Cleaving Expectations: A Review of Proteasome Functional and Catalytic Diversity
Abstract
1. Introduction
2. Tracing the Discovery of Proteasome Catalytic Activities
3. From Uniform to Specialized: Evolution of Proteasome Catalytic Sites
4. Proteasome Variants and Regulatory Particles: Fine-Tuning Degradation for Specialized Functions
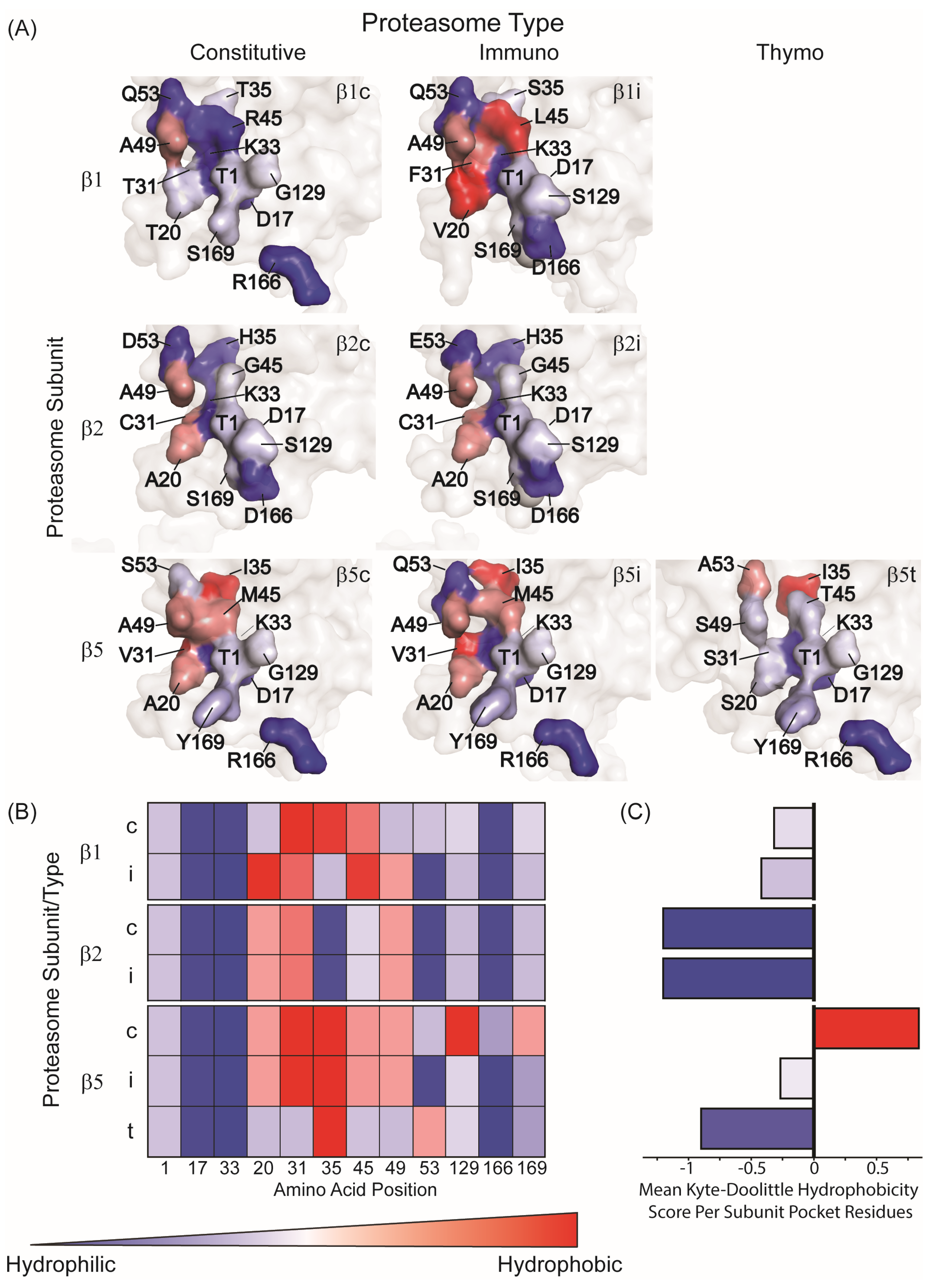
4.1. Immunoproteasome
4.2. Intermediate Proteasome
4.3. Thymoproteasome
4.4. Spermatoproteasome
5. Diversification and Loss in Higher Eukaryotes: Adaptive Evolution of Catalytic Subunits
6. Association with Regulatory Particles
6.1. 19S Regulatory Particle
6.2. PA28αβ
6.3. PA28γ
6.4. PA200
6.5. PI31
7. Tuning Proteasome Activity Through Post-Translational Modifications
8. Proteasome Dynamics Across Physiological Conditions
8.1. Differentiation
8.2. Inflammation
8.3. Oxidative Stress
8.4. Aging
9. Computational Prediction of Proteasomal Cleavage Sites
10. Targeting the Proteasome Catalytic Sites: Chronological Advances and Clinical Applications
11. Summary and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zwickl, P.; Seemüller, E.; Kapelari, B.; Baumeister, W. The proteasome: A supramolecular assembly designed for controlled proteolysis. Adv. Protein Chem. 2001, 59, 187–222. [Google Scholar] [CrossRef]
- Schmidt, M.; Finley, D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 2014, 1843, 13–25. [Google Scholar] [CrossRef]
- Sherman, D.J.; Li, J. Proteasome Inhibitors: Harnessing Proteostasis to Combat Disease. Molecules 2020, 25, 671. [Google Scholar] [CrossRef]
- Brennan, A.; Church, T.R.; Margolis, S.S. Proteasome-derived peptides: Separating the trash from the recycling. Trends Biochem. Sci. 2025, 50, 779–794. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.H. Regulation of Protein Degradation by Proteasomes in Cancer. J. Cancer Prev. 2018, 23, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Turker, F.; Brennan, A.; Margolis, S.S. Neuronal membrane proteasome-derived peptides modulate NMDAR-dependent neuronal signaling to promote changes in gene expression. Mol. Biol. Cell 2024, 35, ar6. [Google Scholar] [CrossRef]
- He, H.Y.; Ahsan, A.; Bera, R.; McLain, N.; Faulkner, R.; Ramachandran, K.V.; Margolis, S.S.; Cline, H.T. Neuronal membrane proteasomes regulate neuronal circuit activity in vivo and are required for learning-induced behavioral plasticity. Proc. Natl. Acad. Sci. USA 2023, 120, e2216537120. [Google Scholar] [CrossRef]
- de Araujo, C.B.; Heimann, A.S.; Remer, R.A.; Russo, L.C.; Colquhoun, A.; Forti, F.L.; Ferro, E.S. Intracellular Peptides in Cell Biology and Pharmacology. Biomolecules 2019, 9, 150. [Google Scholar] [CrossRef]
- Chen, S.-H.; Prakash, S.; Helgason, E.; Gilchrist, C.L.; Kenner, L.R.; Srinivasan, R.; Sterne-Weiler, T.; Hafner, M.; Piskol, R.; Dueber, E.C.; et al. Constitutive protein degradation induces acute cell death via proteolysis products. bioRxiv 2023. [Google Scholar] [CrossRef]
- Silke, J.; Meier, P. Inhibitor of apoptosis (IAP) proteins-modulators of cell death and inflammation. Cold Spring Harb. Perspect. Biol. 2013, 5, a008730. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, K.; Lobov, A.; Antonello, P.; Shmueli, M.D.; Yakir, I.; Weizman, T.; Ulman, A.; Sheban, D.; Laser, E.; Kramer, M.P.; et al. Cell-autonomous innate immunity by proteasome-derived defence peptides. Nature 2025, 639, 1032–1041. [Google Scholar] [CrossRef] [PubMed]
- Kanarek, N.; Ben-Neriah, Y. Regulation of NF-κB by ubiquitination and degradation of the IκBs. Immunol. Rev. 2012, 246, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Baumeister, W.; Walz, J.; Zühl, F.; Seemüller, E. The Proteasome: Paradigm of a Self-Compartmentalizing Protease. Cell 1998, 92, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Kumar Deshmukh, F.; Yaffe, D.; Olshina, M.A.; Ben-Nissan, G.; Sharon, M. The Contribution of the 20S Proteasome to Proteostasis. Biomolecules 2019, 9, 190. [Google Scholar] [CrossRef]
- Ben-Nissan, G.; Sharon, M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules 2014, 4, 862–884. [Google Scholar] [CrossRef]
- Howell, L.A.; Tomko, R.J.; Kusmierczyk, A.R. Putting it all together: Intrinsic and extrinsic mechanisms governing proteasome biogenesis. Front. Biol. 2017, 12, 19–48. [Google Scholar] [CrossRef]
- Kisselev, A.F.; Akopian, T.N.; Castillo, V.; Goldberg, A.L. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol. Cell 1999, 4, 395–402. [Google Scholar] [CrossRef]
- Orlowski, M.; Michaud, C. Pituitary multicatalytic proteinase complex. Specificity of components and aspects of proteolytic activity. Biochemistry 1989, 28, 9270–9278. [Google Scholar] [CrossRef]
- Wilk, S.; Orlowski, M. Evidence that pituitary cation-sensitive neutral endopeptidase is a multicatalytic protease complex. J. Neurochem. 1983, 40, 842–849. [Google Scholar] [CrossRef]
- Stein, R.L.; Melandri, F.; Dick, L. Kinetic Characterization of the Chymotryptic Activity of the 20S Proteasome. Biochemistry 1996, 35, 3899–3908. [Google Scholar] [CrossRef]
- Maupin-Furlow, J.A.; Kaczowka, S.J.; Ou, M.S.; Wilson, H.L. Archaeal proteasomes: Proteolytic nanocompartments of the cell. In Advances in Applied Microbiology; Academic Press: Cambridge, MA, USA, 2001; Volume 50, pp. 279–338. [Google Scholar]
- Cardozo, C.; Kohanski, R.A. Altered Properties of the Branched Chain Amino Acid-preferring Activity Contribute to Increased Cleavages after Branched Chain Residues by the “Immunoproteasome” *. J. Biol. Chem. 1998, 273, 16764–16770. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Heinemeyer, W.; Jäger, S.; Ullrich, T.; Bochtler, M.; Wolf, D.H.; Huber, R. The catalytic sites of 20S proteasomes and their role in subunit maturation: A mutational and crystallographic study. Proc. Natl. Acad. Sci. USA 1999, 96, 10976–10983. [Google Scholar] [CrossRef] [PubMed]
- Löwe, J.; Stock, D.; Jap, B.; Zwickl, P.; Baumeister, W.; Huber, R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 1995, 268, 533–539. [Google Scholar] [CrossRef]
- Groll, M.; Ditzel, L.; Löwe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 1997, 386, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Seemüller, E.; Lupas, A.; Zühl, F.; Zwickl, P.; Baumeister, W. The proteasome from Thermoplasma acidophilum is neither a cysteine nor a serine protease. FEBS Lett. 1995, 359, 173–178. [Google Scholar] [CrossRef]
- Heinemeyer, W.; Fischer, M.; Krimmer, T.; Stachon, U.; Wolf, D.H. The Active Sites of the Eukaryotic 20 S Proteasome and Their Involvement in Subunit Precursor Processing*. J. Biol. Chem. 1997, 272, 25200–25209. [Google Scholar] [CrossRef]
- Winter, M.B.; La Greca, F.; Arastu-Kapur, S.; Caiazza, F.; Cimermancic, P.; Buchholz, T.J.; Anderl, J.L.; Ravalin, M.; Bohn, M.F.; Sali, A.; et al. Immunoproteasome functions explained by divergence in cleavage specificity and regulation. eLife 2017, 6, e27364. [Google Scholar] [CrossRef]
- Sahu, I.; Glickman, M.H. Structural Insights into Substrate Recognition and Processing by the 20S Proteasome. Biomolecules 2021, 11, 148. [Google Scholar] [CrossRef]
- Kisselev, A.F.; Goldberg, A.L. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 2005, 398, 364–378. [Google Scholar] [CrossRef]
- Fajtova, P.; Hurysz, B.M.; Miyamoto, Y.; Serafim, M.S.M.; Jiang, Z.; Vazquez, J.M.; Trujillo, D.F.; Liu, L.J.; Somani, U.; Almaliti, J.; et al. Distinct substrate specificities of the three catalytic subunits of the Trichomonas vaginalis proteasome. Protein Sci. 2024, 33, e5225. [Google Scholar] [CrossRef]
- Rohweder, P.J.; Jiang, Z.; Hurysz, B.M.; O’Donoghue, A.J.; Craik, C.S. Chapter Twelve—Multiplex substrate profiling by mass spectrometry for proteases. In Methods in Enzymology; Shukla, A.K., Ed.; Academic Press: Cambridge, MA, USA, 2023; Volume 682, pp. 375–411. [Google Scholar]
- Wolf-Levy, H.; Javitt, A.; Eisenberg-Lerner, A.; Kacen, A.; Ulman, A.; Sheban, D.; Dassa, B.; Fishbain-Yoskovitz, V.; Carmona-Rivera, C.; Kramer, M.P.; et al. Revealing the cellular degradome by mass spectrometry analysis of proteasome-cleaved peptides. Nat. Biotechnol. 2018, 36, 1110–1116. [Google Scholar] [CrossRef]
- Zittlau, K.I.; Zachor-Movshovitz, D.; Leushkin, Y.; Schimmel Brener, R.; Morgenstern, D.; Ben-Nissan, G.; Sharon, M. Tracking proteasome degradation: A cross-organ analysis via intact degradomics mass spectrometry. Proc. Natl. Acad. Sci. USA 2025, 122, e2419607122. [Google Scholar] [CrossRef]
- Akopian, T.N.; Kisselev, A.F.; Goldberg, A.L. Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J. Biol. Chem. 1997, 272, 1791–1798. [Google Scholar] [CrossRef][Green Version]
- Venkatraman, P.; Wetzel, R.; Tanaka, M.; Nukina, N.; Goldberg, A.L. Eukaryotic Proteasomes Cannot Digest Polyglutamine Sequences and Release Them during Degradation of Polyglutamine-Containing Proteins. Mol. Cell 2004, 14, 95–104. [Google Scholar] [CrossRef]
- Zuhl, F.; Tamura, T.; Dolenc, I.; Cejka, Z.; Nagy, I.; DeMot, R.; Baumeister, W. Subunit topology of the Rhodococcus proteasome. FEBS Lett. 1997, 400, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Nagy, I.; Lupas, A.; Lottspeich, F.; Cejka, Z.; Schoofs, G.; Tanaka, K.; De Mot, R.; Baumeister, W. The first characterization of a eubacterial proteasome: The 20S complex of Rhodococcus. Curr. Biol. 1995, 5, 766–774. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, M.; Flajnik, M.F. Origin and evolution of the specialized forms of proteasomes involved in antigen presentation. Immunogenetics 2019, 71, 251–261. [Google Scholar] [CrossRef]
- Muller, A.U.; Weber-Ban, E. The Bacterial Proteasome at the Core of Diverse Degradation Pathways. Front. Mol. Biosci. 2019, 6, 23. [Google Scholar] [CrossRef]
- Bos, D.H. Natural selection during functional divergence to LMP7 and proteasome subunit X (PSMB5) following gene duplication. J. Mol. Evol. 2005, 60, 221–228. [Google Scholar] [CrossRef]
- van Deventer, S.; Neefjes, J. The Immunoproteasome Cleans up after Inflammation. Cell 2010, 142, 517–518. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sutoh, Y.; Kondo, M.; Ohta, Y.; Ota, T.; Tomaru, U.; Flajnik, M.F.; Kasahara, M. Comparative genomic analysis of the proteasome beta5t subunit gene: Implications for the origin and evolution of thymoproteasomes. Immunogenetics 2012, 64, 49–58. [Google Scholar] [CrossRef]
- Basler, M.; Groettrup, M. On the Role of the Immunoproteasome in Protein Homeostasis. Cells 2021, 10, 3216. [Google Scholar] [CrossRef] [PubMed]
- Mishto, M.; Liepe, J.; Textoris-Taube, K.; Keller, C.; Henklein, P.; Weberruss, M.; Dahlmann, B.; Enenkel, C.; Voigt, A.; Kuckelkorn, U.; et al. Proteasome isoforms exhibit only quantitative differences in cleavage and epitope generation. Eur. J. Immunol. 2014, 44, 3508–3521. [Google Scholar] [CrossRef] [PubMed]
- Ferrington, D.A.; Gregerson, D.S. Immunoproteasomes: Structure, function, and antigen presentation. Prog. Mol. Biol. Transl. Sci. 2012, 109, 75–112. [Google Scholar] [CrossRef]
- Huber, E.M.; Basler, M.; Schwab, R.; Heinemeyer, W.; Kirk, C.J.; Groettrup, M.; Groll, M. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 2012, 148, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Abi Habib, J.; De Plaen, E.; Stroobant, V.; Zivkovic, D.; Bousquet, M.P.; Guillaume, B.; Wahni, K.; Messens, J.; Busse, A.; Vigneron, N.; et al. Efficiency of the four proteasome subtypes to degrade ubiquitinated or oxidized proteins. Sci. Rep. 2020, 10, 15765. [Google Scholar] [CrossRef]
- Kincaid, E.Z.; Che, J.W.; York, I.; Escobar, H.; Reyes-Vargas, E.; Delgado, J.C.; Welsh, R.M.; Karow, M.L.; Murphy, A.J.; Valenzuela, D.M.; et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat. Immunol. 2011, 13, 129–135. [Google Scholar] [CrossRef]
- Chen, B.; Zhu, H.; Yang, B.; Cao, J. The dichotomous role of immunoproteasome in cancer: Friend or foe? Acta Pharm. Sin. B 2023, 13, 1976–1989. [Google Scholar] [CrossRef]
- Angeles, A.; Fung, G.; Luo, H. Immune and non-immune functions of the immunoproteasome. Front. Biosci. (Landmark Ed.) 2012, 17, 1904–1916. [Google Scholar] [CrossRef]
- Ding, Q.; Martin, S.; Dimayuga, E.; Bruce-Keller, A.J.; Keller, J.N. LMP2 Knock-Out Mice Have Reduced Proteasome Activities and Increased Levels of Oxidatively Damaged Proteins. Antioxid. Redox Signal. 2006, 8, 130–135. [Google Scholar] [CrossRef]
- Kitamura, A.; Maekawa, Y.; Uehara, H.; Izumi, K.; Kawachi, I.; Nishizawa, M.; Toyoshima, Y.; Takahashi, H.; Standley, D.M.; Tanaka, K.; et al. A mutation in the immunoproteasome subunit PSMB8 causes autoinflammation and lipodystrophy in humans. J. Clin. Investig. 2011, 121, 4150–4160. [Google Scholar] [CrossRef]
- Muchamuel, T.; Basler, M.; Aujay, M.A.; Suzuki, E.; Kalim, K.W.; Lauer, C.; Sylvain, C.; Ring, E.R.; Shields, J.; Jiang, J.; et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat. Med. 2009, 15, 781–787. [Google Scholar] [CrossRef]
- Moebius, J.; van den Broek, M.; Groettrup, M.; Basler, M. Immunoproteasomes are essential for survival and expansion of T cells in virus-infected mice. Eur. J. Immunol. 2010, 40, 3439–3449. [Google Scholar] [CrossRef] [PubMed]
- Astakhova, T.M.; Karpov, N.S.; Dashenkova, N.O.; Alpeeva, E.V.; Nesterchuk, M.V.; Akopov, S.B.; Mikaelyan, A.S.; Ryabchenko, A.S.; Erokhov, P.A.; Sharova, N.P. Inhibition of Proteasome LMP2 Activity Suppresses Chil3 Expression in Mouse Colon Adenocarcinoma Tissue and Restrains Tumor Growth. Oncol. Res. 2025, 33, 2573–2595. [Google Scholar] [CrossRef] [PubMed]
- Johnston-Carey, H.K.; Pomatto, L.C.D.; Davies, K.J.A. The Immunoproteasome in oxidative stress, aging, and disease. Crit. Rev. Biochem. Mol. 2016, 51, 268–281. [Google Scholar] [CrossRef]
- Watanabe, A.; Yashiroda, H.; Ishihara, S.; Lo, M.; Murata, S. The Molecular Mechanisms Governing the Assembly of the Immuno- and Thymoproteasomes in the Presence of Constitutive Proteasomes. Cells 2022, 11, 1580. [Google Scholar] [CrossRef]
- Klare, N.; Seeger, M.; Janek, K.; Jungblut, P.R.; Dahlmann, B. Intermediate-type 20 S proteasomes in HeLa cells: “asymmetric” subunit composition, diversity and adaptation. J. Mol. Biol. 2007, 373, 1–10. [Google Scholar] [CrossRef]
- van den Eshof, B.L.; Medfai, L.; Nolfi, E.; Wawrzyniuk, M.; Sijts, A. The Function of Immunoproteasomes-An Immunologists’ Perspective. Cells 2021, 10, 3360. [Google Scholar] [CrossRef]
- Fabre, B.; Lambour, T.; Garrigues, L.; Ducoux-Petit, M.; Amalric, F.; Monsarrat, B.; Burlet-Schiltz, O.; Bousquet-Dubouch, M.P. Label-free quantitative proteomics reveals the dynamics of proteasome complexes composition and stoichiometry in a wide range of human cell lines. J. Proteome Res. 2014, 13, 3027–3037. [Google Scholar] [CrossRef] [PubMed]
- Guillaume, B.; Chapiro, J.; Stroobant, V.; Colau, D.; Van Holle, B.; Parvizi, G.; Bousquet-Dubouch, M.P.; Théate, I.; Parmentier, N.; Van den Eynde, B.J. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc. Natl. Acad. Sci. USA 2010, 107, 18599–18604. [Google Scholar] [CrossRef]
- Freudenburg, W.; Gautam, M.; Chakraborty, P.; James, J.; Richards, J.; Salvatori, A.S.; Baldwin, A.; Schriewer, J.; Buller, R.M.; Corbett, J.A.; et al. Reduction in ATP levels triggers immunoproteasome activation by the 11S (PA28) regulator during early antiviral response mediated by IFNβ in mouse pancreatic β-cells. PLoS ONE 2013, 8, e52408. [Google Scholar] [CrossRef]
- Vigneron, N.; Abi Habib, J.; Van den Eynde, B.J. The capture proteasome assay: A method to measure proteasome activity in vitro. Anal. Biochem. 2015, 482, 7–15. [Google Scholar] [CrossRef]
- Khilji, M.S.; Verstappen, D.; Dahlby, T.; Burstein Prause, M.C.; Pihl, C.; Bresson, S.E.; Bryde, T.H.; Keller Andersen, P.A.; Klindt, K.; Zivkovic, D.; et al. The intermediate proteasome is constitutively expressed in pancreatic beta cells and upregulated by stimulatory, low concentrations of interleukin 1 beta. PLoS ONE 2020, 15, e0222432. [Google Scholar] [CrossRef]
- Tanaka, K. The proteasome: From basic mechanisms to emerging roles. Keio J. Med. 2013, 62, 1–12. [Google Scholar] [CrossRef]
- Murata, S.; Sasaki, K.; Kishimoto, T.; Niwa, S.; Hayashi, H.; Takahama, Y.; Tanaka, K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 2007, 316, 1349–1353. [Google Scholar] [CrossRef]
- Kamber Kaya, H.E.; Radhakrishnan, S.K. Trash Talk: Mammalian Proteasome Regulation at the Transcriptional Level. Trends Genet. 2021, 37, 160–173. [Google Scholar] [CrossRef]
- Murata, S.; Takahama, Y.; Tanaka, K. Thymoproteasome: Probable role in generating positively selecting peptides. Curr. Opin. Immunol. 2008, 20, 192–196. [Google Scholar] [CrossRef]
- Bai, M.; Zhao, X.; Sahara, K.; Ohte, Y.; Hirano, Y.; Kaneko, T.; Yashiroda, H.; Murata, S. Assembly Mechanisms of Specialized Core Particles of the Proteasome. Biomolecules 2014, 4, 662–677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.H.; Jiang, T.X.; Chen, L.B.; Zhou, W.; Liu, Y.; Gao, F.; Qiu, X.B. Proteasome subunit alpha4s is essential for formation of spermatoproteasomes and histone degradation during meiotic DNA repair in spermatocytes. J. Biol. Chem. 2021, 296, 100130. [Google Scholar] [CrossRef] [PubMed]
- Zivkovic, D.; Sanchez Dafun, A.; Menneteau, T.; Schahl, A.; Lise, S.; Kervarrec, C.; Toste Rego, A.; da Fonseca, P.C.A.; Chavent, M.; Pineau, C.; et al. Proteasome complexes experience profound structural and functional rearrangements throughout mammalian spermatogenesis. Proc. Natl. Acad. Sci. USA 2022, 119, e2116826119. [Google Scholar] [CrossRef] [PubMed]
- Zivkovic, D.; Dafun, A.; Menneteau, T.; Schahl, A.; Lise, S.; Kervarrec, C.; Toste-Rego, A.; Fonseca, P.; Chavent, M.; Pineau, C.; et al. Interactome analyses reveal profound proteasome structural and functional rearrangements throughout mammalian spermatogenesis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Sato, B.; Kim, J.; Morohoshi, K.; Kang, W.; Miyado, K.; Tsuruta, F.; Kawano, N.; Chiba, T. Proteasome-Associated Proteins, PA200 and ECPAS, Are Essential for Murine Spermatogenesis. Biomolecules 2023, 13, 586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ji, S.-Y.; Busayavalasa, K.; Shao, J.; Yu, C. Meiosis I progression in spermatogenesis requires a type of testis-specific 20S core proteasome. Nat. Commun. 2019, 10, 3387. [Google Scholar] [CrossRef] [PubMed]
- Gomez, H.L.; Felipe-Medina, N.; Condezo, Y.B.; Garcia-Valiente, R.; Ramos, I.; Suja, J.A.; Barbero, J.L.; Roig, I.; Sanchez-Martin, M.; de Rooij, D.G.; et al. The PSMA8 subunit of the spermatoproteasome is essential for proper meiotic exit and mouse fertility. PLoS Genet. 2019, 15, e1008316. [Google Scholar] [CrossRef]
- Khor, B.; Bredemeyer, A.L.; Huang, C.Y.; Turnbull, I.R.; Evans, R.; Maggi, L.B., Jr.; White, J.M.; Walker, L.M.; Carnes, K.; Hess, R.A.; et al. Proteasome activator PA200 is required for normal spermatogenesis. Mol. Cell. Biol. 2006, 26, 2999–3007. [Google Scholar] [CrossRef] [PubMed]
- Kniepert, A.; Groettrup, M. The unique functions of tissue-specific proteasomes. Trends Biochem. Sci. 2014, 39, 17–24. [Google Scholar] [CrossRef]
- Nonaka, M.; Yamada-Namikawa, C.; Flajnik, M.F.; Du Pasquier, L. Trans-species polymorphism of the major histocompatibility complex-encoded proteasome subunit LMP7 in an amphibian genus. Immunogenetics 2000, 51, 186–192. [Google Scholar] [CrossRef]
- Miura, F.; Tsukamoto, K.; Mehta, R.B.; Naruse, K.; Magtoon, W.; Nonaka, M. Transspecies dimorphic allelic lineages of the proteasome subunit β-type 8 gene (PSMB8) in the teleost genus Oryzias. Proc. Natl. Acad. Sci. USA 2010, 107, 21599–21604. [Google Scholar] [CrossRef]
- Fujito, N.T.; Nonaka, M. Highly divergent dimorphic alleles of the proteasome subunit beta type-8 (PSMB8) gene of the bichir Polypterus senegalus: Implication for evolution of the PSMB8 gene of jawed vertebrates. Immunogenetics 2012, 64, 447–453. [Google Scholar] [CrossRef]
- Kandil, E.; Namikawa, C.; Nonaka, M.; Greenberg, A.S.; Flajnik, M.F.; Ishibashi, T.; Kasahara, M. Isolation of low molecular mass polypeptide complementary DNA clones from primitive vertebrates—Implications for the origin of MHC class I-restricted antigen presentation. J. Immunol. 1996, 156, 4245–4253. [Google Scholar] [CrossRef]
- Belote, J.M.; Zhong, L. Duplicated proteasome subunit genes in Drosophila and their roles in spermatogenesis. Heredity 2009, 103, 23–31. [Google Scholar] [CrossRef]
- Hua, Z.H.; Yu, P.F. Diversifying Evolution of the Ubiquitin-26S Proteasome System in Brassicaceae and Poaceae. Int. J. Mol. Sci. 2019, 20, 3226. [Google Scholar] [CrossRef] [PubMed]
- Kovács, J.; Poór, P.; Kaschani, F.; Chandrasekar, B.; Hong, T.N.; Misas-Villamil, J.C.; Xin, B.T.; Kaiser, M.; Overkleeft, H.S.; Tari, I.; et al. Proteasome Activity Profiling Uncovers Alteration of Catalytic β2 and β5 Subunits of the Stress-Induced Proteasome during Salinity Stress in Tomato Roots. Front. Plant Sci. 2017, 8, 107. [Google Scholar] [CrossRef]
- Emmerich, N.P.; Nussbaum, A.K.; Stevanovic, S.; Priemer, M.; Toes, R.E.; Rammensee, H.G.; Schild, H. The human 26 S and 20 S proteasomes generate overlapping but different sets of peptide fragments from a model protein substrate. J. Biol. Chem. 2000, 275, 21140–21148. [Google Scholar] [CrossRef]
- Thomas, T.; Salcedo-Tacuma, D.; Smith, D.M. Structure, Function, and Allosteric Regulation of the 20S Proteasome by the 11S/PA28 Family of Proteasome Activators. Biomolecules 2023, 13, 1326. [Google Scholar] [CrossRef]
- Ma, C.P.; Slaughter, C.A.; Demartino, G.N. Purification and Characterization of a Protein Inhibitor of the 20s Proteasome (Macropain). Biochim. Biophys. Acta 1992, 1119, 303–311. [Google Scholar] [CrossRef]
- Raule, M.; Cerruti, F.; Benaroudj, N.; Migotti, R.; Kikuchi, J.; Bachi, A.; Navon, A.; Dittmar, G.; Cascio, P. PA28αβ reduces size and increases hydrophilicity of 20S immunoproteasome peptide products. Chem. Biol. 2014, 21, 470–480. [Google Scholar] [CrossRef]
- Schmidtke, G.; Schregle, R.; Alvarez, G.; Huber, E.M.; Groettrup, M. The 20S immunoproteasome and constitutive proteasome bind with the same affinity to PA28alphabeta and equally degrade FAT10. Mol. Immunol. 2019, 113, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Cascio, P. PA28alphabeta: The enigmatic magic ring of the proteasome? Biomolecules 2014, 4, 566–584. [Google Scholar] [CrossRef]
- Inholz, K.; Bader, U.; Mundt, S.; Basler, M. The Significant Role of PA28alphabeta in CD8(+) T Cell-Mediated Graft Rejection Contrasts with Its Negligible Impact on the Generation of MHC-I Ligands. Int. J. Mol. Sci. 2024, 25, 5649. [Google Scholar] [CrossRef]
- Cascio, P. PA28γ: New Insights on an Ancient Proteasome Activator. Biomolecules 2021, 11, 228. [Google Scholar] [CrossRef]
- Noda, C.; Tanahashi, N.; Shimbara, N.; Hendil, K.B.; Tanaka, K. Tissue distribution of constitutive proteasomes, immunoproteasomes, and PA28 in rats. Biochem. Biophys. Res. Commun. 2000, 277, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Frayssinhes, J.Y.A.; Cerruti, F.; Laulin, J.; Cattaneo, A.; Bachi, A.; Apcher, S.; Coux, O.; Cascio, P. PA28γ-20S proteasome is a proteolytic complex committed to degrade unfolded proteins. Cell. Mol. Life Sci. 2022, 79, 45. [Google Scholar] [CrossRef]
- Thomas, T.A.; Smith, D.M. Proteasome activator 28γ (PA28γ) allosterically activates trypsin-like proteolysis by binding to the α-ring of the 20S proteasome. J. Biol. Chem. 2022, 298, 102140. [Google Scholar] [CrossRef] [PubMed]
- Fort, P.; Kajava, A.V.; Delsuc, F.; Coux, O. Evolution of Proteasome Regulators in Eukaryotes. Genome Biol. Evol. 2015, 7, 1363–1379. [Google Scholar] [CrossRef]
- Guan, H.X.; Wang, Y.W.; Yu, T.; Huang, Y.N.; Li, M.H.; Saeed, A.F.U.H.; Perculija, V.; Li, D.L.; Xiao, J.; Wang, D.M.; et al. Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures. PLoS Biol. 2020, 18, e3000654. [Google Scholar] [CrossRef]
- Toste Rêgo, A.; da Fonseca, P.C.A. Characterization of Fully Recombinant Human 20S and 20S-PA200 Proteasome Complexes. Mol. Cell 2019, 76, 138–147.e135. [Google Scholar] [CrossRef]
- Wang, J.S.; Kjellgren, A.; DeMartino, G.N. PI31 is a positive regulator of 20S immunoproteasome assembly. J. Cell Sci. 2025, 138, jcs263887. [Google Scholar] [CrossRef] [PubMed]
- Kors, S.; Geijtenbeek, K.; Reits, E.; Schipper-Krom, S. Regulation of Proteasome Activity by (Post-)transcriptional Mechanisms. Front. Mol. Biosci. 2019, 6, 48. [Google Scholar] [CrossRef]
- Wang, D.; Fang, C.; Zong, N.C.; Liem, D.A.; Cadeiras, M.; Scruggs, S.B.; Yu, H.; Kim, A.K.; Yang, P.; Deng, M.; et al. Regulation of acetylation restores proteolytic function of diseased myocardium in mouse and human. Mol. Cell. Proteom. 2013, 12, 3793–3802. [Google Scholar] [CrossRef]
- Ishii, T.; Sakurai, T.; Usami, H.; Uchida, K. Oxidative modification of proteasome: Identification of an oxidation-sensitive subunit in 26 S proteasome. Biochemistry 2005, 44, 13893–13901. [Google Scholar] [CrossRef] [PubMed]
- Bulteau, A.L.; Lundberg, K.C.; Humphries, K.M.; Sadek, H.A.; Szweda, P.A.; Friguet, B.; Szweda, L.I. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J. Biol. Chem. 2001, 276, 30057–30063. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.M.; Netto, L.E.; Simoes, V.; Santos, L.F.; Gozzo, F.C.; Demasi, M.A.; Oliveira, C.L.; Bicev, R.N.; Klitzke, C.F.; Sogayar, M.C.; et al. Redox control of 20S proteasome gating. Antioxid. Redox Signal 2012, 16, 1183–1194. [Google Scholar] [CrossRef]
- Ben-Nissan, G.; Katzir, N.; Füzesi-Levi, M.G.; Sharon, M. Biology of the Extracellular Proteasome. Biomolecules 2022, 12, 619. [Google Scholar] [CrossRef] [PubMed]
- Leushkin, Y.; Morgenstern, D.; Ben-Dor, S.; Haffner-Krausz, R.; Zittlau, K.; Ben-Nissan, G.; Sharon, M. Molecular insights into the unique properties of the blood-circulating proteasome. J. Extracell. Biol. 2025, 4, e70034. [Google Scholar] [CrossRef]
- Vilchez, D.; Boyer, L.; Morantte, I.; Lutz, M.; Merkwirth, C.; Joyce, D.; Spencer, B.; Page, L.; Masliah, E.; Berggren, W.T.; et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 2012, 489, 304–308. [Google Scholar] [CrossRef]
- Semren, N.; Welk, V.; Korfei, M.; Keller, I.E.; Fernandez, I.E.; Adler, H.; Gunther, A.; Eickelberg, O.; Meiners, S. Regulation of 26S Proteasome Activity in Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2015, 192, 1089–1101. [Google Scholar] [CrossRef]
- Cui, Z.; Hwang, S.M.; Gomes, A.V. Identification of the immunoproteasome as a novel regulator of skeletal muscle differentiation. Mol. Cell. Biol. 2014, 34, 96–109. [Google Scholar] [CrossRef]
- Hernebring, M.; Fredriksson, A.; Liljevald, M.; Cvijovic, M.; Norrman, K.; Wiseman, J.; Semb, H.; Nystrom, T. Removal of damaged proteins during ES cell fate specification requires the proteasome activator PA28. Sci. Rep. 2013, 3, 1381. [Google Scholar] [CrossRef]
- Welk, V.; Meul, T.; Lukas, C.; Kammerl, I.E.; Mulay, S.R.; Schamberger, A.C.; Semren, N.; Fernandez, I.E.; Anders, H.J.; Gunther, A.; et al. Proteasome activator PA200 regulates myofibroblast differentiation. Sci. Rep. 2019, 9, 15224. [Google Scholar] [CrossRef]
- Chen, Q.; Wu, Z.; Shi, Y.; Li, Z.; Yang, J.; Qu, M.; Zhang, S.; Wang, Z.; Ji, N.; Li, J.; et al. Loss of PA28gamma exacerbates imbalanced differentiation of bone marrow stromal cells during bone formation and bone healing in mice. J. Bone Miner. Res. 2024, 39, 326–340. [Google Scholar] [CrossRef]
- Aki, M.; Shimbara, N.; Takashina, M.; Akiyama, K.; Kagawa, S.; Tamura, T.; Tanahashi, N.; Yoshimura, T.; Tanaka, K.; Ichihara, A. Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J. Biochem. 1994, 115, 257–269. [Google Scholar] [CrossRef]
- Hallermalm, K.; Seki, K.; Wei, C.; Castelli, C.; Rivoltini, L.; Kiessling, R.; Levitskaya, J. Tumor necrosis factor-alpha induces coordinated changes in major histocompatibility class I presentation pathway, resulting in increased stability of class I complexes at the cell surface. Blood 2001, 98, 1108–1115. [Google Scholar] [CrossRef]
- Kotamraju, S.; Matalon, S.; Matsunaga, T.; Shang, T.; Hickman-Davis, J.M.; Kalyanaraman, B. Upregulation of immunoproteasomes by nitric oxide: Potential antioxidative mechanism in endothelial cells. Free Radic. Biol. Med. 2006, 40, 1034–1044. [Google Scholar] [CrossRef]
- Reis, J.; Guan, X.Q.; Kisselev, A.F.; Papasian, C.J.; Qureshi, A.A.; Morrison, D.C.; Van Way, C.W., 3rd; Vogel, S.N.; Qureshi, N. LPS-induced formation of immunoproteasomes: TNF-alpha and nitric oxide production are regulated by altered composition of proteasome-active sites. Cell Biochem. Biophys. 2011, 60, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Macagno, A.; Gilliet, M.; Sallusto, F.; Lanzavecchia, A.; Nestle, F.O.; Groettrup, M. Dendritic cells up-regulate immunoproteasomes and the proteasome regulator PA28 during maturation. Eur. J. Immunol. 1999, 29, 4037–4042. [Google Scholar] [CrossRef]
- Ahn, J.Y.; Tanahashi, N.; Akiyama, K.; Hisamatsu, H.; Noda, C.; Tanaka, K.; Chung, C.H.; Shibmara, N.; Willy, P.J.; Mott, J.D.; et al. Primary structures of two homologous subunits of PA28, a gamma-interferon-inducible protein activator of the 20S proteasome. FEBS Lett. 1995, 366, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Heink, S.; Ludwig, D.; Kloetzel, P.M.; Krüger, E. IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc. Natl. Acad. Sci. USA 2005, 102, 9241–9246. [Google Scholar] [CrossRef]
- Wang, X.; Chemmama, I.E.; Yu, C.; Huszagh, A.; Xu, Y.; Viner, R.; Block, S.A.; Cimermancic, P.; Rychnovsky, S.D.; Ye, Y.; et al. The proteasome-interacting Ecm29 protein disassembles the 26S proteasome in response to oxidative stress. J. Biol. Chem. 2017, 292, 16310–16320. [Google Scholar] [CrossRef] [PubMed]
- Haratake, K.; Sato, A.; Tsuruta, F.; Chiba, T. KIAA0368-deficiency affects disassembly of 26S proteasome under oxidative stress condition. J. Biochem. 2016, 159, 609–618. [Google Scholar] [CrossRef]
- Reinheckel, T.; Sitte, N.; Ullrich, O.; Kuckelkorn, U.; Davies, K.J.; Grune, T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem. J. 1998, 335 Pt 3, 637–642. [Google Scholar] [CrossRef]
- Pickering, A.M.; Koop, A.L.; Teoh, C.Y.; Ermak, G.; Grune, T.; Davies, K.J. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 2010, 432, 585–594. [Google Scholar] [CrossRef]
- Meng, F.; Yao, D.; Shi, Y.; Kabakoff, J.; Wu, W.; Reicher, J.; Ma, Y.; Moosmann, B.; Masliah, E.; Lipton, S.A.; et al. Oxidation of the cysteine-rich regions of parkin perturbs its E3 ligase activity and contributes to protein aggregation. Mol. Neurodegener. 2011, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Jahngen-Hodge, J.; Obin, M.S.; Gong, X.; Shang, F.; Nowell, T.R., Jr.; Gong, J.; Abasi, H.; Blumberg, J.; Taylor, A. Regulation of ubiquitin-conjugating enzymes by glutathione following oxidative stress. J. Biol. Chem. 1997, 272, 28218–28226. [Google Scholar] [CrossRef] [PubMed]
- Kastle, M.; Reeg, S.; Rogowska-Wrzesinska, A.; Grune, T. Chaperones, but not oxidized proteins, are ubiquitinated after oxidative stress. Free Radic. Biol. Med. 2012, 53, 1468–1477. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, B.S.; Belenghi, B.; Levine, A. Oxidative stress increased respiration and generation of reactive oxygen species, resulting in ATP depletion, opening of mitochondrial permeability transition, and programmed cell death. Plant Physiol. 2002, 128, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Pickering, A.M.; Davies, K.J. Differential roles of proteasome and immunoproteasome regulators Pa28αβ, Pa28γ and Pa200 in the degradation of oxidized proteins. Arch. Biochem. Biophys. 2012, 523, 181–190. [Google Scholar] [CrossRef]
- Seifert, U.; Bialy, L.P.; Ebstein, F.; Bech-Otschir, D.; Voigt, A.; Schroter, F.; Prozorovski, T.; Lange, N.; Steffen, J.; Rieger, M.; et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell 2010, 142, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Pickering, A.M.; Linder, R.A.; Zhang, H.; Forman, H.J.; Davies, K.J.A. Nrf2-dependent induction of proteasome and Pa28alphabeta regulator are required for adaptation to oxidative stress. J. Biol. Chem. 2012, 287, 10021–10031. [Google Scholar] [CrossRef] [PubMed]
- Kelmer Sacramento, E.; Kirkpatrick, J.M.; Mazzetto, M.; Baumgart, M.; Bartolome, A.; Di Sanzo, S.; Caterino, C.; Sanguanini, M.; Papaevgeniou, N.; Lefaki, M.; et al. Reduced proteasome activity in the aging brain results in ribosome stoichiometry loss and aggregation. Mol. Syst. Biol. 2020, 16, e9596. [Google Scholar] [CrossRef]
- Nago, N.; Murata, S.; Tanaka, K.; Tanahashi, N. Changes in brain proteasome dynamics associated with aging. Genes Cells 2024, 29, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Dasuri, K.; Zhang, L.; Ebenezer, P.; Liu, Y.; Fernandez-Kim, S.O.; Keller, J.N. Aging and dietary restriction alter proteasome biogenesis and composition in the brain and liver. Mech. Ageing Dev. 2009, 130, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Shibatani, T.; Nazir, M.; Ward, W.F. Alteration of rat liver 20S proteasome activities by age and food restriction. J. Gerontol. A Biol. Sci. Med. Sci. 1996, 51, B316–B322. [Google Scholar] [CrossRef] [PubMed]
- Bulteau, A.L.; Szweda, L.I.; Friguet, B. Age-dependent declines in proteasome activity in the heart. Arch. Biochem. Biophys. 2002, 397, 298–304. [Google Scholar] [CrossRef]
- Husom, A.D.; Peters, E.A.; Kolling, E.A.; Fugere, N.A.; Thompson, L.V.; Ferrington, D.A. Altered proteasome function and subunit composition in aged muscle. Arch. Biochem. Biophys. 2004, 421, 67–76. [Google Scholar] [CrossRef]
- Bulteau, A.L.; Petropoulos, I.; Friguet, B. Age-related alterations of proteasome structure and function in aging epidermis. Exp. Gerontol. 2000, 35, 767–777. [Google Scholar] [CrossRef]
- Ponnappan, U.; Zhong, M.; Trebilcock, G.U. Decreased proteasome-mediated degradation in T cells from the elderly: A role in immune senescence. Cell. Immunol. 1999, 192, 167–174. [Google Scholar] [CrossRef]
- Vernace, V.A.; Arnaud, L.; Schmidt-Glenewinkel, T.; Figueiredo-Pereira, M.E. Aging perturbs 26S proteasome assembly in Drosophila melanogaster. FASEB J. 2007, 21, 2672–2682. [Google Scholar] [CrossRef]
- Munkacsy, E.; Chocron, E.S.; Quintanilla, L.; Gendron, C.M.; Pletcher, S.D.; Pickering, A.M. Neuronal-specific proteasome augmentation via Prosbeta5 overexpression extends lifespan and reduces age-related cognitive decline. Aging Cell 2019, 18, e13005. [Google Scholar] [CrossRef]
- Tonoki, A.; Kuranaga, E.; Tomioka, T.; Hamazaki, J.; Murata, S.; Tanaka, K.; Miura, M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol. Cell. Biol. 2009, 29, 1095–1106. [Google Scholar] [CrossRef]
- Hegde, A.N.; Duke, L.M.; Timm, L.E.; Nobles, H. The Proteasome and Ageing. Subcell. Biochem. 2023, 102, 99–112. [Google Scholar] [CrossRef]
- Holzhütter, H.G.; Frömmel, C.; Kloetzel, P.M. A theoretical approach towards the identification of cleavage-determining amino acid motifs of the 20 S proteasome. J. Mol. Biol. 1999, 286, 1251–1265. [Google Scholar] [CrossRef]
- Holzhütter, H.G.; Kloetzel, P.M. A kinetic model of vertebrate 20S proteasome accounting for the generation of major proteolytic fragments from oligomeric peptide substrates. Biophys. J. 2000, 79, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Kuttler, C.; Nussbaum, A.K.; Dick, T.P.; Rammensee, H.G.; Schild, H.; Hadeler, K.P. An algorithm for the prediction of proteasomal cleavages. J. Mol. Biol. 2000, 298, 417–429. [Google Scholar] [CrossRef]
- Nussbaum, A.K.; Kuttler, C.; Hadeler, K.P.; Rammensee, H.G.; Schild, H. PAProC: A prediction algorithm for proteasomal cleavages available on the WWW. Immunogenetics 2001, 53, 87–94. [Google Scholar] [CrossRef]
- Keşmir, C.; Nussbaum, A.K.; Schild, H.; Detours, V.; Brunak, S. Prediction of proteasome cleavage motifs by neural networks. Protein Eng. 2002, 15, 287–296. [Google Scholar] [CrossRef]
- Nielsen, M.; Lundegaard, C.; Lund, O.; Keşmir, C. The role of the proteasome in generating cytotoxic T-cell epitopes: Insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 2005, 57, 33–41. [Google Scholar] [CrossRef]
- Saxová, P.; Buus, S.; Brunak, S.; Keşmir, C. Predicting proteasomal cleavage sites: A comparison of available methods. Int. Immunol. 2003, 15, 781–787. [Google Scholar] [CrossRef]
- Singh, H.; Raghava, G.P. ProPred1: Prediction of promiscuous MHC Class-I binding sites. Bioinformatics 2003, 19, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Toes, R.E.; Nussbaum, A.K.; Degermann, S.; Schirle, M.; Emmerich, N.P.; Kraft, M.; Laplace, C.; Zwinderman, A.; Dick, T.P.; Müller, J.; et al. Discrete cleavage motifs of constitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J. Exp. Med. 2001, 194, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bhasin, M.; Raghava, G.P. A hybrid approach for predicting promiscuous MHC class I restricted T cell epitopes. J. Biosci. 2007, 32, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Bhasin, M.; Raghava, G.P. Pcleavage: An SVM based method for prediction of constitutive proteasome and immunoproteasome cleavage sites in antigenic sequences. Nucleic Acids Res. 2005, 33, W202–W207. [Google Scholar] [CrossRef]
- Gomez-Perosanz, M.; Ras-Carmona, A.; Lafuente, E.M.; Reche, P.A. Identification of CD8(+) T cell epitopes through proteasome cleavage site predictions. BMC Bioinform. 2020, 21, 484. [Google Scholar] [CrossRef]
- Weeder, B.R.; Wood, M.A.; Li, E.; Nellore, A.; Thompson, R.F. Pepsickle rapidly and accurately predicts proteasomal cleavage sites for improved neoantigen identification. Bioinformatics 2021, 37, 3723–3733. [Google Scholar] [CrossRef]
- Fenteany, G.; Standaert, R.F.; Lane, W.S.; Choi, S.; Corey, E.J.; Schreiber, S.L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 1995, 268, 726–731. [Google Scholar] [CrossRef]
- Adams, J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell 2004, 5, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Moreau, P.; Richardson, P.G.; Cavo, M.; Orlowski, R.Z.; San Miguel, J.F.; Palumbo, A.; Harousseau, J.L. Proteasome inhibitors in multiple myeloma: 10 years later. Blood 2012, 120, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Kraus, M.; Bader, J.; Geurink, P.P.; Weyburne, E.S.; Mirabella, A.C.; Silzle, T.; Shabaneh, T.B.; van der Linden, W.A.; de Bruin, G.; Haile, S.R.; et al. The novel β2-selective proteasome inhibitor LU-102 synergizes with bortezomib and carfilzomib to overcome proteasome inhibitor resistance of myeloma cells. Haematologica 2015, 100, 1350–1360. [Google Scholar] [CrossRef]
- Britton, M.; Lucas, M.M.; Downey, S.L.; Screen, M.; Pletnev, A.A.; Verdoes, M.; Tokhunts, R.A.; Amir, O.; Goddard, A.L.; Pelphrey, P.M.; et al. Selective inhibitor of proteasome’s caspase-like sites sensitizes cells to specific inhibition of chymotrypsin-like sites. Chem. Biol. 2009, 16, 1278–1289. [Google Scholar] [CrossRef]
- Kuhn, D.J.; Hunsucker, S.A.; Chen, Q.; Voorhees, P.M.; Orlowski, M.; Orlowski, R.Z. Targeted inhibition of the immunoproteasome is a potent strategy against models of multiple myeloma that overcomes resistance to conventional drugs and nonspecific proteasome inhibitors. Blood 2009, 113, 4667–4676. [Google Scholar] [CrossRef]
- Wehenkel, M.; Ban, J.O.; Ho, Y.K.; Carmony, K.C.; Hong, J.T.; Kim, K.B. A selective inhibitor of the immunoproteasome subunit LMP2 induces apoptosis in PC-3 cells and suppresses tumour growth in nude mice. Br. J. Cancer 2012, 107, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Muchamuel, T.; Fan, R.A.; Anderl, J.L.; Bomba, D.J.; Johnson, H.W.B.; Lowe, E.; Tuch, B.B.; McMinn, D.L.; Millare, B.; Kirk, C.J. Zetomipzomib (KZR-616) attenuates lupus in mice via modulation of innate and adaptive immune responses. Front. Immunol. 2023, 14, 1043680. [Google Scholar] [CrossRef] [PubMed]
- Kingwell, K. Proteasome target to tackle non-replicating TB. Nat. Rev. Drug Discov. 2009, 8, 845. [Google Scholar] [CrossRef]
- Khare, S.; Nagle, A.S.; Biggart, A.; Lai, Y.H.; Liang, F.; Davis, L.C.; Barnes, S.W.; Mathison, C.J.N.; Myburgh, E.; Gao, M.-Y.; et al. Proteasome inhibition for treatment of leishmaniasis, Chagas disease and sleeping sickness. Nature 2016, 537, 229–233. [Google Scholar] [CrossRef]
- Drugs for Neglected Diseases. LXE408 for Treatment of Visceral Leishmaniasis in Ethiopia, a Proof of Concept Study. Available online: https://clinicaltrials.gov/study/NCT05957978 (accessed on 15 September 2025).
- Zhan, W.; Zhang, H.; Ginn, J.; Leung, A.; Liu, Y.J.; Michino, M.; Toita, A.; Okamoto, R.; Wong, T.T.; Imaeda, T.; et al. Development of a Highly Selective Plasmodium falciparum Proteasome Inhibitor with Anti-malaria Activity in Humanized Mice. Angew. Chem. Int. Ed. Engl. 2021, 60, 9279–9283. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zachor-Movshovitz, D.; Leushkin, Y.; Zittlau, K.I.; Ben-Nissan, G.; Sharon, M. Cleaving Expectations: A Review of Proteasome Functional and Catalytic Diversity. Biomolecules 2025, 15, 1524. https://doi.org/10.3390/biom15111524
Zachor-Movshovitz D, Leushkin Y, Zittlau KI, Ben-Nissan G, Sharon M. Cleaving Expectations: A Review of Proteasome Functional and Catalytic Diversity. Biomolecules. 2025; 15(11):1524. https://doi.org/10.3390/biom15111524
Chicago/Turabian StyleZachor-Movshovitz, Daniel, Yegor Leushkin, Katharina I. Zittlau, Gili Ben-Nissan, and Michal Sharon. 2025. "Cleaving Expectations: A Review of Proteasome Functional and Catalytic Diversity" Biomolecules 15, no. 11: 1524. https://doi.org/10.3390/biom15111524
APA StyleZachor-Movshovitz, D., Leushkin, Y., Zittlau, K. I., Ben-Nissan, G., & Sharon, M. (2025). Cleaving Expectations: A Review of Proteasome Functional and Catalytic Diversity. Biomolecules, 15(11), 1524. https://doi.org/10.3390/biom15111524





