Dimerization of the β-Hairpin Membrane-Active Cationic Antimicrobial Peptide Capitellacin from Marine Polychaeta: An NMR Structural and Thermodynamic Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression and Purification of 15N-Labeled Capitellacin
2.2. NMR Experiments and Data Analysis
2.3. Accessing Codes
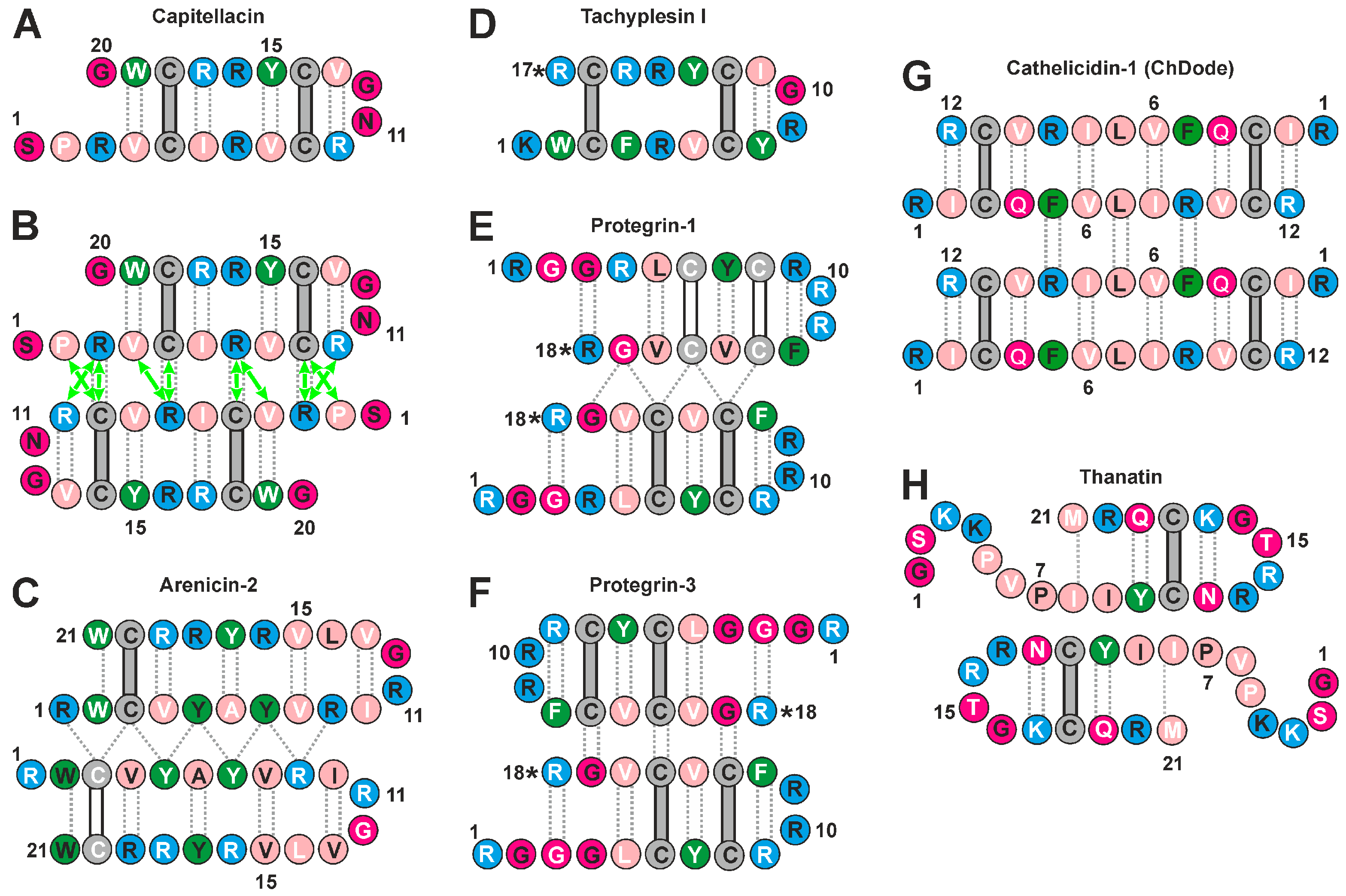
2.4. Analisys of Physicochemical Properties of β-Hairpin AMPs
2.5. Study of Capitellacin Dimerization
3. Results
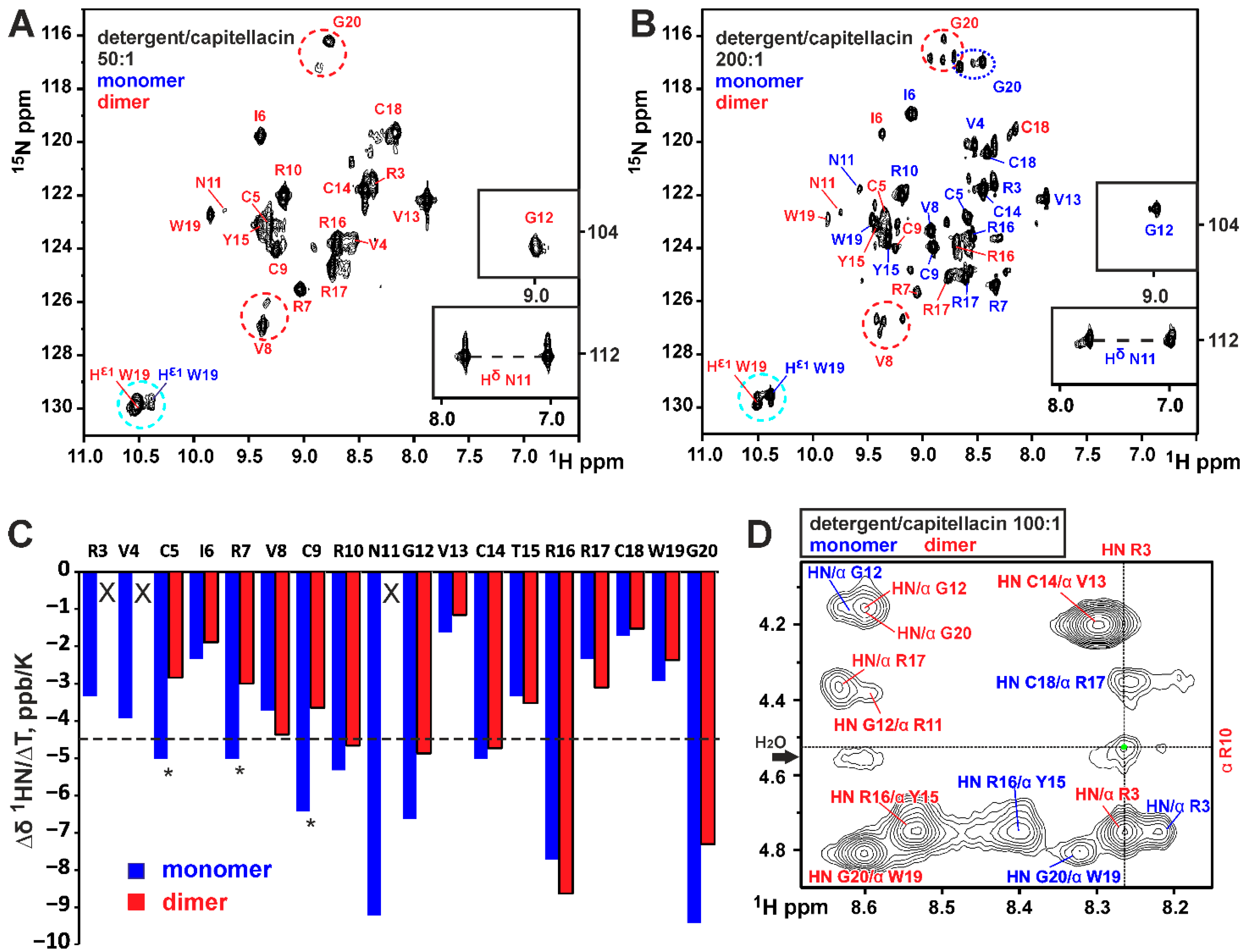
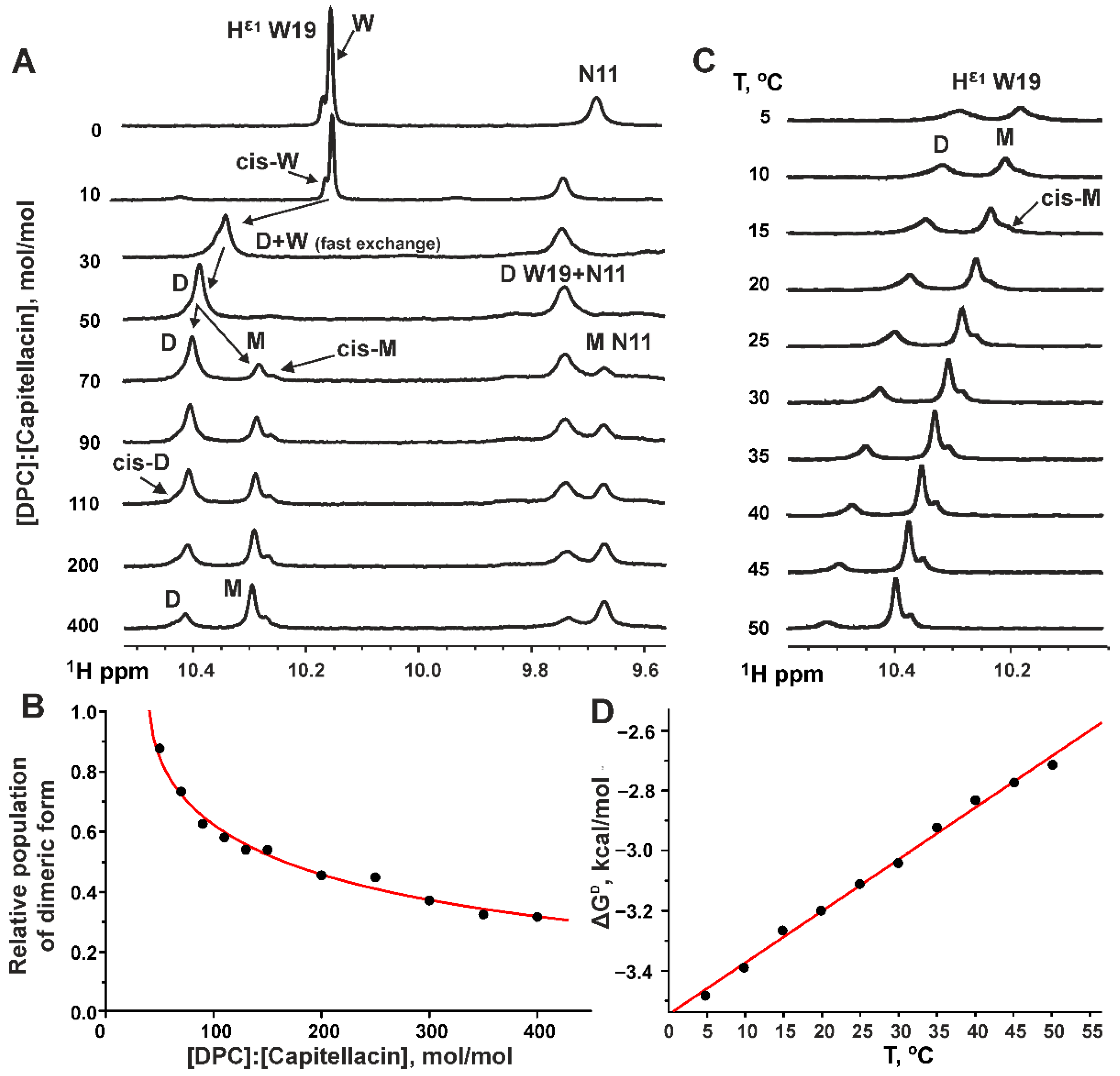
3.1. Dimerization of Capitellacin in DPC Micelles
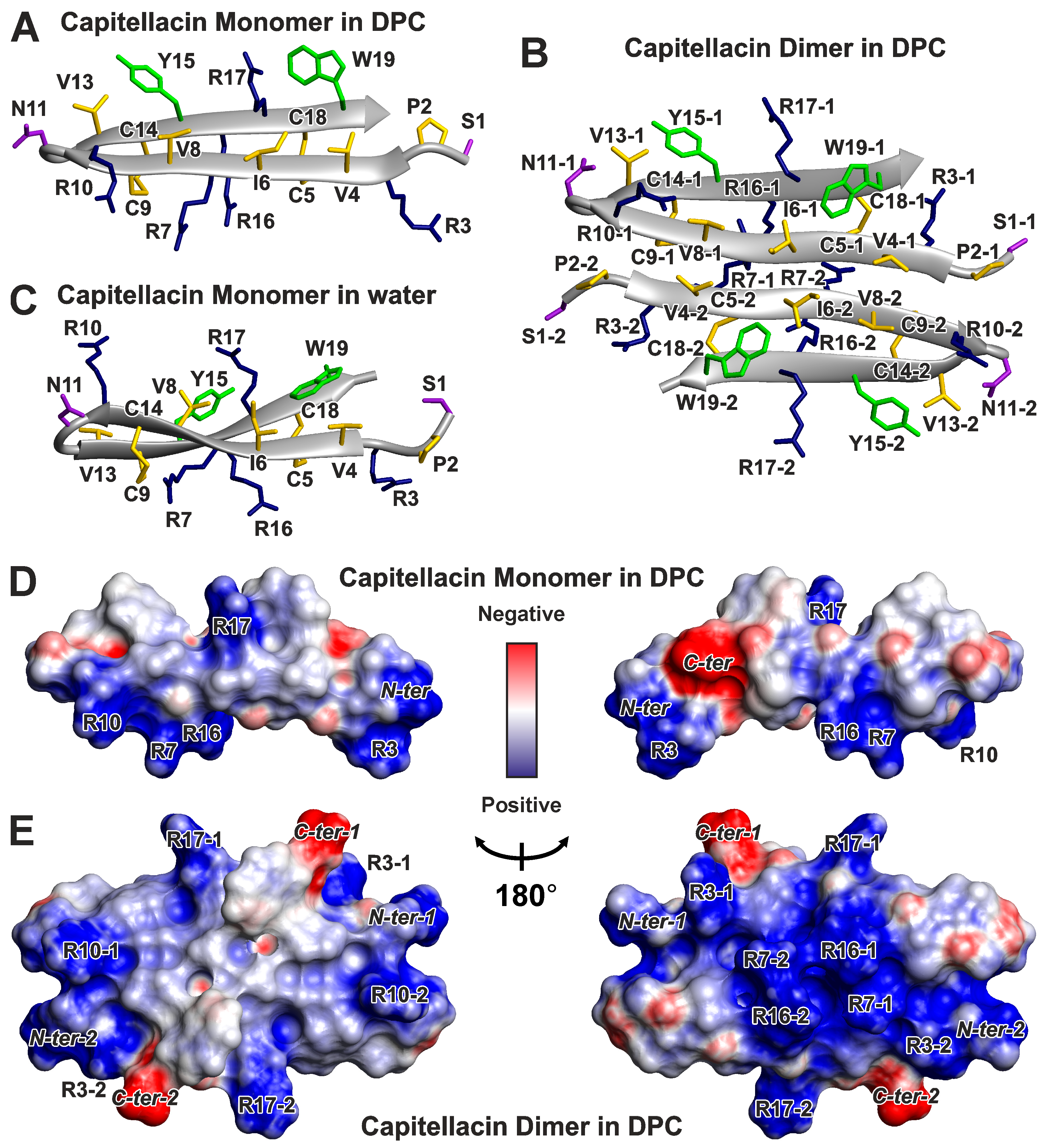
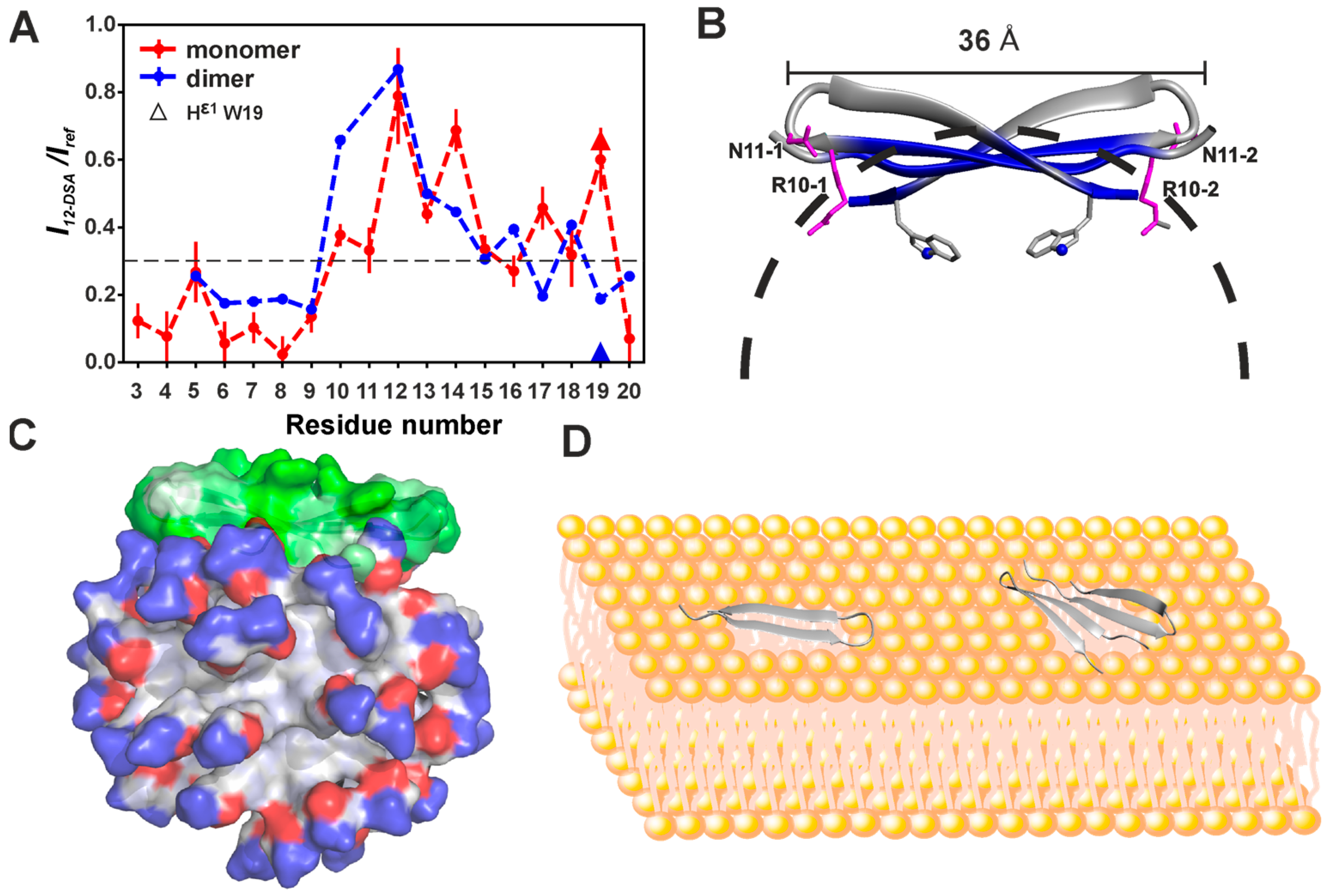
3.2. Spatial Structures of Monomer and Dimer of Capitellacin in DPC Micelles
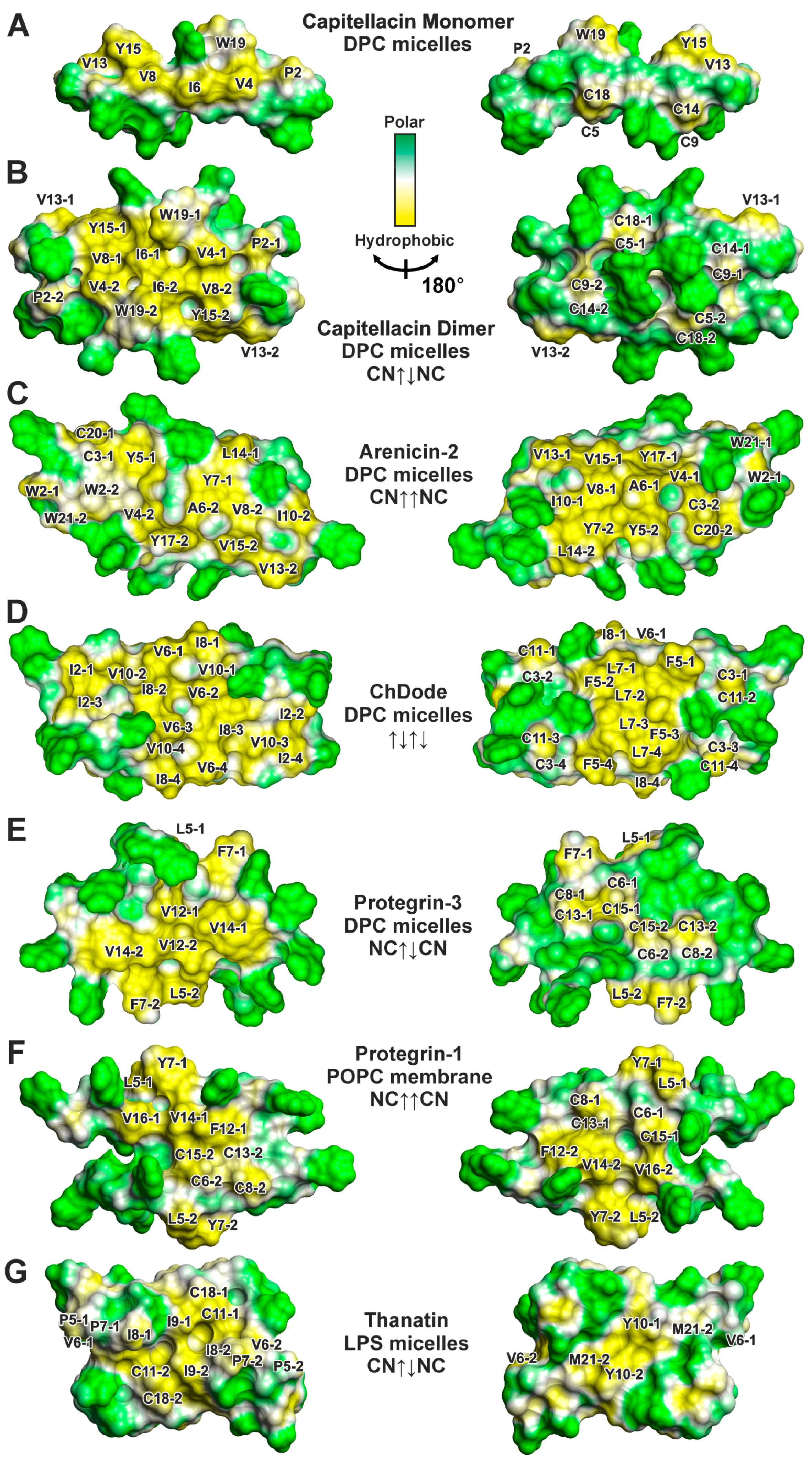
3.3. Topology of the Capitellacin Interaction with DPC Micelles
3.4. Thermodynamics of the Capitellacin Dimerization in DPC Micelles
4. Discussion
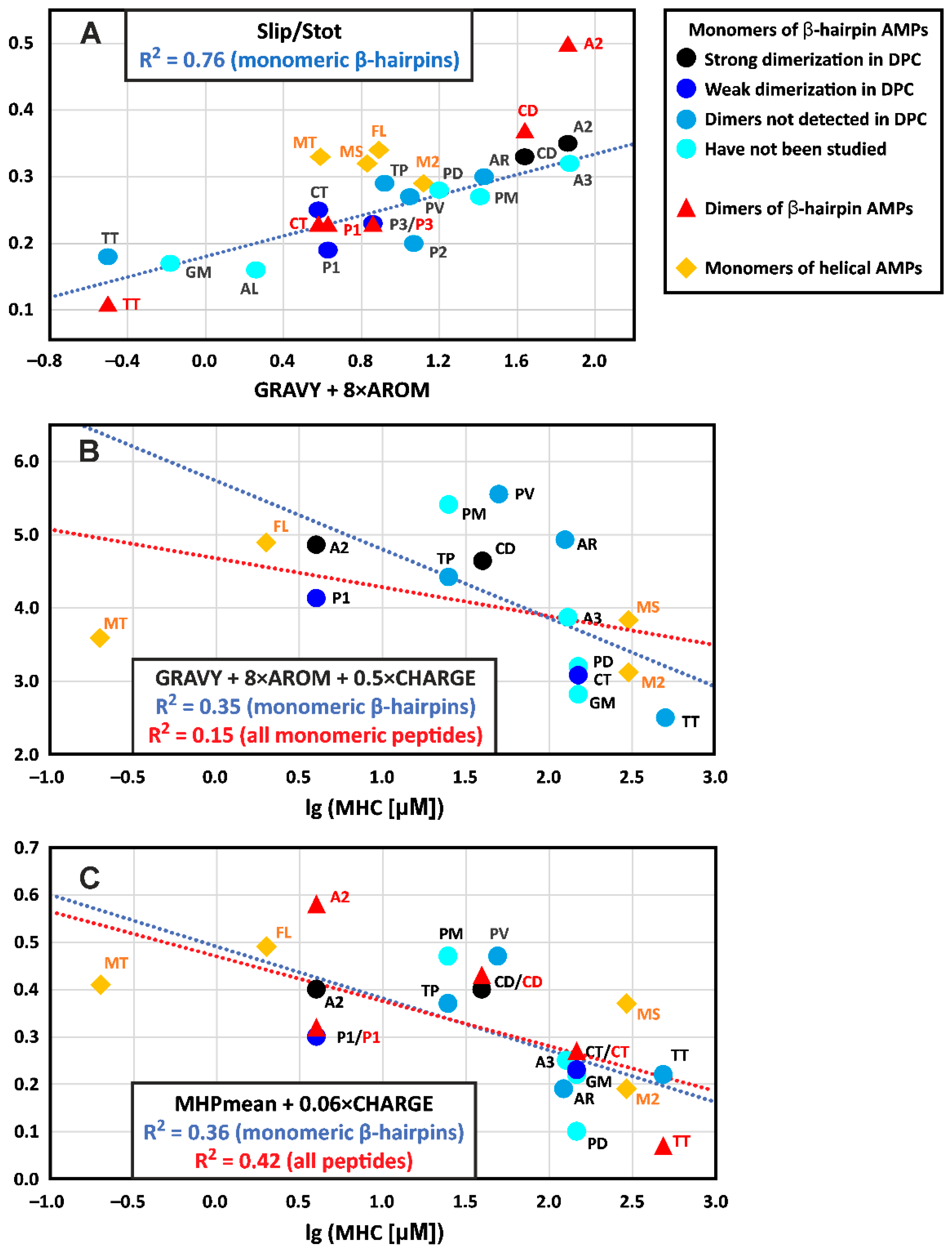
4.1. Physicochemical Properties of β-Hairpin AMPs and Their Change upon Incorporation into a Membrane-like Environment and Dimerization
4.2. Hydrophobicity of the β-Hairpin AMPs Correlates with the Stability of Dimers in a Membrane-like Environment
4.3. The Hydrophobicity and Total Charge of β-Hairpin AMPs, but Not the Ability to Form Dimers, Are Positively Correlated with Hemolytic Activity
4.4. The Distribution of Hydrophobic and Polar Regions on the Capitellacin Surface Determines the Surface Mode of the Peptide/Micelle Interaction and the Carpet Mechanism of Action on Membranes
4.5. The Dimerization of Capitellacin in DPC Micelles Is an Exothermic- and Enthalpy-Driven Process
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheesman, M.; Ilanko, A.; Blonk, B.; Cock, I. Pharmacognosy Reviews Developing New Antimicrobial Therapies: Are Synergistic Combinations of Plant Extracts/Compounds with Conventional Antibiotics the Solution? Pharmacogn. Rev. 2017, 11, 57–72. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Brogden, K. Antimicrobial Peptides: Pore Formers or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Lorenzon, E.N.; Piccoli, J.P.; Santos-Filho, N.A.; Cilli, E.M. Dimerization of Antimicrobial Peptides: A Promising Strategy to Enhance Antimicrobial Peptide Activity. Protein Pept. Lett. 2019, 26, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Panteleev, P.V.; Balandin, S.V.; Ivanov, V.T.; Ovchinnikova, T.V. A Therapeutic Potential of Animal β-Hairpin Antimicrobial Peptides. Curr. Med. Chem. 2017, 24, 1724–1746. [Google Scholar] [CrossRef] [PubMed]
- Safronova, V.N.; Panteleev, P.V.; Sukhanov, S.V.; Toropygin, I.Y.; Bolosov, I.A.; Ovchinnikova, T.V. Mechanism of Action and Therapeutic Potential of the β-Hairpin Antimicrobial Peptide Capitellacin from the Marine Polychaeta Capitella Teleta. Mar. Drugs 2022, 20, 167. [Google Scholar] [CrossRef] [PubMed]
- Fehlbaum, P.; Bulet, P.; Chernysh, S.; Briand, J.P.; Roussel, J.P.; Letellier, L.; Hetru, C.; Hoffmann, J.A. Structure-Activity Analysis of Thanatin, a 21-Residue Inducible Insect Defense Peptide with Sequence Homology to Frog Skin Antimicrobial Peptides. Proc. Natl. Acad. Sci. USA 1996, 93, 1221–1225. [Google Scholar] [CrossRef] [PubMed]
- Tamamura, H.; Ishihara, T.; Otaka, A.; Murakami, T.; Ibuka, T.; Waki, M.; Matsumoto, A.; Yamamoto, N.; Fujii, N. Analysis of the Interaction of an Anti-HIV Peptide, T22 ([Tyr5,12, Lys7]-Polyphemusin II), with Gp 120 and CD4 by Surface Plasmon Resonance. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 1996, 1298, 37–44. [Google Scholar] [CrossRef]
- Radermacher, S.W.; Schoop, V.M.; Schluesener, H.J. Bactenecin, a Leukocytic Antimicrobial Peptide, Is Cytotoxic to Neuronal and Glial Cells. J. Neurosci. Res. 1993, 36, 657–662. [Google Scholar] [CrossRef]
- Hou, Z.; Da, F.; Liu, B.; Xue, X.; Xu, X.; Zhou, Y.; Li, M.; Li, Z.; Ma, X.; Meng, J.; et al. R-Thanatin Inhibits Growth and Biofilm Formation of Methicillin-Resistant Staphylococcus Epidermidis in Vivo and in Vitro. Antimicrob. Agents Chemother. 2013, 57, 5045–5052. [Google Scholar] [CrossRef]
- Mani, R.; Cady, S.D.; Tang, M.; Waring, A.J.; Lehrer, R.I.; Hong, M. Membrane-Dependent Oligomeric Structure and Pore Formation of a β-Hairpin Antimicrobial Peptide in Lipid Bilayers from Solid-State NMR. Proc. Natl. Acad. Sci. USA 2006, 103, 16242–16247. [Google Scholar] [CrossRef] [PubMed]
- Roumestand, C.; Louis, V.; Aumelas, A.; Grassy, G.; Calas, B.; Chavanieu, A. Oligomerization of Protegrin-1 in the Presence of DPC Micelles. A Proton High-Resolution NMR Study. FEBS Lett. 1998, 421, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Usachev, K.S.; Efimov, S.V.; Kolosova, O.A.; Klochkova, E.A.; Aganov, A.V.; Klochkov, V.V. Antimicrobial Peptide Protegrin-3 Adopt an Antiparallel Dimer in the Presence of DPC Micelles: A High-Resolution NMR Study. J. Biomol. NMR 2015, 62, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Mani, R.; Tang, M.; Wu, X.; Buffy, J.J.; Waring, A.J.; Sherman, M.A.; Hong, M. Membrane-Bound Dimer Structure of a β-Hairpin Antimicrobial Peptide from Rotational-Echo Double-Resonance Solid-State NMR. Biochemistry 2006, 45, 8341–8349. [Google Scholar] [CrossRef] [PubMed]
- Shenkarev, Z.O.; Balandin, S.V.; Trunov, K.I.; Paramonov, A.S.; Sukhanov, S.V.; Barsukov, L.I.; Arseniev, A.S.; Ovchinnikova, T.V. Molecular Mechanism of Action of β-Hairpin Antimicrobial Peptide Arenicin: Oligomeric Structure in Dodecylphosphocholine Micelles and Pore Formation in Planar Lipid Bilayers. Biochemistry 2011, 50, 6255–6265. [Google Scholar] [CrossRef] [PubMed]
- Bolosov, I.A.; Panteleev, P.V.; Sychev, S.V.; Sukhanov, S.V.; Mironov, P.A.; Myshkin, M.Y.; Shenkarev, Z.O.; Ovchinnikova, T.V. Dodecapeptide Cathelicidins of Cetartiodactyla: Structure, Mechanism of Antimicrobial Action, and Synergistic Interaction With Other Cathelicidins. Front. Microbiol. 2021, 12, 2249. [Google Scholar] [CrossRef]
- Sinha, S.; Zheng, L.; Mu, Y.; Ng, W.J.; Bhattacharjya, S. Structure and Interactions of A Host Defense Antimicrobial Peptide Thanatin in Lipopolysaccharide Micelles Reveal Mechanism of Bacterial Cell Agglutination. Sci. Rep. 2017, 7, 17795. [Google Scholar] [CrossRef]
- Laederach, A.; Andreotti, A.H.; Fulton, D.B. Solution and Micelle-Bound Structures of Tachyplesin I and Its Active Aromatic Linear Derivatives. Biochemistry 2002, 41, 12359–12368. [Google Scholar] [CrossRef]
- Powers, J.-P.S.; Tan, A.; Ramamoorthy, A.; Hancock, R.E.W. Solution Structure and Interaction of the Antimicrobial Polyphemusins with Lipid Membranes. Biochemistry 2005, 44, 15504–15513. [Google Scholar] [CrossRef]
- Usachev, K.S.; Efimov, S.V.; Kolosova, O.A.; Filippov, A.V.; Klochkov, V.V. High-Resolution NMR Structure of the Antimicrobial Peptide Protegrin-2 in the Presence of DPC Micelles. J. Biomol. NMR 2015, 61, 227–234. [Google Scholar] [CrossRef]
- Panteleev, P.V.; Myshkin, M.Y.; Shenkarev, Z.O.; Ovchinnikova, T.V. Dimerization of the Antimicrobial Peptide Arenicin Plays a Key Role in the Cytotoxicity but Not in the Antibacterial Activity. Biochem. Biophys. Res. Commun. 2017, 482, 1320–1326. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Ng, W.J.; Bhattacharjya, S. NMR Structure and Localization of the Host Defense Antimicrobial Peptide Thanatin in Zwitterionic Dodecylphosphocholine Micelle: Implications in Antimicrobial Activity. Biochim. Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183432. [Google Scholar] [CrossRef] [PubMed]
- Panteleev, P.V.; Tsarev, A.V.; Safronova, V.N.; Reznikova, O.V.; Bolosov, I.A.; Sychev, S.V.; Shenkarev, Z.O.; Ovchinnikova, T.V. Structure Elucidation and Functional Studies of a Novel β-Hairpin Antimicrobial Peptide from the Marine Polychaeta Capitella Teleta. Mar. Drugs 2020, 18, 620. [Google Scholar] [CrossRef]
- Schmidt, E.; Güntert, P. Automated Structure Determination from NMR Spectra. In Structural Proteomics; Owens, R.J., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; Volume 1261, pp. 303–329. ISBN 978-1-4939-2229-1. [Google Scholar]
- Shen, Y.; Bax, A. Protein Backbone and Sidechain Torsion Angles Predicted from NMR Chemical Shifts Using Artificial Neural Networks. J. Biomol. NMR 2013, 56, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Stavrakoudis, A.; Tsoulos, I.G.; Shenkarev, Z.O.; Ovchinnikova, T.V. Molecular Dynamics Simulation of Antimicrobial Peptide Arenicin-2: β-Hairpin Stabilization by Noncovalent Interactions. Biopolymers 2009, 92, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Pyrkov, T.V.; Chugunov, A.O.; Krylov, N.A.; Nolde, D.E.; Efremov, R.G. PLATINUM: A Web Tool for Analysis of Hydrophobic/Hydrophilic Organization of Biomolecular Complexes. Bioinformatics 2009, 25, 1201–1202. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Fahrner, R.L.; Dieckmann, T.; Harwig, S.S.L.; Lehrer, R.I.; Eisenberg, D.; Feigon, J. Solution Structure of Protegrin-1, a Broad-Spectrum Antimicrobial Peptide from Porcine Leukocytes. Chem. Biol. 1996, 3, 543–550. [Google Scholar] [CrossRef]
- Ovchinnikova, T.V.; Shenkarev, Z.O.; Nadezhdin, K.D.; Balandin, S.V.; Zhmak, M.N.; Kudelina, I.A.; Finkina, E.I.; Kokryakov, V.N.; Arseniev, A.S. Recombinant Expression, Synthesis, Purification, and Solution Structure of Arenicin. Biochem. Biophys. Res. Commun. 2007, 360, 156–162. [Google Scholar] [CrossRef]
- Elliott, A.G.; Huang, J.X.; Neve, S.; Zuegg, J.; Edwards, I.A.; Cain, A.K.; Boinett, C.J.; Barquist, L.; Lundberg, C.V.; Steen, J.; et al. An Amphipathic Peptide with Antibiotic Activity against Multidrug-Resistant Gram-Negative Bacteria. Nat. Commun. 2020, 11, 3184. [Google Scholar] [CrossRef]
- Powers, J.-P.S.; Rozek, A.; Hancock, R.E.W. Structure-Activity Relationships for the Beta-Hairpin Cationic Antimicrobial Peptide Polyphemusin I. Biochim. Biophys. Acta 2004, 1698, 239–250. [Google Scholar] [CrossRef]
- Tasiemski, A.; Jung, S.; Boidin-Wichlacz, C.; Jollivet, D.; Cuvillier-Hot, V.; Pradillon, F.; Vetriani, C.; Hecht, O.; Sönnichsen, F.D.; Gelhaus, C.; et al. Characterization and Function of the First Antibiotic Isolated from a Vent Organism: The Extremophile Metazoan Alvinella Pompejana. PLoS ONE 2014, 9, e95737. [Google Scholar] [CrossRef]
- Deplazes, E.; Chin, Y.K.-Y.; King, G.F.; Mancera, R.L. The Unusual Conformation of Cross-Strand Disulfide Bonds Is Critical to the Stability of β-Hairpin Peptides. Proteins 2020, 88, 485–502. [Google Scholar] [CrossRef]
- Xiao, Y.; Dai, H.; Bommineni, Y.R.; Soulages, J.L.; Gong, Y.-X.; Prakash, O.; Zhang, G. Structure–Activity Relationships of Fowlicidin-1, a Cathelicidin Antimicrobial Peptide in Chicken. FEBS J. 2006, 273, 2581–2593. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Eisenberg, D. The Structure of Melittin. II. Interpretation of the Structure. J. Biol. Chem. 1982, 257, 6016–6022. [Google Scholar] [CrossRef] [PubMed]
- Gesell, J.; Zasloff, M.; Opella, S.J. Two-Dimensional 1H NMR Experiments Show That the 23-Residue Magainin Antibiotic Peptide Is an α-Helix in Dodecylphosphocholine Micelles, Sodium Dodecylsulfate Micelles, and Trifluoroethanol/Water Solution. J. Biomol. NMR 1997, 9, 127–135. [Google Scholar] [CrossRef]
- Bhunia, A.; Ramamoorthy, A.; Bhattacharjya, S. Helical Hairpin Structure of a Potent Antimicrobial Peptide MSI-594 in Lipopolysaccharide Micelles by NMR Spectroscopy. Chem.—A Eur. J. 2009, 15, 2036–2040. [Google Scholar] [CrossRef] [PubMed]
- Edwards, I.A.; Elliott, A.G.; Kavanagh, A.M.; Zuegg, J.; Blaskovich, M.A.T.; Cooper, M.A. Contribution of Amphipathicity and Hydrophobicity to the Antimicrobial Activity and Cytotoxicity of β-Hairpin Peptides. ACS Infect. Dis. 2016, 2, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Scott, M.G.; Yan, H.; Mayer, L.D.; Hancock, R.E. Interaction of Polyphemusin I and Structural Analogs with Bacterial Membranes, Lipopolysaccharide, and Lipid Monolayers. Biochemistry 2000, 39, 14504–14514. [Google Scholar] [CrossRef]
- Xiao, Y.; Cai, Y.; Bommineni, Y.R.; Fernando, S.C.; Prakash, O.; Gilliland, S.E.; Zhang, G. Identification and Functional Characterization of Three Chicken Cathelicidins with Potent Antimicrobial Activity*. J. Biol. Chem. 2006, 281, 2858–2867. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Sugishita, K.; Fujii, N.; Miyajima, K. Molecular Basis for Membrane Selectivity of an Antimicrobial Peptide, Magainin 2. Biochemistry 1995, 34, 3423–3429. [Google Scholar] [CrossRef] [PubMed]
- Mineev, K.S.; Lesovoy, D.M.; Usmanova, D.R.; Goncharuk, S.A.; Shulepko, M.A.; Lyukmanova, E.N.; Kirpichnikov, M.P.; Bocharov, E.V.; Arseniev, A.S. NMR-Based Approach to Measure the Free Energy of Transmembrane Helix-Helix Interactions. Biochim. Biophys. Acta 2014, 1838, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Josse, D.; Ebel, C.; Stroebel, D.; Fontaine, A.; Borges, F.; Echalier, A.; Baud, D.; Renault, F.; le Maire, M.; Chabrières, E.; et al. Oligomeric States of the Detergent-Solubilized Human Serum Paraoxonase (PON1). J. Biol. Chem. 2002, 277, 33386–33397. [Google Scholar] [CrossRef]
- Fisher, L.E.; Engelman, D.M.; Sturgis, J.N. Effect of Detergents on the Association of the Glycophorin A Transmembrane Helix. Biophys. J. 2003, 85, 3097–3105. [Google Scholar] [CrossRef][Green Version]
- Lauterwein, J.; Bösch, C.; Brown, L.R.; Wüthrich, K. Physicochemical Studies of the Protein-Lipid Interactions in Melittin-Containing Micelles. Biochim. Biophys. Acta 1979, 556, 244–264. [Google Scholar] [CrossRef] [PubMed]
- Fleming, K.G. Standardizing the Free Energy Change of Transmembrane Helix–Helix Interactions. J. Mol. Biol. 2002, 323, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Strandberg, E.; Ulrich, A. NMR Methods for Studying Membrane-active Antimicrobial Peptides. Concepts Magn. Reson. Part A 2004, 23A, 89–120. [Google Scholar] [CrossRef]
- Brown, L.R.; Bösch, C.; Wüthrich, K. Location and Orientation Relative to the Micelle Surface for Glucagon in Mixed Micelles with Dodecylphosphocholine: EPR and NMR Studies. Biochim. Biophys. Acta 1981, 642, 296–312. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Porcelli, F.; Buck-Koehntop, B.A.; Thennarasu, S.; Ramamoorthy, A.; Veglia, G. Structures of the Dimeric and Monomeric Variants of Magainin Antimicrobial Peptides (MSI-78 and MSI-594) in Micelles and Bilayers, Determined by NMR Spectroscopy. Biochemistry 2006, 45, 5793–5799. [Google Scholar] [CrossRef]
- Saravanan, R.; Bhattacharjya, S. Oligomeric Structure of a Cathelicidin Antimicrobial Peptide in Dodecylphosphocholine Micelle Determined by NMR Spectroscopy. Biochim. Biophys. Acta (BBA)-Biomembr. 2011, 1808, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Vetterli, S.U.; Zerbe, K.; Müller, M.; Urfer, M.; Mondal, M.; Wang, S.-Y.; Moehle, K.; Zerbe, O.; Vitale, A.; Pessi, G.; et al. Thanatin Targets the Intermembrane Protein Complex Required for Lipopolysaccharide Transport in Escherichia Coli. Sci. Adv. 2018, 4, eaau2634. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Fang, C.; Lu, L.; Wang, M.; Xue, X.; Zhou, Y.; Li, M.; Hu, Y.; Luo, X.; Hou, Z. The Antimicrobial Peptide Thanatin Disrupts the Bacterial Outer Membrane and Inactivates the NDM-1 Metallo-β-Lactamase. Nat. Commun. 2019, 10, 3517. [Google Scholar] [CrossRef] [PubMed]
- Buffy, J.J.; Waring, A.J.; Hong, M. Determination of Peptide Oligomerization in Lipid Bilayers Using 19F Spin Diffusion NMR. J. Am. Chem. Soc. 2005, 127, 4477–4483. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of Peptide Hydrophobicity in the Mechanism of Action of Alpha-Helical Antimicrobial Peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Hirakura, Y.; Kobayashi, S.; Matsuzaki, K. Specific Interactions of the Antimicrobial Peptide Cyclic Beta-Sheet Tachyplesin I with Lipopolysaccharides. Biochim. Biophys. Acta 2002, 1562, 32–36. [Google Scholar] [CrossRef]
- Giangaspero, A.; Sandri, L.; Tossi, A. Amphipathic Alpha Helical Antimicrobial Peptides. Eur. J. Biochem. 2001, 268, 5589–5600. [Google Scholar] [CrossRef]
- Li, J.; Koh, J.-J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Yoneyama, S.; Fujii, N.; Miyajima, K.; Yamada, K.; Kirino, Y.; Anzai, K. Membrane Permeabilization Mechanisms of a Cyclic Antimicrobial Peptide, Tachyplesin I, and Its Linear Analog. Biochemistry 1997, 36, 9799–9806. [Google Scholar] [CrossRef]
- Imura, Y.; Nishida, M.; Ogawa, Y.; Takakura, Y.; Matsuzaki, K. Action Mechanism of Tachyplesin I and Effects of PEGylation. Biochim. Biophys. Acta (BBA)-Biomembr. 2007, 1768, 1160–1169. [Google Scholar] [CrossRef]
- Lee, D.-K.; Bhunia, A.; Kotler, S.A.; Ramamoorthy, A. Detergent-Type Membrane Fragmentation by MSI-78, MSI-367, MSI-594, and MSI-843 Antimicrobial Peptides and Inhibition by Cholesterol: A Solid-State Nuclear Magnetic Resonance Study. Biochemistry 2015, 54, 1897–1907. [Google Scholar] [CrossRef]
- Wimley, W.C.; Hristova, K.; Ladokhin, A.S.; Silvestro, L.; Axelsen, P.H.; White, S.H. Folding of β-Sheet Membrane Proteins: A Hydrophobic Hexapeptide Model. J. Mol. Biol. 1998, 277, 1091–1110. [Google Scholar] [CrossRef]
- Fleming, K.G. Energetics of Membrane Protein Folding. Annu. Rev. Biophys. 2014, 43, 233–255. [Google Scholar] [CrossRef]
- Chen, L.; Novicky, L.; Merzlyakov, M.; Hristov, T.; Hristova, K. Measuring the Energetics of Membrane Protein Dimerization in Mammalian Membranes. J. Am. Chem. Soc. 2010, 132, 3628–3635. [Google Scholar] [CrossRef]
- Hong, H.; Blois, T.M.; Cao, Z.; Bowie, J.U. Method to Measure Strong Protein–Protein Interactions in Lipid Bilayers Using a Steric Trap. Proc. Natl. Acad. Sci. USA 2010, 107, 19802–19807. [Google Scholar] [CrossRef]
- Bocharov, E.V.; Mineev, K.S.; Goncharuk, M.V.; Arseniev, A.S. Structural and Thermodynamic Insight into the Process of “Weak” Dimerization of the ErbB4 Transmembrane Domain by Solution NMR. Biochim. Biophys. Acta 2012, 1818, 2158–2170. [Google Scholar] [CrossRef]
- Myers, J.K.; Nick Pace, C.; Martin Scholtz, J. Denaturant m Values and Heat Capacity Changes: Relation to Changes in Accessible Surface Areas of Protein Unfolding. Protein Sci. 1995, 4, 2138–2148. [Google Scholar] [CrossRef]
- Gómez, J.; Hilser, V.J.; Xie, D.; Freire, E. The Heat Capacity of Proteins. Proteins Struct. Funct. Bioinform. 1995, 22, 404–412. [Google Scholar] [CrossRef]
- Mogensen, J.E.; Tapadar, D.; Schmidt, M.A.; Otzen, D.E. Barriers to Folding of the Transmembrane Domain of the Escherichia Coli Autotransporter Adhesin Involved in Diffuse Adherence. Biochemistry 2005, 44, 4533–4545. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mironov, P.A.; Paramonov, A.S.; Reznikova, O.V.; Safronova, V.N.; Panteleev, P.V.; Bolosov, I.A.; Ovchinnikova, T.V.; Shenkarev, Z.O. Dimerization of the β-Hairpin Membrane-Active Cationic Antimicrobial Peptide Capitellacin from Marine Polychaeta: An NMR Structural and Thermodynamic Study. Biomolecules 2024, 14, 332. https://doi.org/10.3390/biom14030332
Mironov PA, Paramonov AS, Reznikova OV, Safronova VN, Panteleev PV, Bolosov IA, Ovchinnikova TV, Shenkarev ZO. Dimerization of the β-Hairpin Membrane-Active Cationic Antimicrobial Peptide Capitellacin from Marine Polychaeta: An NMR Structural and Thermodynamic Study. Biomolecules. 2024; 14(3):332. https://doi.org/10.3390/biom14030332
Chicago/Turabian StyleMironov, Pavel A., Alexander S. Paramonov, Olesya V. Reznikova, Victoria N. Safronova, Pavel V. Panteleev, Ilia A. Bolosov, Tatiana V. Ovchinnikova, and Zakhar O. Shenkarev. 2024. "Dimerization of the β-Hairpin Membrane-Active Cationic Antimicrobial Peptide Capitellacin from Marine Polychaeta: An NMR Structural and Thermodynamic Study" Biomolecules 14, no. 3: 332. https://doi.org/10.3390/biom14030332
APA StyleMironov, P. A., Paramonov, A. S., Reznikova, O. V., Safronova, V. N., Panteleev, P. V., Bolosov, I. A., Ovchinnikova, T. V., & Shenkarev, Z. O. (2024). Dimerization of the β-Hairpin Membrane-Active Cationic Antimicrobial Peptide Capitellacin from Marine Polychaeta: An NMR Structural and Thermodynamic Study. Biomolecules, 14(3), 332. https://doi.org/10.3390/biom14030332







