PlastiCRISPR: Genome Editing-Based Plastic Waste Management with Implications in Polyethylene Terephthalate (PET) Degradation
Abstract
1. Introduction
2. Genome Editing in Plastic Waste Management
3. PlastiCRISPR Technology
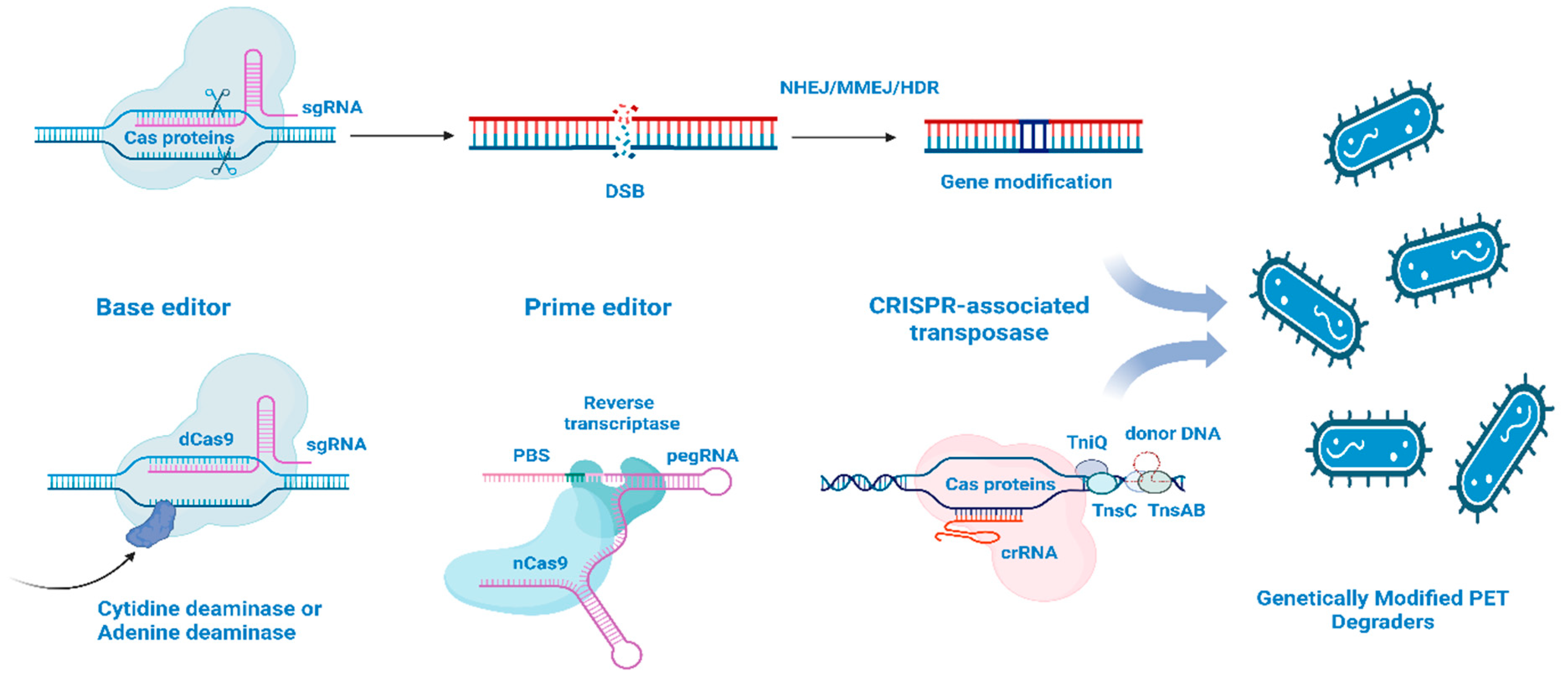
4. Molecular Mechanisms of Genome Editing for Enhanced Plastic Degradation
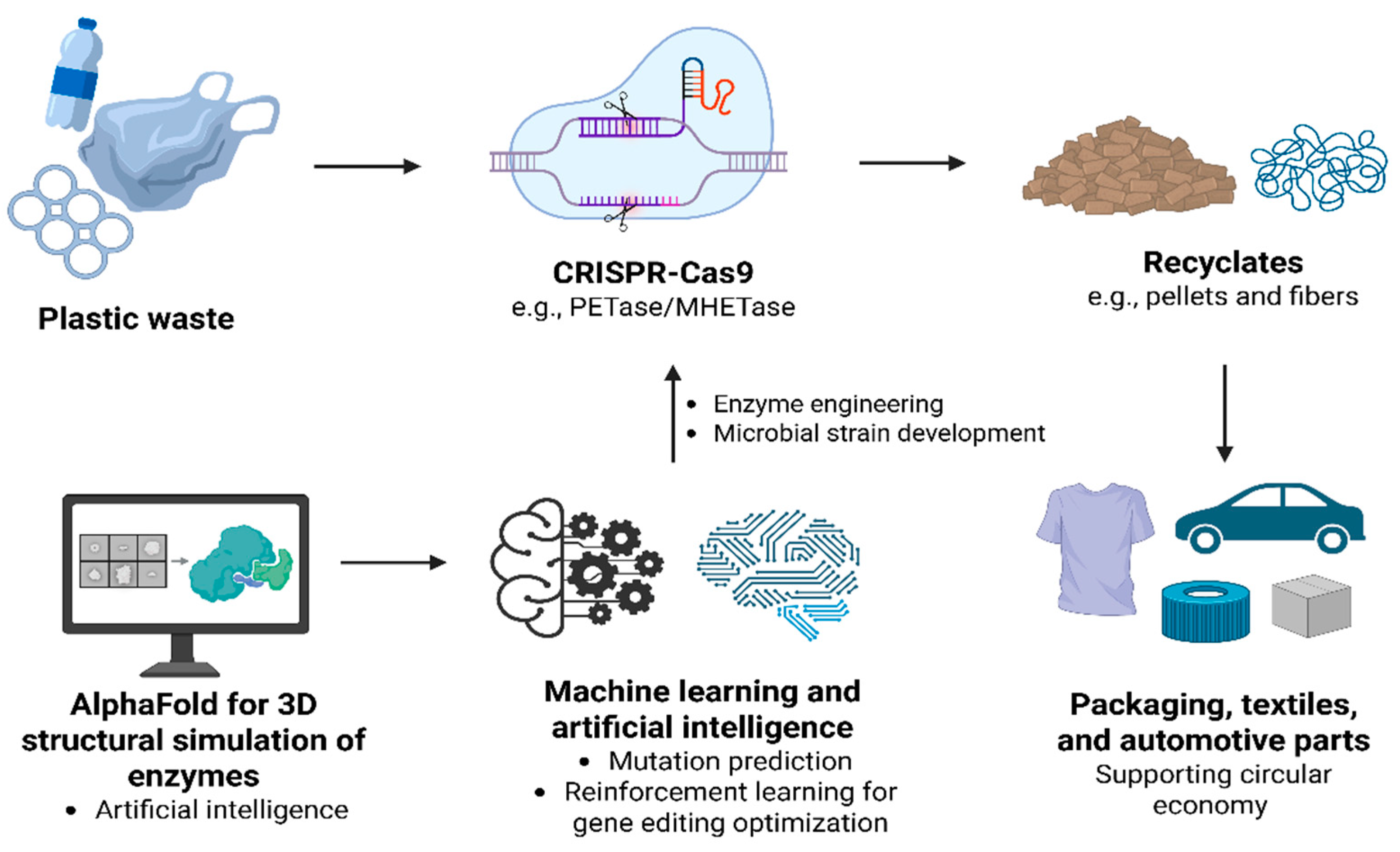
5. PlastiCRISPR and Artificial Intelligence: Plastic Waste Management in the Circular Economy
6. Potential Applications and Scalability
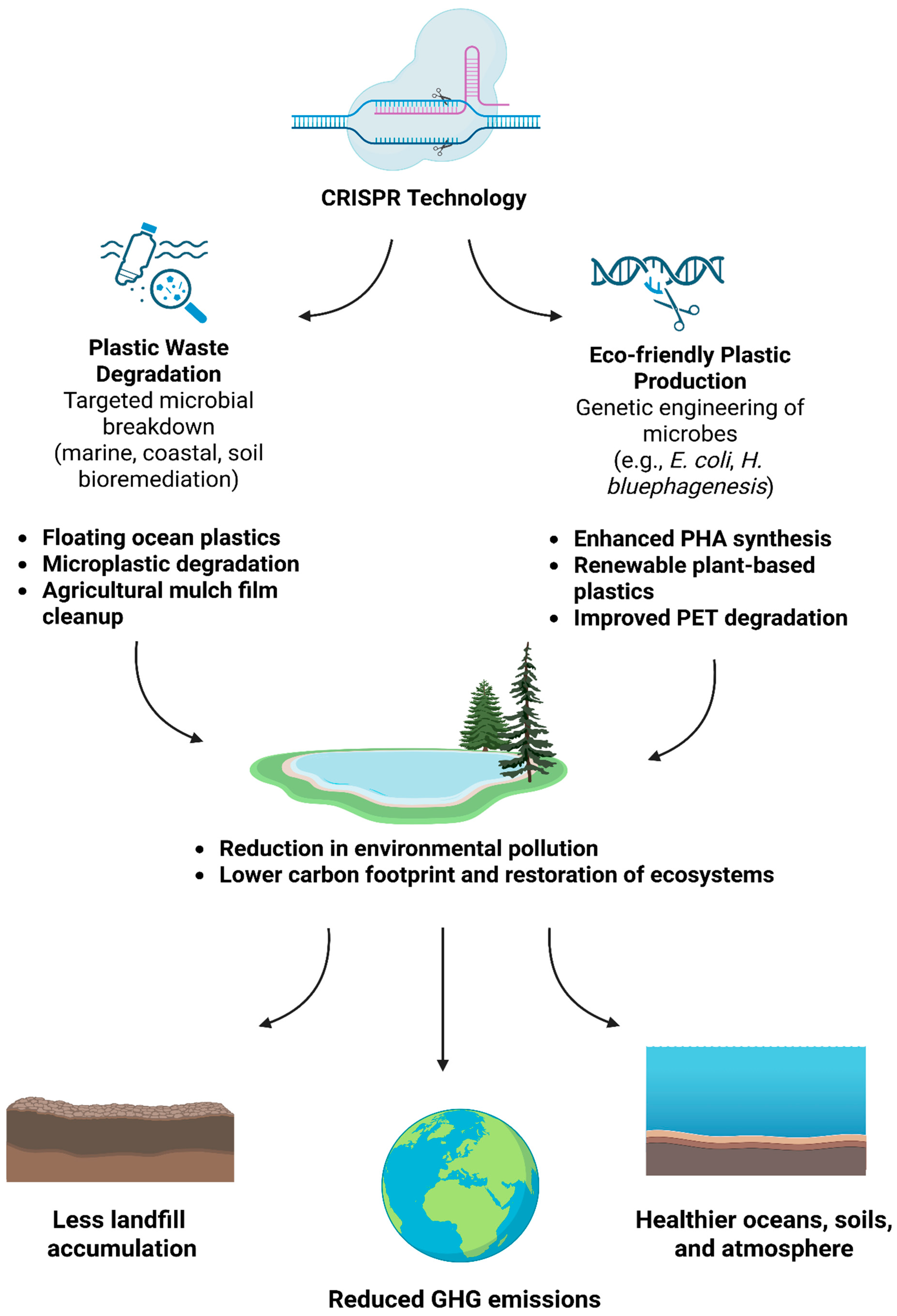
7. Environmental Benefits of Using PlastiCRISPR for Enhanced Plastic Degradation
8. PlastiCRISPR-Based Bioplastic Production and Its Comparison with Traditional Methods
9. Ethical Implications
10. Conclusions and Future Direction
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xia, B. Potential Solution for Plastic Degradation—Genetical Modification. In Proceedings of the 2021 International Conference on Social Development and Media Communication (SDMC 2021); Atlantis Press: Dordrecht, The Netherlands, 2022; Volume 631, pp. 1076–1079. [Google Scholar] [CrossRef]
- Anand, U.; Dey, S.; Bontempi, E.; Ducoli, S.; Vethaak, A.D.; Dey, A.; Federici, S. Biotechnological methods to remove microplastics: A review. Environ. Chem. Lett. 2023, 21, 1787–1810. [Google Scholar] [CrossRef] [PubMed]
- Gambarini, V.; Pantos, O.; Kingsbury, J.M.; Weaver, L.; Handley, K.M.; Lear, G. Phylogenetic Distribution of Plastic-Degrading Microorganisms. mSystems 2021, 6, e01112-20. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Dhiman, M.; Kansal, A.; Subudhi, S.P. Plastic waste management for sustainable environment: Techniques and approaches. Waste Dispos. Sustain. Energy 2023, 5, 205–222. [Google Scholar] [CrossRef]
- Nidhi, S.; Anand, U.; Oleksak, P.; Tripathi, P.; Lal, J.A.; Thomas, G.; Kuca, K.; Tripathi, V. Novel CRISPR–Cas Systems: An Updated Review of the Current Achievements, Applications, and Future Research Perspectives. Int. J. Mol. Sci. 2021, 22, 3327. [Google Scholar] [CrossRef]
- Romero-Orejon, K.; Karbalaei-Heidari, H.R.; Budisa, N.; Levin, D. Antibiotic-free whole-cell biocatalytic fermentation: Escherichia coli with surface-displayed PETases for sustainable plastic degradation. bioRxiv 2024. [Google Scholar] [CrossRef]
- Zhou, Y.; Lin, L.; Wang, H.; Zhang, Z.; Zhou, J.; Jiao, N. Development of a CRISPR/Cas9n-based tool for metabolic engineering of Pseudomonas putida for ferulic acid-to-polyhydroxyalkanoate bioconversion. Commun. Biol. 2020, 3, 98. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Gao, P.; Li, B.; Feng, L.; Liu, Y.; Du, Z.; Zhang, L. Occurrence and migration of microplastics and plasticizers in different wastewater and sludge treatment units in municipal wastewater treatment plant. Front. Environ. Sci. Eng. 2022, 16, 142. [Google Scholar] [CrossRef]
- Wang, Y.; Qian, H. Phthalates and Their Impacts on Human Health. Healthcare 2021, 9, 603. [Google Scholar] [CrossRef]
- Hennebert, P. Hazardous properties of plasticisers that may hinder the recycling of plastics. Detritus 2022, 21, 35. [Google Scholar] [CrossRef]
- Zhang, Y.; Pedersen, J.N.; Eser, B.E.; Guo, Z. Biodegradation of polyethylene and polystyrene: From microbial deterioration to enzyme discovery. Biotechnol. Adv. 2022, 60, 107991. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- Cai, Z.; Li, M.; Zhu, Z.; Wang, X.; Huang, Y.; Li, T.; Gong, H.; Yan, M. Biological Degradation of Plastics and Microplastics: A Recent Perspective on Associated Mechanisms and Influencing Factors. Microorganisms 2023, 11, 1661. [Google Scholar] [CrossRef] [PubMed]
- Vigouroux, A.; Bikard, D. CRISPR Tools To Control Gene Expression in Bacteria. Microbiol. Mol. Biol. Rev. 2020, 84, e00077-19. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Olarte, R.D.; Bravo Rodríguez, R.; Morales-Ríos, E. Genome Editing in Bacteria: CRISPR-Cas and Beyond. Microorganisms 2021, 9, 844. [Google Scholar] [CrossRef]
- Liu, C.; Yue, Y.; Xue, Y.; Zhou, C.; Ma, Y. CRISPR-Cas9 assisted non-homologous end joining genome editing system of Halomonas bluephagenesis for large DNA fragment deletion. Microb. Cell Fact. 2023, 22, 211. [Google Scholar] [CrossRef]
- Orlando, M.; Molla, G.; Castellani, P.; Pirillo, V.; Torretta, V.; Ferronato, N. Microbial Enzyme Biotechnology to Reach Plastic Waste Circularity: Current Status, Problems and Perspectives. Int. J. Mol. Sci. 2023, 24, 3877. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Long, R.; Lin, X.; Liu, W.; Zhu, L.; Jiang, L. Development and characterization of a bacterial enzyme cascade reaction system for efficient and stable PET degradation. J. Hazard. Mater. 2024, 472, 134480. [Google Scholar] [CrossRef]
- Kushwaha, A.; Goswami, L.; Kim, B.S. Advancement in innovative strategies for poly (ethylene terephthalate) biodegradation. Curr. Opin. Chem. Eng. 2025, 48, 101121. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, F.; Yan, W.; Dai, Z.; Dong, W.; Zhou, J.; Zhang, W.; Xin, F.; Jiang, M. Recent Advances of CRISPR/Cas9-Based Genetic Engineering and Transcriptional Regulation in Industrial Biology. Front. Bioeng. Biotechnol. 2020, 7, 459. [Google Scholar] [CrossRef]
- Jeong, S.H.; Lee, H.J.; Lee, S.J. Recent Advances in CRISPR-Cas Technologies for Synthetic Biology. J. Microbiol. 2023, 61, 13. [Google Scholar] [CrossRef]
- Mao, J.; Zhang, H.; Chen, Y.; Wei, L.; Liu, J.; Nielsen, J.; Xu, N. Relieving metabolic burden to improve robustness and bioproduction by industrial microorganisms. Biotechnol. Adv. 2024, 74, 108401. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, W.; Shao, S.; Wang, Q. Gene Silencing Through CRISPR Interference in Bacteria: Current Advances and Future Prospects. Front. Microbiol. 2021, 12, 635227. [Google Scholar] [CrossRef] [PubMed]
- Yeom, S.J.; Le, T.K.; Yun, C.H. P450-driven plastic-degrading synthetic bacteria. Trends Biotechnol. 2022, 40, 166–179. [Google Scholar] [CrossRef]
- Valamontes, A. Genetic Engineering and Metabolic Pathway Optimization of Ideonella sakaiensis for Enhanced PET Biodegradation; 2024; Available online: https://www.researchgate.net/publication/380889467_Genetic_Engineering_and_Metabolic_Pathway_Optimization_of_Ideonella_sakaiensis_for_Enhanced_PET_Biodegradation (accessed on 18 March 2025). [CrossRef]
- Najmi, S.M.; Schneider, D.A. Quorum sensing regulates rRNA synthesis in Saccharomyces cerevisiae. Gene 2021, 776, 145442. [Google Scholar] [CrossRef] [PubMed]
- Rafeeq, H.; Afsheen, N.; Rafique, S.; Arshad, A.; Intisar, M.; Hussain, A.; Bilal, M.; Iqbal, H.M. Genetically engineered microorganisms for environmental remediation. Chemosphere 2023, 310, 136751. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, M.; Li, Y.; Wang, J.; Chen, G.; Wang, J. Microbial bioremediation techniques of microplastics and nanoplastics in the marine environment. TrAC Trends Anal. Chem. 2024, 180, 117971. [Google Scholar] [CrossRef]
- Joho, Y.; Vongsouthi, V.; Gomez, C.; Larsen, J.S.; Ardevol, A.; Jackson, C.J. Improving plastic degrading enzymes via directed evolution. Protein Eng. Des. Sel. 2024, 37, gzae009. [Google Scholar] [CrossRef]
- Liu, S.; Narancic, T.; Davis, C.; O’Connor, K.E. CRISPR-Cas9 Editing of the Synthesis of Biodegradable Polyesters Polyhydroxyalkanaotes (PHA) in Pseudomonas Putida KT2440; Springer: Berlin/Heidelberg, Germany, 2022; pp. 341–358. [Google Scholar] [CrossRef]
- Geissdoerfer, M.; Savaget, P.; Bocken, N.M.P.; Hultink, E.J. The Circular Economy—A new sustainability paradigm? J. Clean. Prod. 2017, 143, 757–768. [Google Scholar] [CrossRef]
- Danso, D.; Chow, J.; Streita, W.R. Plastics: Environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 2019, 85, e01095-19. [Google Scholar] [CrossRef]
- Wei, R.; Oeser, T.; Schmidt, J.; Meier, R.; Barth, M.; Then, J.; Zimmermann, W. Engineered bacterial polyester hydrolases efficiently degrade polyethylene terephthalate due to relieved product inhibition. Biotechnol. Bioeng. 2016, 113, 1658–1665. [Google Scholar] [CrossRef]
- Ruggieri, A.; Braccini, A.M.; Poponi, S.; Mosconi, E.M. A meta-model of inter-organisational cooperation for the transition to a circular economy. Sustainability 2016, 8, 1153. [Google Scholar] [CrossRef]
- Alley, E.C.; Khimulya, G.; Biswas, S.; AlQuraishi, M.; Church, G.M. Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods 2019, 16, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Radivojević, T.; Costello, Z.; Workman, K.; Garcia Martin, H. A machine learning Automated Recommendation Tool for synthetic biology. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Alaghemandi, M. Sustainable Solutions Through Innovative Plastic Waste Recycling Technologies. Sustainability 2024, 16, 10401. [Google Scholar] [CrossRef]
- Yadav, K.; Nikalje, G.C. Comprehensive analysis of bioplastics: Life cycle assessment, waste management, biodiversity impact, and sustainable mitigation strategies. PeerJ 2024, 12, e18013. [Google Scholar] [CrossRef]
- Rüthi, J.; Cerri, M.; Brunner, I.; Stierli, B.; Sander, M.; Frey, B. Discovery of plastic-degrading microbial strains isolated from the alpine and Arctic terrestrial plastisphere. Front. Microbiol. 2023, 14, 1178474. [Google Scholar] [CrossRef]
- Qiao, Y.; Hu, R.; Chen, D.; Wang, L.; Wang, Z.; Yu, H.; Fu, Y.; Li, C.; Dong, Z.; Weng, Y.-X.; et al. Fluorescence-activated droplet sorting of PET degrading microorganisms. J. Hazard. Mater. 2022, 424, 127417. [Google Scholar] [CrossRef]
- Charnock, C. Norwegian Soils and Waters Contain Mesophilic, Plastic-Degrading Bacteria. Microorganisms 2021, 9, 94. [Google Scholar] [CrossRef]
- Viel, T.; Manfra, L.; Zupo, V.; Libralato, G.; Cocca, M.; Costantini, M. Biodegradation of Plastics Induced by Marine Organisms: Future Perspectives for Bioremediation Approaches. Polymers 2023, 15, 2673. [Google Scholar] [CrossRef]
- Zhai, X.; Zhang, X.H.; Yu, M. Microbial colonization and degradation of marine microplastics in the plastisphere: A review. Front. Microbiol. 2023, 14, 1127308. [Google Scholar] [CrossRef]
- Salinas, J.; Carpena, V.; Martínez-Gallardo, M.R.; Segado, M.; Estrella-González, M.J.; Toribio, A.J.; Jurado, M.M.; López-González, J.A.; Suárez-Estrella, F.; López, M.J. Development of plastic-degrading microbial consortia by induced selection in microcosms. Front. Microbiol. 2023, 14, 1143769. [Google Scholar] [CrossRef] [PubMed]
- Bao, T.; Qian, Y.; Xin, Y.; Collins, J.J.; Lu, T. Engineering microbial division of labor for plastic upcycling. Nat. Commun. 2023, 14, 5712. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Ye, Q.; Seo, Y.; Wei, N. Enzymatic Degradation of Polyethylene Terephthalate Plastics by Bacterial Curli Display PETase. Environ. Sci. Technol. Lett. 2022, 9, 650–657. [Google Scholar] [CrossRef]
- Ogunbayo, A.O.; Olanipekun, O.O.; Adamu, I.A.; Ogunbayo, A.O.; Olanipekun, O.O.; Adamu, I.A. Preliminary Studies on the Microbial Degradation of Plastic Waste Using Aspergillus niger and Pseudomonas sp. J. Environ. Prot. 2019, 10, 625–631. [Google Scholar] [CrossRef]
- Liu, J.; Wu, K. The Application of CRISPR Gene Editing Technology in PET Biodegradation. Highlights Sci. Eng. Technol. 2024, 91, 283–288. [Google Scholar] [CrossRef]
- Jung, H.-R.; Yang, S.-Y.; Moon, Y.-M.; Choi, T.-R.; Song, H.-S.; Bhatia, S.K.; Gurav, R.; Kim, E.-J.; Kim, B.-G.; Yang, Y.-H. Construction of Efficient Platform Escherichia coli Strains for Polyhydroxyalkanoate Production by Engineering Branched Pathway. Polymers 2019, 11, 509. [Google Scholar] [CrossRef]
- Maurya, A.; Bhattacharya, A.; Khare, S.K. Enzymatic Remediation of Polyethylene Terephthalate (PET)–Based Polymers for Effective Management of Plastic Wastes: An Overview. Front. Bioeng. Biotechnol. 2020, 8, 602325. [Google Scholar] [CrossRef] [PubMed]
- Yeung, C.W.S.; Teo, J.Y.Q.; Loh, X.J.; Lim, J.Y.C. Polyolefins and Polystyrene as Chemical Resources for a Sustainable Future: Challenges, Advances, and Prospects. ACS Mater. Lett. 2021, 3, 1660–1676. [Google Scholar] [CrossRef]
- (PDF) Bioplastics and CRISPR/Cas 9 Mediated Gene Replacement to Overcome the Limitations of Bioplastics. Available online: https://www.researchgate.net/publication/344161522_Bioplastics_and_CRISPRCas_9_mediated_gene_replacement_to_overcome_the_limitations_of_bioplastics (accessed on 17 February 2025).
- Arora, Y.; Sharma, S.; Sharma, V. Microalgae in Bioplastic Production: A Comprehensive Review. Arab. J. Sci. Eng. 2023, 48, 7225–7241. [Google Scholar] [CrossRef]
- Zhang, B.; Cao, J. Improving and Streamlining Gene Editing in Yarrowia lipolytica via Integration of Engineered Cas9 Protein. J. Fungi 2024, 10, 63. [Google Scholar] [CrossRef]
- Hou, X.; Guo, X.; Zhang, Y.; Zhang, Q. CRISPR/Cas genome editing system and its application in potato. Front. Genet. 2023, 14, 1017388. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wang, X.; Chen, N.; Ji, B.; Qu, L. Trash to treasure: Converting plastic waste into a useful graphene foil. Nanoscale 2017, 9, 9089–9094. [Google Scholar] [CrossRef] [PubMed]
- Culligan, E.P.; Sleator, R.D. Editorial: From genes to species: Novel insights from metagenomics. Front. Microbiol. 2016, 7, 213940. [Google Scholar] [CrossRef] [PubMed]
- Shinwari, Z.K.; Tanveer, F.; Khalil, A.T. Ethical Issues Regarding CRISPR Mediated Genome Editing. Curr. Issues Mol. Biol. 2018, 26, 103–110. [Google Scholar] [CrossRef]
- Yang, Z.; Zeng, X.; Zhao, Y.; Chen, R. AlphaFold2 and its applications in the fields of biology and medicine. Signal Transduct. Target. Ther. 2023, 8, 115. [Google Scholar] [CrossRef]
- Fang, Y.; Wen, Y.; Dai, L.; Wang, C.H.; You, S.; Li, W. Artificial intelligence in plastic recycling and conversion: A review. Resour. Conserv. Recycl. 2025, 215, 108090. [Google Scholar] [CrossRef]

| Organism | Product | Pathway | Substrate | Reference |
|---|---|---|---|---|
| Streptomyces spp. | Polyhydroxyalkanoates (PHA) | Biodegradation of PET | Pre-treated post-consumer PET | [39] |
| Pseudomonas spp. | Biopolymers | Hydrolysis and fermentation | Various plastic-related substrates | [3] |
| Escherichia coli | Degradation products | Enzymatic hydrolysis | Polyethylene, polystyrene | [13] |
| Ideonella sakaiensis | Monomers of PET | PET degradation | Polyethylene terephthalate (PET) | [12] |
| Staphylococcus epidermidis Un-C2-8 | PET-degrading enzyme | Genetic engineering and expression | Polyethylene terephthalate (PET) | [40] |
| Kineococcus endophyticus Un-5 | PET-degrading enzyme | Genetic engineering and expression | Polyethylene terephthalate (PET) | [40] |
| Rhodococcus spp. | Hydrolytic enzymes | Biodegradation | Various plastics | [41] |
| Vibrio alginolyticus | Hydrolytic enzymes | Biodegradation | Polyvinyl alcohol, LDPE | [42] |
| Vibrio parahemolyticus | Hydrolytic enzymes | Biodegradation | Polyvinyl alcohol, LDPE | [42] |
| Bacillus pumilus | Hydrolytic enzymes | Biodegradation | Low-density polyethylene (LDPE) | [43] |
| Pseudomonas aeruginosa | Hydrolytic enzymes | Biodegradation | Linear low-density polyethylene (LLDPE) | [44] |
| Pseudomonas alloputida | Hydrolytic enzymes | Biodegradation | Linear low-density polyethylene (LLDPE) | [44] |
| Castellaniella denitrificans | Enzymatic degradation products | Ligninolytic degradation | Linear low-density polyethylene (LLDPE) | [44] |
| Debaryomyces hansenii | Enzymatic degradation products | Ligninolytic degradation | Linear low-density polyethylene (LLDPE) | [44] |
| Thermobifida fusca | Enzymatic degradation products | Ligninolytic degradation | Polyethylene terephthalate (PET) | [45] |
| Saccharomonospora viridis | Hydrolytic enzymes | Biodegradation | Polyethylene terephthalate (PET) | [45] |
| BIND-PETase | TPA and MHET | Enzymatic hydrolysis | Polyethylene terephthalate (PET) | [46] |
| Bacillus cereus | Hydrolytic enzymes | Biodegradation | Low-density polyethylene (LDPE) | [47] |
| Aspect | PlastiCRISPR-Based Bioplastic Production and Degradation | Traditional Methods (Petrochemical Plastics and Waste Management) |
|---|---|---|
| Source Material | Renewable sources (e.g., plant biomass, bacteria) | Fossil fuels (petroleum, natural gas) |
| Technology Core | CRISPR-Cas9 genetic engineering of microbes and plants | Chemical polymerization for plastics; mechanical or thermal processes for recycling |
| Production Efficiency | Enhanced through gene editing (e.g., increased yield of PHAs in E. coli) | Limited by the physical and chemical processes; resource-intensive |
| Biodegradability | High: Engineered microbes improve degradation; bioplastics are compostable | Low: Most synthetic plastics persist for centuries in the environment |
| Environmental Impact | Reduced: Less carbon emission, less plastic pollution, targeted degradation | High: Pollution, microplastic contamination, greenhouse gas emissions |
| Customization Flexibility | High: Genetic edits allow tuning for resilience, malleability, and degradation speed | Low: Chemical properties fixed post-production |
| Waste Management Role | Proactive: Microbes genetically enhanced to degrade plastics | Reactive: Relies on incineration, landfill, or recycling |
| Scalability | Emerging: Lab-scale and pilot projects growing; scaling requires bioengineering infrastructure | Mature infrastructure but costly and energy-intensive |
| Economic Feasibility | Currently higher costs; potential for decrease with innovation and policy support | Economically efficient (currently) but with hidden environmental costs |
| Sustainability Alignment | Strong: Supports circular economy and green transitions | Weak: Linear economy model (produce–consume–dispose) |
| Public and Regulatory Acceptance | Developing: Concerns about GMOs and biosafety in open ecosystems | Well established but facing increasing regulation due to environmental harm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palit, P.; Minkara, M.; Abida, M.; Marwa, S.; Sen, C.; Roy, A.; Pasha, M.R.; Mosae, P.S.; Saha, A.; Ferdoush, J. PlastiCRISPR: Genome Editing-Based Plastic Waste Management with Implications in Polyethylene Terephthalate (PET) Degradation. Biomolecules 2025, 15, 684. https://doi.org/10.3390/biom15050684
Palit P, Minkara M, Abida M, Marwa S, Sen C, Roy A, Pasha MR, Mosae PS, Saha A, Ferdoush J. PlastiCRISPR: Genome Editing-Based Plastic Waste Management with Implications in Polyethylene Terephthalate (PET) Degradation. Biomolecules. 2025; 15(5):684. https://doi.org/10.3390/biom15050684
Chicago/Turabian StylePalit, Puja, Maya Minkara, Maisha Abida, Safa Marwa, Chandrima Sen, Ayan Roy, Md Ridoan Pasha, Paulraj Selvakumar Mosae, Ayan Saha, and Jannatul Ferdoush. 2025. "PlastiCRISPR: Genome Editing-Based Plastic Waste Management with Implications in Polyethylene Terephthalate (PET) Degradation" Biomolecules 15, no. 5: 684. https://doi.org/10.3390/biom15050684
APA StylePalit, P., Minkara, M., Abida, M., Marwa, S., Sen, C., Roy, A., Pasha, M. R., Mosae, P. S., Saha, A., & Ferdoush, J. (2025). PlastiCRISPR: Genome Editing-Based Plastic Waste Management with Implications in Polyethylene Terephthalate (PET) Degradation. Biomolecules, 15(5), 684. https://doi.org/10.3390/biom15050684






