Unveiling the Genetic Diversity and Population Structure of the Endangered Fern Angiopteris fokiensis Through Genome Survey and Genomic SSR Markers
Abstract
1. Introduction
2. Materials and Methods
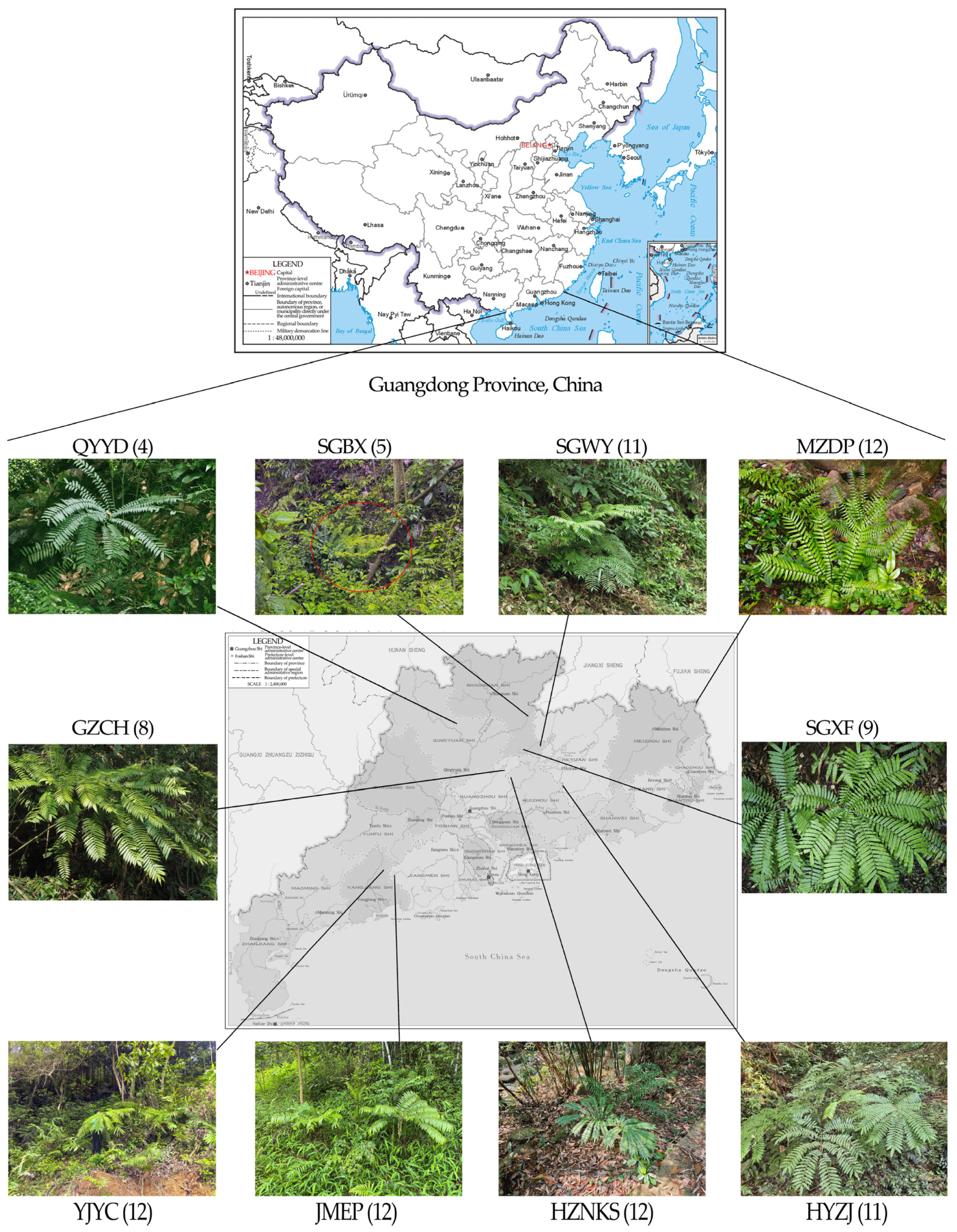
2.1. Sampling of Plant Materials
2.2. Genome Sequencing and Preliminary Assembly
2.3. Genome-Wide SSR Identification and Primer Design
2.4. SSR-PCR Analysis
2.5. Genetic Diversity Analysis
3. Results
3.1. Genome Survey and Preliminary Assembly
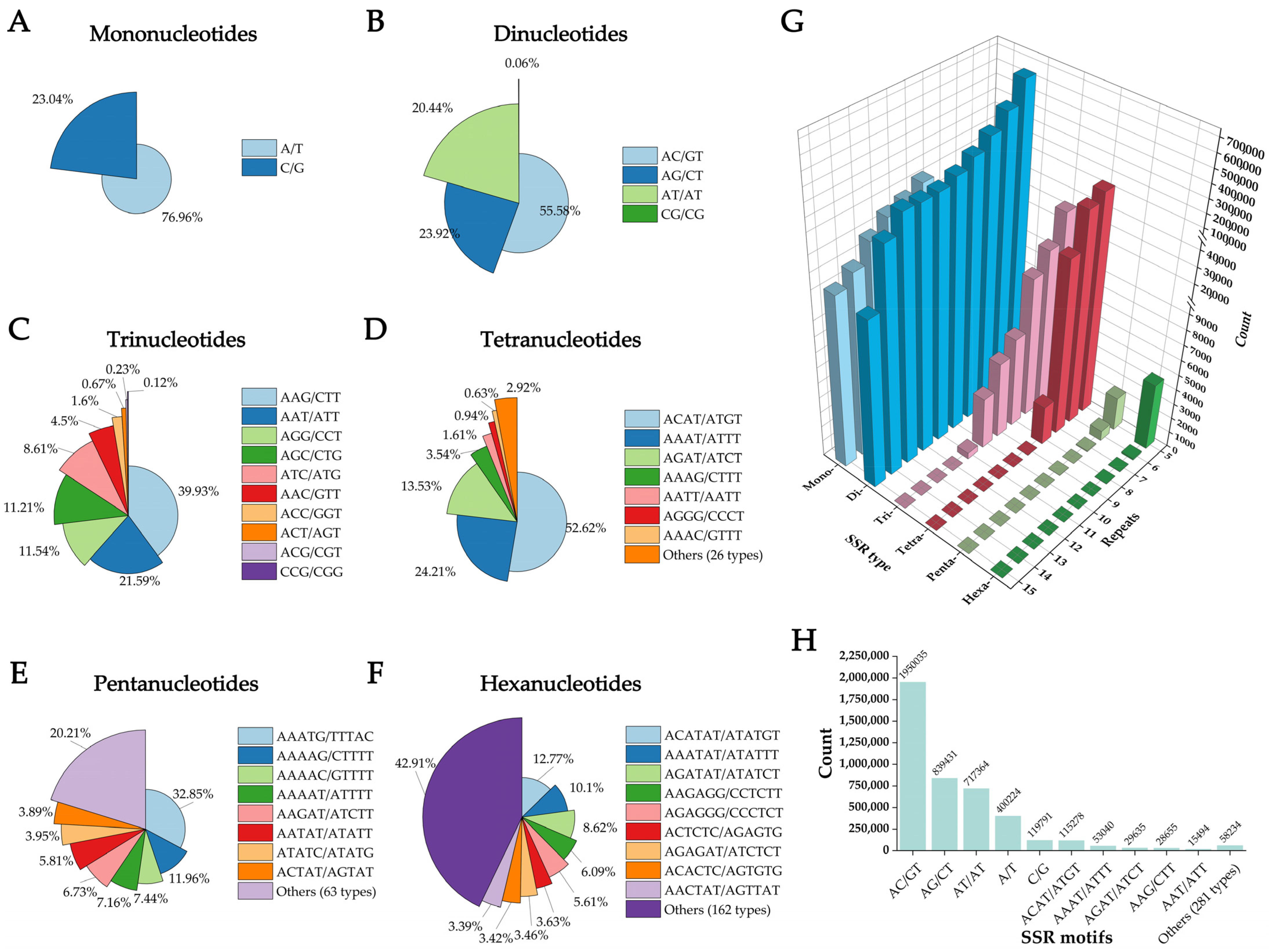
3.2. SSR Mining in the Genome of Angiopteris fokiensis
3.3. Population Genetic Diversity Based on Polymorphic SSR Markers
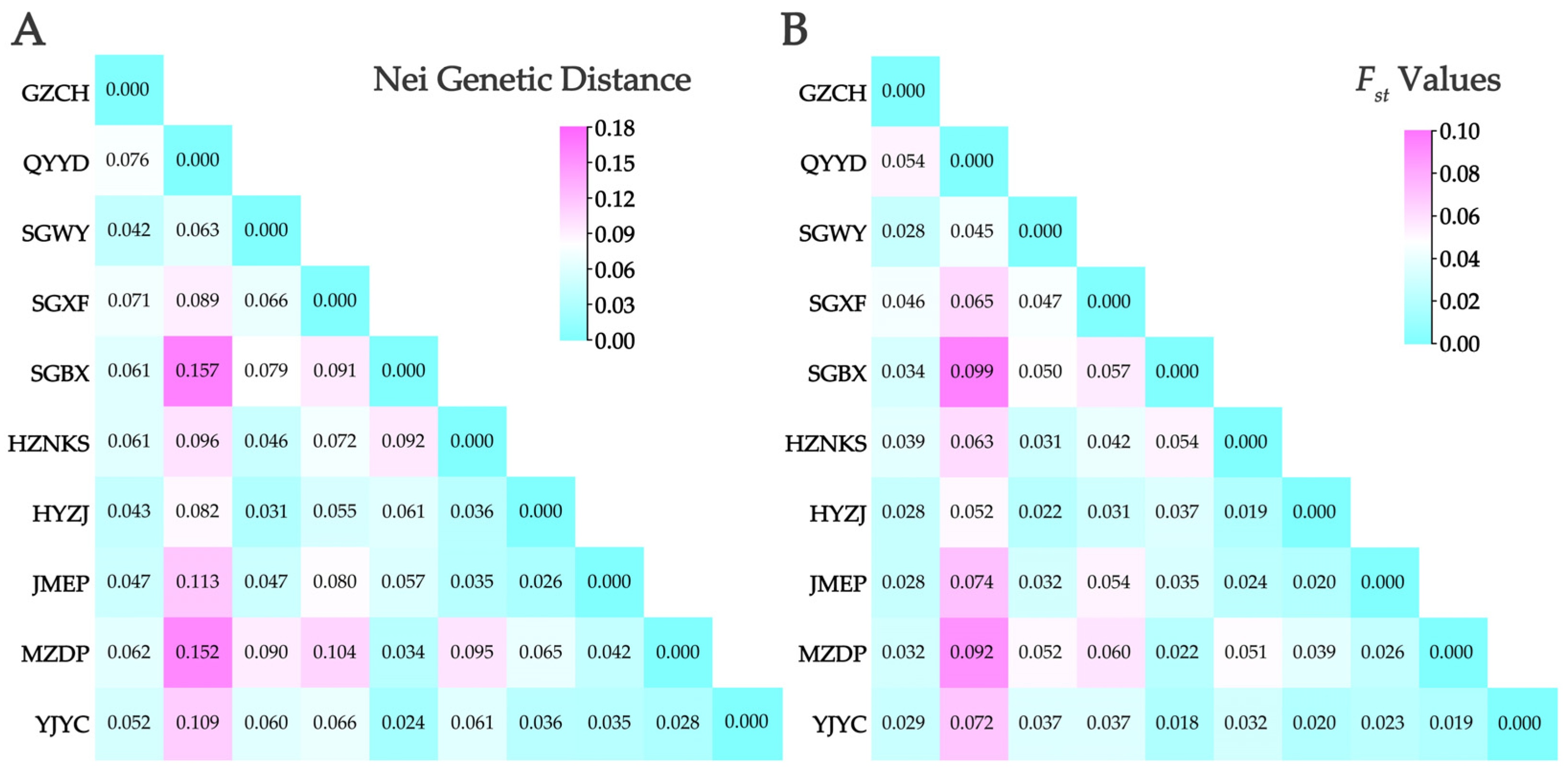
3.4. Population Genetic Differentiation
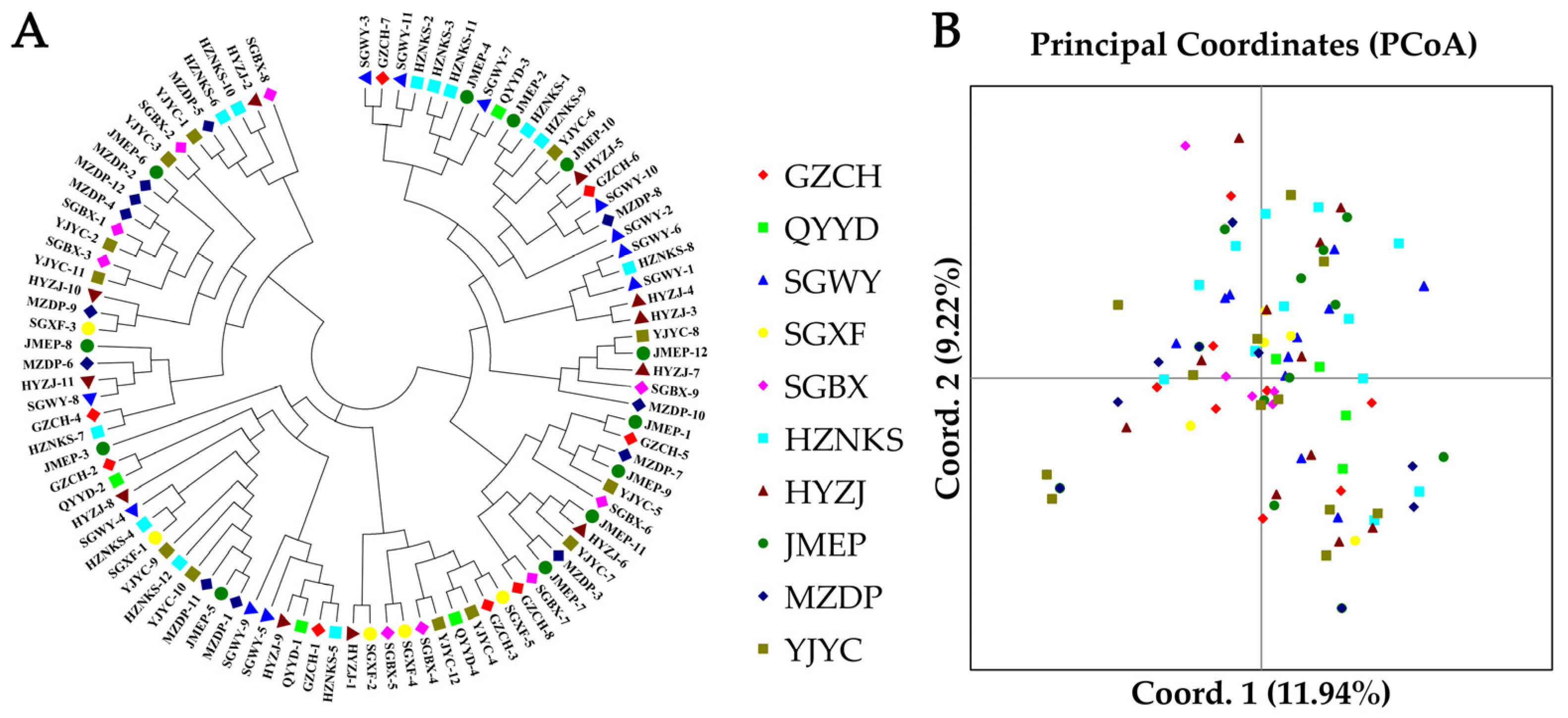
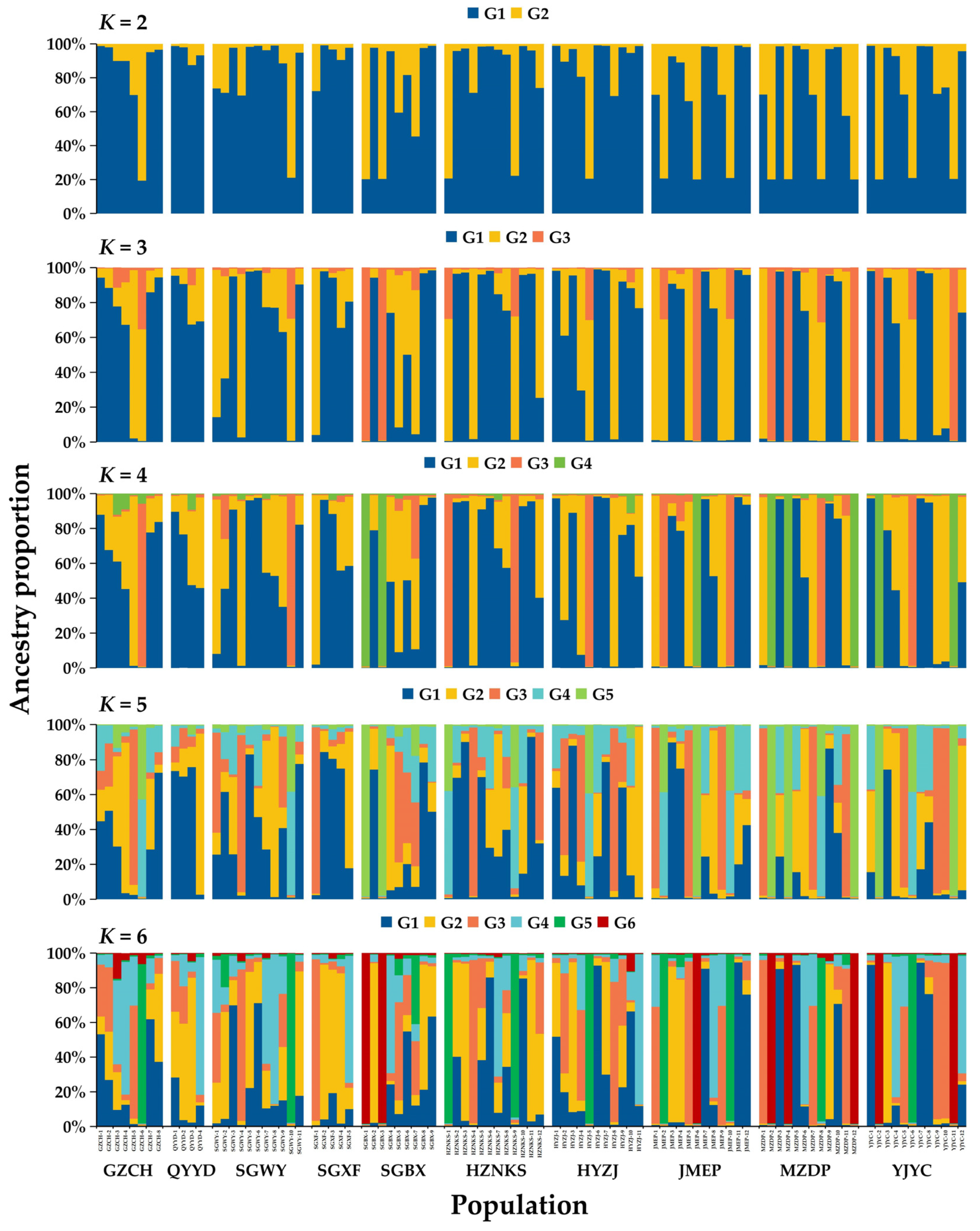
3.5. NJ Clustering, PCoA, and Genetic Structure Analysis
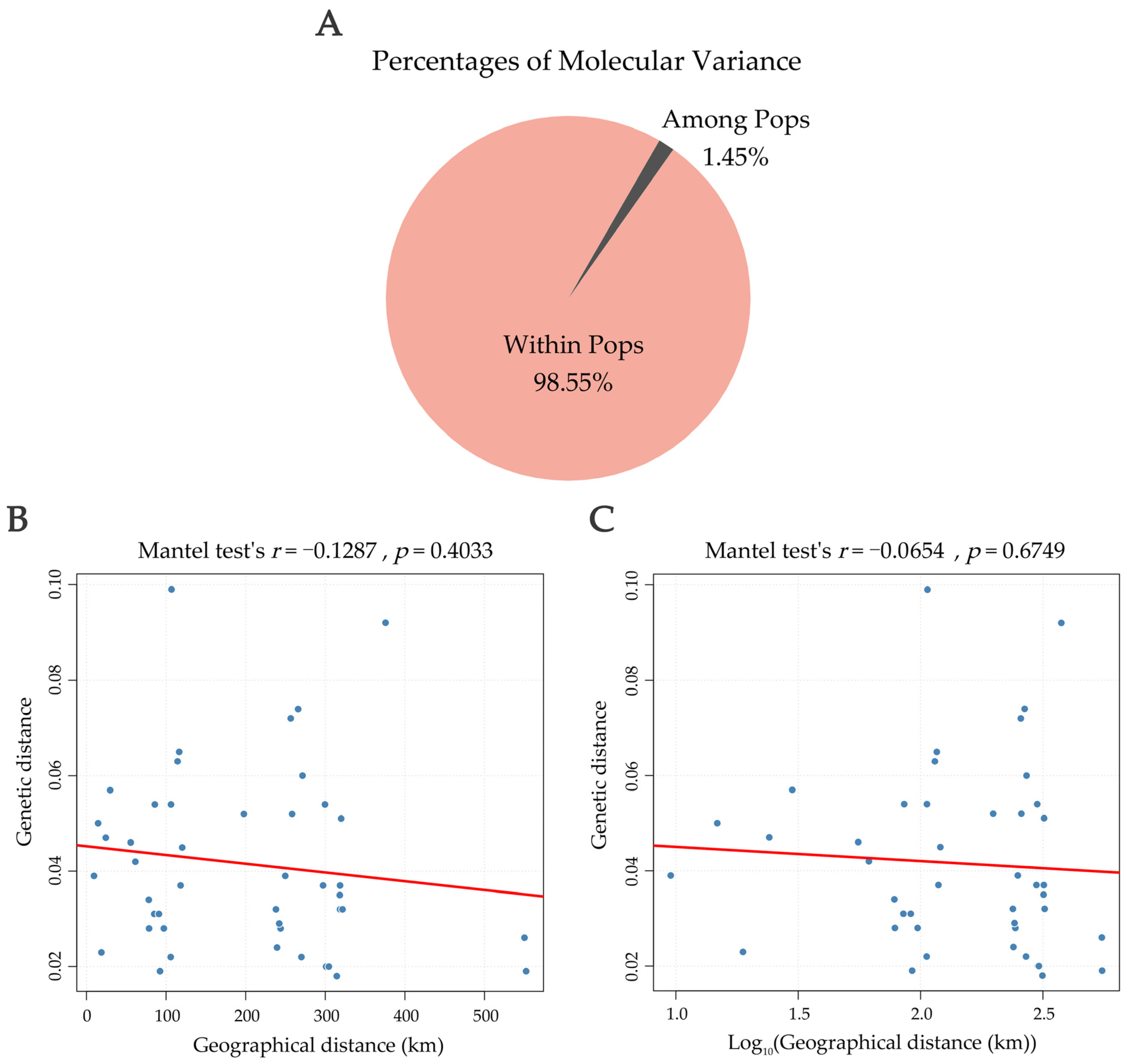
3.6. AMOVA and Correlation Between Genetic and Geographic Distances
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, T.; Xiao, B.; Liu, E.; Nguyen, K.S.; Duan, J.; Wang, K.; Yan, Y.; Xiang, J. Rediscovery of Angiopteris tonkinensis (Marattiaceae) after 100 years, and its revision. PhytoKeys 2020, 161, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.-H.; Chang, Y.-H.; Chiou, W.-L.; Huang, Y.-M. Chromosome numbers of five species of the Marattiaceae in Taiwan. Taiwan J. For. Sci. 2008, 23, 377–381. Available online: https://www.tfri.gov.tw/en/News_Content2.aspx?n=7589&s=15000 (accessed on 10 November 2025).
- Tsai, J.L.; Shieh, W.C. A cytotaxonomic survey of the Pteridophytes in Taiwan. J. Sci. Engin 1983, 20, 137–158. Available online: https://tpl.ncl.edu.tw/NclService/JournalContentDetail?SysId=A11043005 (accessed on 10 November 2025).
- Dong, S.-R.; Vasques, D.T. Notes on the typification of Angiopteris tamdaoensis and A. tonkinensis (Marattiaceae). Phytotaxa 2023, 622, 298–300. [Google Scholar] [CrossRef]
- Li, C.L.S. Phylogeny and divergence of Chinese Angiopteridaceae based on chloroplast dna sequence data (rbcl and trnl-f). Chin. Sci. Bull. 2007, 52, 91–97. [Google Scholar] [CrossRef]
- Yoshinaga, K.; Kubota, Y.; Ishii, T.; Wada, K. Nucleotide sequence of atpb, rbcl, trnr, dedb and psai chloroplast genes from a fern Angiopteris lygodiifolia: A possible emergence of spermatophyta lineage before the separation of bryophyta and pteridophyta. Plant Mol. Biol. 1992, 18, 79–82. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, X.; Fang, S.; Zhu, Z.; Li, Y.; Yu, H.; He, Z. Transcriptome-based study on the phylogeny and hybridization of marattialean ferns (Marattiaceae). Plants 2023, 12, 2237. [Google Scholar] [CrossRef]
- Jiang, Q.; Schneider, H.; Liu, H. Complete chloroplast genome of Angiopteris yunnanensis (Marattiaceae). Mitochondrial DNA. Part B Resour. 2019, 4, 3912–3913. [Google Scholar] [CrossRef]
- Roper, J.M.; Kellon Hansen, S.; Wolf, P.G.; Karol, K.G.; Mandoli, D.F.; Everett, K.D.E.; Kuehl, J.; Boore, J.L. The complete plastid genome sequence of Angiopteris evecta (G. Forst.) Hoffm. (Marattiaceae). Am. Fern J. 2007, 97, 95–106. [Google Scholar] [CrossRef]
- Chen, C.R.; Liao, Y.W.; Wu, H.T.; Shih, W.L.; Tzeng, C.Y.; Yang, S.Z.; Hernandez, C.E.; Chang, C.I. Triterpenoids from angiopteris palmiformis. Chem. Pharm. Bull. 2010, 58, 408–411. [Google Scholar] [CrossRef]
- Yu, Y.M.; Yang, J.S.; Peng, C.Z.; Caer, V.; Cong, P.Z.; Zou, Z.M.; Lu, Y.; Yang, S.Y.; Gu, Y.C. Lactones from Angiopteris caudatiformis. J. Nat. Prod. 2009, 72, 921–924. [Google Scholar] [CrossRef]
- Chen, Y.; Tao, Y.; Lian, X.; Wang, L.; Zhao, Y.; Jiang, J.; Zhang, Y. Chemical constituents of Angiopteris esculenta including two new natural lactones. Food Chem. 2010, 122, 1173–1175. [Google Scholar] [CrossRef]
- Jang, W.Y.; Lee, H.P.; Kim, S.A.; Huang, L.; Yoon, J.H.; Shin, C.Y.; Mitra, A.; Kim, H.G.; Cho, J.Y. Angiopteris cochinchinensis de vriese ameliorates lps-induced acute lung injury via src inhibition. Plants 2022, 11, 1306. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.R.; Omoloso, A.D. Antibacterial and antifungal activities of Angiopteris evecta. Fitoterapia 2008, 79, 366–369. [Google Scholar] [CrossRef]
- Lamichhane, R.; Pandeya, P.R.; Lee, K.H.; Kim, S.G.; Devkota, H.P.; Jung, H.J. Anti-adipogenic and anti-inflammatory activities of (-)-epi-osmundalactone and angiopteroside from Angiopteris helferiana C. Presl. Molecules 2020, 25, 1337. [Google Scholar] [CrossRef]
- Pandey, B.; Thapa, S.; Biradar, M.S.; Singh, B.; Ghale, J.B.; Kharel, P.; Jha, P.K.; Yadav, R.K.; Dawadi, S.; V, P. Lc-ms profiling and cytotoxic activity of angiopteris helferiana against hepg2 cell line: Molecular insight to investigate anticancer agent. PLoS ONE 2024, 19, e309797. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, G.L.; Yan, Y.H.; Sun, D.K.; Shu, J.-P.; Duan, J.-Q.; Zhao, G.-H.; Zhang, G.-S.; Xiang, J.-Y.; Wang, K.-L. Angiopteris sugongii (Marattiaceae), a new diploid species with transitional morphology between Angiopteris (sen. str.) and Archangiopteris. Phytotaxa 2021, 516, 276–282. [Google Scholar] [CrossRef]
- Wang, T.; Yang, T.; Zhang, J.; Zhang, G.; Yao, S.; Xiang, J.; Yan, Y.; Chen, H. Angiopteris nodosipetiolata (marattiaceae), a new fern species from yunnan, china. PhytoKeys 2024, 241, 177–189. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, S.H.; Lin, D.D.; Zhuang, Y.B.; Zhang, S.Y.; Xie, H.S. Analysis of the content of components in different extraction parts of Angiopteris fokiensis Hieron leaf extracts and their correlation with antioxidant activity. Sci. Technol. Food Ind. 2021, 42, 49–54. [Google Scholar]
- Shi, X.Y.; Feng, Y.Q.; Li, Y.P.; Luo, G.Y.; Zhang, J.J. Study on extraction optimization of antioxidants from roots of Angiopteris fokiensis Hieron. Sci. Technol. Food Ind. 2022, 43, 235–243. [Google Scholar]
- Yu, D.H.; Tang, X.J.; Xie, Z.G.; Liao, J.; Wen, C.R.; Wu, Q.F.; Li, P.; Yang, S.Y. Resource status and habitat of rare fern species Cibotium barometz and Angiopteris fokiensis in Leigong Mountain, Fujian Province. J. Anhui Agric. Sci. 2019, 47, 1–27. [Google Scholar]
- Vieira, M.L.; Santini, L.; Diniz, A.L.; Munhoz, C.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Geethanjali, S.; Kadirvel, P.; Anumalla, M.; Hemanth, S.N.; Annamalai, A.; Ali, J. Streamlining of simple sequence repeat data mining methodologies and pipelines for crop scanning. Plants 2024, 13, 2619. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wei, X.; Abbas, F.; Yu, Y.; Yu, R.; Fan, Y. Genome-wide identification of simple sequence repeats and assessment of genetic diversity in hedychium. J. Appl. Res. Med. Aromat. Plants 2021, 24, 100312. [Google Scholar] [CrossRef]
- Ye, Y.; Tan, J.; Lin, J.; Zhang, Y.; Zhu, G.; Nie, C.; Huang, L.; Zhou, Y.; Xu, Y. Genome-wide identification of ssr markers for curcuma alismatifolia gagnep., And their potential for wider application in this genus. J. Appl. Res. Med. Aromat. Plants 2024, 42, 100572. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one fastq preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Marcais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef]
- Liu, B.; Shi, Y.; Yuan, J.; Hu, X.; Zhang, H.; Li, N.; Li, Z.; Chen, Y.; Mu, D.; Fan, W. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. arXiv 2013. [Google Scholar] [CrossRef]
- Chikhi, R.; Rizk, G. Space-efficient and exact de bruijn graph representation based on a bloom filter. Algorithms Mol. Biol. 2013, 8, 22. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. Quast: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Munch, T.; Scholz, U.; Mascher, M. Misa-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ye, Y.; Zhu, G.; Xu, Y.; Tan, J.; Liu, J. Diversity, classification, and EST-SSR-based association analysis of caladium ornamental traits. Physiol. Plant. 2023, 175, e13841. [Google Scholar] [CrossRef] [PubMed]
- VAN Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Chapuis, M.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. Genalex 6.5: Genetic analysis in excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. Tassel: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. Powermarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B.M. Structure harvester: A website and program for visualizing structure output and implementing the evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. Clumpp: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Marchant, D.B.; Chen, G.; Cai, S.; Chen, F.; Schafran, P.; Jenkins, J.; Shu, S.; Plott, C.; Webber, J.; Lovell, J.T.; et al. Dynamic genome evolution in a model fern. Nat. Plants 2022, 8, 1038–1051. [Google Scholar] [CrossRef]
- Shu, J.; Zhang, Y.; Huang, T.; Yan, Y. The chromosome-level genome assembly of broad-leaf fern (Dipteris shenzhenensis). Sci. Data 2025, 12, 475–477. [Google Scholar] [CrossRef]
- Qin, G.; Pan, D.; Long, Y.; Lan, H.; Guan, D.; Song, J. Chromosome-scale genome of the fern Cibotium barometz unveils a genetic resource of medicinal value. Horticulturae 2024, 10, 1191. [Google Scholar] [CrossRef]
- Fang, Y.; Qin, X.; Liao, Q.; Du, R.; Luo, X.; Zhou, Q.; Li, Z.; Chen, H.; Jin, W.; Yuan, Y.; et al. The genome of homosporous maidenhair fern sheds light on the euphyllophyte evolution and defences. Nat. Plants 2022, 8, 1024–1037. [Google Scholar] [CrossRef]
- Huang, X.; Wang, W.; Gong, T.; Wickell, D.; Kuo, L.; Zhang, X.; Wen, J.; Kim, H.; Lu, F.; Zhao, H.; et al. Author correction: The flying spider-monkey tree fern genome provides insights into fern evolution and arborescence. Nat. Plants 2024, 10, 344. [Google Scholar] [CrossRef]
- Zhong, Y.; Liu, Y.; Wu, W.; Chen, J.; Sun, C.; Liu, H.; Shu, J.; Ebihara, A.; Yan, Y.; Zhou, R.; et al. Genomic insights into genetic diploidization in the homosporous fern Adiantum nelumboides. Genome Biol. Evol. 2022, 14, evac127. [Google Scholar] [CrossRef]
- Rahmatpour, N.; Kuo, L.Y.; Kang, J.; Herman, E.; Lei, L.; Li, M.; Srinivasan, S.; Zipper, R.; Wolniak, S.M.; Delwiche, C.F.; et al. Analyses of Marsilea vestita genome and transcriptomes do not support widespread intron retention during spermatogenesis. New Phytol. 2023, 237, 1490–1494. [Google Scholar] [CrossRef]
- Wu, Q.; Zang, F.; Ma, Y.; Zheng, Y.; Zang, D. Analysis of genetic diversity and population structure in endangered populus wulianensis based on 18 newly developed est-ssr markers. Glob. Ecol. Conserv. 2020, 24, e1329. [Google Scholar] [CrossRef]
- Khan, S.; Al-Qurainy, F.; Nadeem, M. Biotechnological approaches for conservation and improvement of rare and endangered plants of saudi arabia. Saudi J. Biol. Sci. 2012, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Zhu, L.; Zhao, M.; Li, C.; Zhang, Y.; Li, L. The first transcriptome sequencing and analysis of the endangered plant species picea neoveitchii mast. And potential EST-SSR markers development. Biotechnol. Biotechnol. Equip. 2019, 33, 967–973. [Google Scholar] [CrossRef]
- Tian, Z.Q.; Duan, F.; Li, S.; Gan, X. The development of ssr markers from the endangered plant tetracentron sinense oliv. (Tetracentraceae) based on rad–seq technique. Biológia 2023, 78, 15–22. [Google Scholar] [CrossRef]
- Yan, Z.; Zhu, S.; Wang, C.; Feng, Y.; Lei, Z.; Liu, X.; Zheng, F.; Jiang, W. Development of EST-SSR markers and population genetic analysis of Hemsleya zhejiangensis, an endangered species endemic to eastern china. Plant Mol. Biol. Rep. 2024, 42, 550–556. [Google Scholar] [CrossRef]
- Majeed, A.; Singh, A.; Choudhary, S.; Bhardwaj, P. Transcriptome characterization and development of functional polymorphic ssr marker resource for himalayan endangered species, Taxus contorta (Griff). Ind. Crop. Prod. 2019, 140, 111600. [Google Scholar] [CrossRef]
- Booy, G.; Hendriks, R.J.J.; Smulders, M.J.M.; Van Groenendael, J.M.; Vosman, B. Genetic diversity and the survival of populations. Plant Biol. 2000, 2, 379–395. [Google Scholar] [CrossRef]
- Hu, L.; Wang, J.; Wang, X.; Zhang, D.; Sun, Y.; Lu, T.; Shi, W. Development of SSR markers and evaluation of genetic diversity of endangered plant Saussurea involucrata. Biomolecules 2024, 14, 1010. [Google Scholar] [CrossRef]
- Storfer, A. Gene flow and endangered species translocations: A topic revisited. Biol. Conserv. 1999, 87, 173–180. [Google Scholar] [CrossRef]
- Szczecinska, M.; Sramko, G.; Wolosz, K.; Sawicki, J. Genetic diversity and population structure of the rare and endangered plant species Pulsatilla patens (L.) Mill in east central europe. PLoS ONE 2016, 11, e0151730. [Google Scholar] [CrossRef]
- Spielman, D.; Brook, B.W.; Frankham, R.; Schaal, B.A. Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. USA 2004, 101, 15261–15264. [Google Scholar] [CrossRef]
| SSR Marker | Repeat Motif | Na | Ne | I | Ho | He | PIC |
|---|---|---|---|---|---|---|---|
| AfgSSR-3 | (GA)9 | 2.900 | 2.477 | 0.960 | 0.988 | 0.593 | 0.534 |
| AfgSSR-17 | (TC)6 | 2.200 | 1.412 | 0.423 | 0.055 | 0.245 | 0.263 |
| AfgSSR-21 | (AAG)9 | 6.100 | 4.474 | 1.603 | 0.878 | 0.769 | 0.794 |
| AfgSSR-35 | (AC)8 | 1.400 | 1.061 | 0.101 | 0.022 | 0.052 | 0.059 |
| AfgSSR-47 | (CT)6 | 3.800 | 1.844 | 0.807 | 0.262 | 0.419 | 0.446 |
| AfgSSR-55 | (AC)9 | 9.700 | 7.439 | 2.096 | 0.639 | 0.855 | 0.898 |
| AfgSSR-59 | (AC)8 | 2.300 | 1.897 | 0.691 | 0.424 | 0.463 | 0.397 |
| AfgSSR-93 | (CT)7 | 3.700 | 2.168 | 0.911 | 0.519 | 0.494 | 0.494 |
| AfgSSR-95 | (GA)9 | 4.000 | 2.275 | 0.973 | 0.484 | 0.524 | 0.512 |
| AfgSSR-131 | (TC)6 | 4.000 | 1.827 | 0.863 | 0.501 | 0.446 | 0.448 |
| AfgSSR-139 | (TC)7 | 3.400 | 2.549 | 1.011 | 0.752 | 0.595 | 0.609 |
| AfgSSR-141 | (CA)8 | 2.200 | 1.295 | 0.359 | 0.185 | 0.200 | 0.221 |
| AfgSSR-145 | (CT)6 | 3.200 | 2.351 | 0.940 | 0.584 | 0.567 | 0.482 |
| AfgSSR-185 | (TC)6 | 2.500 | 1.394 | 0.449 | 0.154 | 0.242 | 0.232 |
| AfgSSR-189 | (AC)6 | 2.200 | 1.319 | 0.371 | 0.174 | 0.210 | 0.223 |
| Mean | 3.573 | 2.385 | 0.837 | 0.441 | 0.445 | 0.441 | |
| Population | Na | Ne | I | Ho | He |
|---|---|---|---|---|---|
| GZCH | 3.333 | 2.287 | 0.802 | 0.417 | 0.433 |
| QYYD | 2.533 | 2.003 | 0.666 | 0.467 | 0.381 |
| SGWY | 3.533 | 2.270 | 0.799 | 0.412 | 0.425 |
| SGXF | 3.067 | 2.455 | 0.815 | 0.550 | 0.456 |
| SGBX | 3.467 | 2.218 | 0.811 | 0.452 | 0.433 |
| HZNKS | 3.933 | 2.348 | 0.879 | 0.396 | 0.454 |
| HYZJ | 3.933 | 2.661 | 0.924 | 0.425 | 0.479 |
| JMEP | 4.000 | 2.713 | 0.882 | 0.430 | 0.447 |
| MZDP | 4.067 | 2.620 | 0.926 | 0.428 | 0.482 |
| YJYC | 3.867 | 2.278 | 0.870 | 0.436 | 0.459 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Tan, J.; Huang, L.; Luo, Y.; Huang, S.; Ye, Y.; Xu, Y. Unveiling the Genetic Diversity and Population Structure of the Endangered Fern Angiopteris fokiensis Through Genome Survey and Genomic SSR Markers. Biomolecules 2025, 15, 1649. https://doi.org/10.3390/biom15121649
Zhou Y, Tan J, Huang L, Luo Y, Huang S, Ye Y, Xu Y. Unveiling the Genetic Diversity and Population Structure of the Endangered Fern Angiopteris fokiensis Through Genome Survey and Genomic SSR Markers. Biomolecules. 2025; 15(12):1649. https://doi.org/10.3390/biom15121649
Chicago/Turabian StyleZhou, Yiwei, Jianjun Tan, Lishan Huang, Yanyu Luo, Shaoli Huang, Yuanjun Ye, and Yechun Xu. 2025. "Unveiling the Genetic Diversity and Population Structure of the Endangered Fern Angiopteris fokiensis Through Genome Survey and Genomic SSR Markers" Biomolecules 15, no. 12: 1649. https://doi.org/10.3390/biom15121649
APA StyleZhou, Y., Tan, J., Huang, L., Luo, Y., Huang, S., Ye, Y., & Xu, Y. (2025). Unveiling the Genetic Diversity and Population Structure of the Endangered Fern Angiopteris fokiensis Through Genome Survey and Genomic SSR Markers. Biomolecules, 15(12), 1649. https://doi.org/10.3390/biom15121649







