | this study |
| Antibody | Host | Dilution | Source |
|---|---|---|---|
| anti-HA (3F10) | rat | 1:2000 | Roche |
| anti-HA (3F10)-peroxidase | rat | 1:2000 | Roche |
| anti-FLAG (M2) | mouse | 1:1000 | Sigma-Aldrich |
| anti-Pre10/a7 | rabbit | 1:1000 | [12] |
| anti-Pup2/a5 | rabbit | 1:1000 | this study |
| anti-Pre5/a6 | rabbit | 1:1000 | this study |
| anti-mouse-IgG-HRP | goat | 1:5000 | Sigma-Aldrich |
| anti-rabbit-IgG-HRP | donkey | 1:5000 | GE healthcare |
| anti-mouse Alexa Fluor Plus 680 | goat | 1:5000 | Thermo Fisher |
| anti-rabbit Alexa Fluor Plus 800 | goat | 1:5000 | Thermo Fisher |
| anti-rat Alexa Fluor 680 | goat | 1.5000 | Thermo Fisher |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matias, A.C.; Tiago, M.N.; Zimmermann, J.; Dohmen, R.J.; Ramos, P.C. The α5-α6-α7-Pba3-Pba4 Complex: A Starting Unit in Proteasome Core Particle Assembly. Biomolecules 2025, 15, 683. https://doi.org/10.3390/biom15050683
Matias AC, Tiago MN, Zimmermann J, Dohmen RJ, Ramos PC. The α5-α6-α7-Pba3-Pba4 Complex: A Starting Unit in Proteasome Core Particle Assembly. Biomolecules. 2025; 15(5):683. https://doi.org/10.3390/biom15050683
Chicago/Turabian StyleMatias, Ana C., Margarida N. Tiago, Jessica Zimmermann, R. Jürgen Dohmen, and Paula C. Ramos. 2025. "The α5-α6-α7-Pba3-Pba4 Complex: A Starting Unit in Proteasome Core Particle Assembly" Biomolecules 15, no. 5: 683. https://doi.org/10.3390/biom15050683
APA StyleMatias, A. C., Tiago, M. N., Zimmermann, J., Dohmen, R. J., & Ramos, P. C. (2025). The α5-α6-α7-Pba3-Pba4 Complex: A Starting Unit in Proteasome Core Particle Assembly. Biomolecules, 15(5), 683. https://doi.org/10.3390/biom15050683








