Single-Cell RNA-Seq Analysis Links DNMT3B and PFKFB4 Transcriptional Profiles with Metastatic Traits in Hepatoblastoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Public Hepatoblastoma Tumor Transcriptome Dataset
2.2. Expression of Metabolic Program
2.3. Machine Learning ElasticNet Model of Metabolic Markers
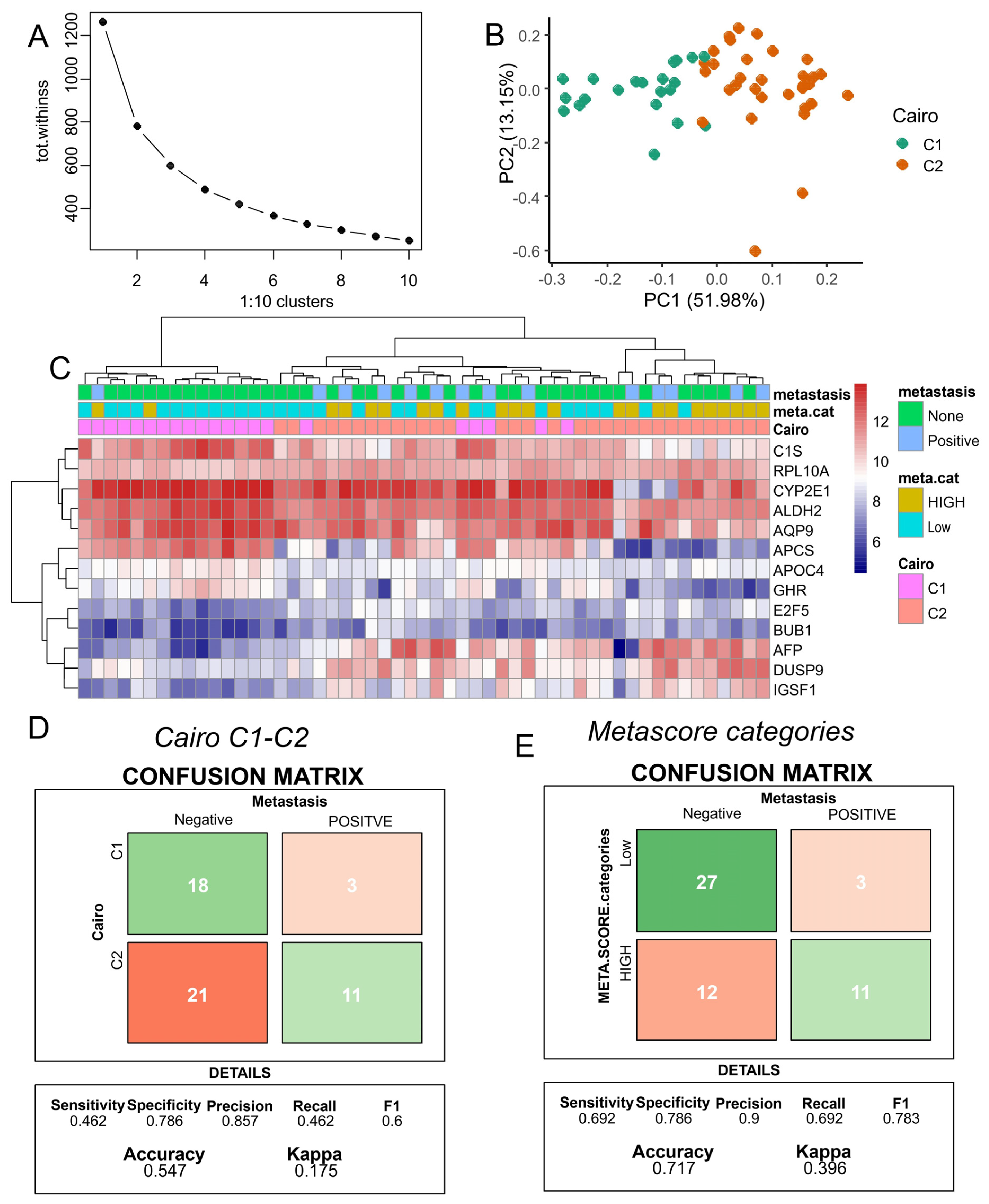
2.4. C1/C2 Classifier
3. Results
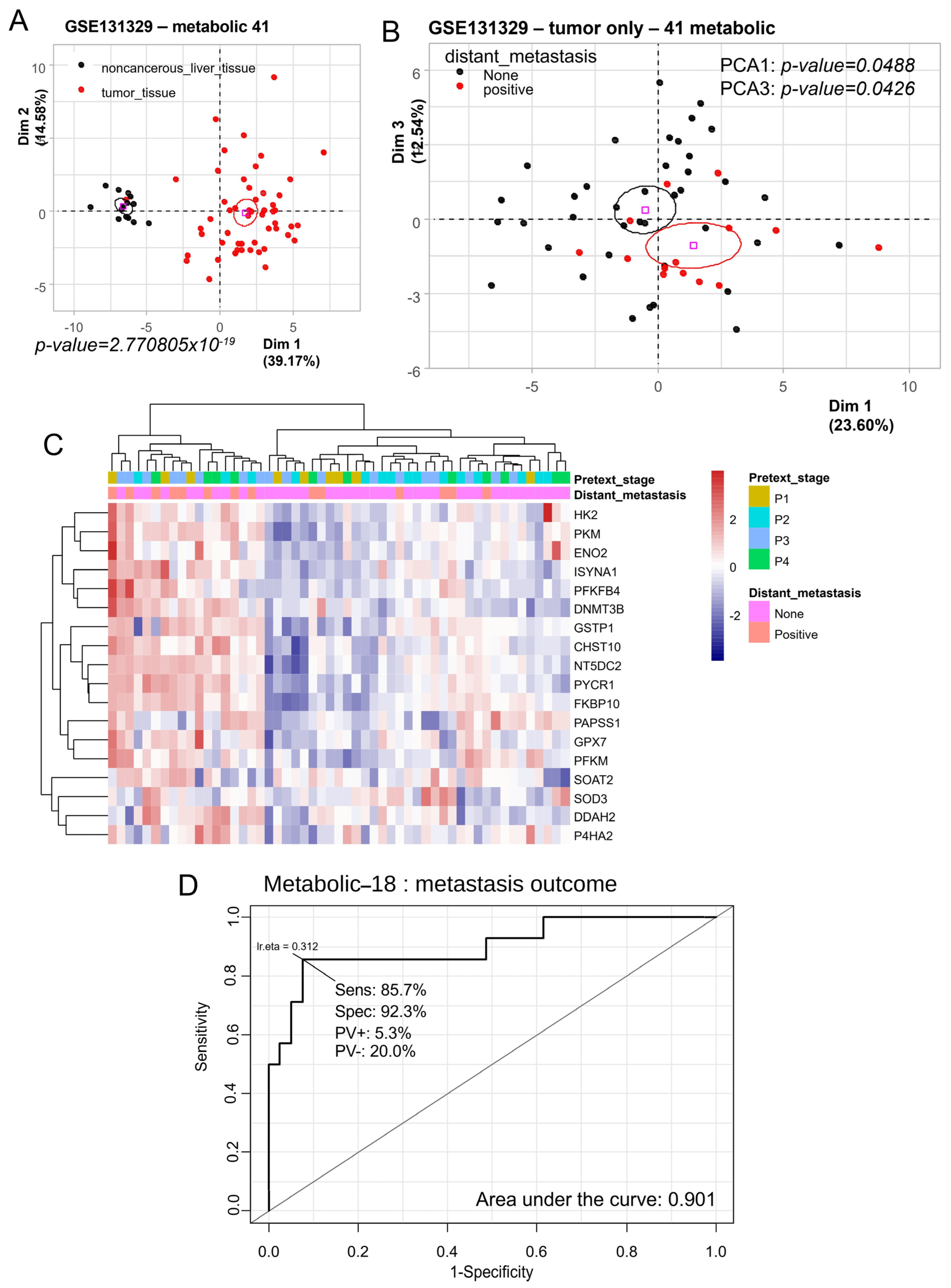
3.1. Metabolic Program of Hepatoblastoma Tumors Predicts Distant Metastasis Status
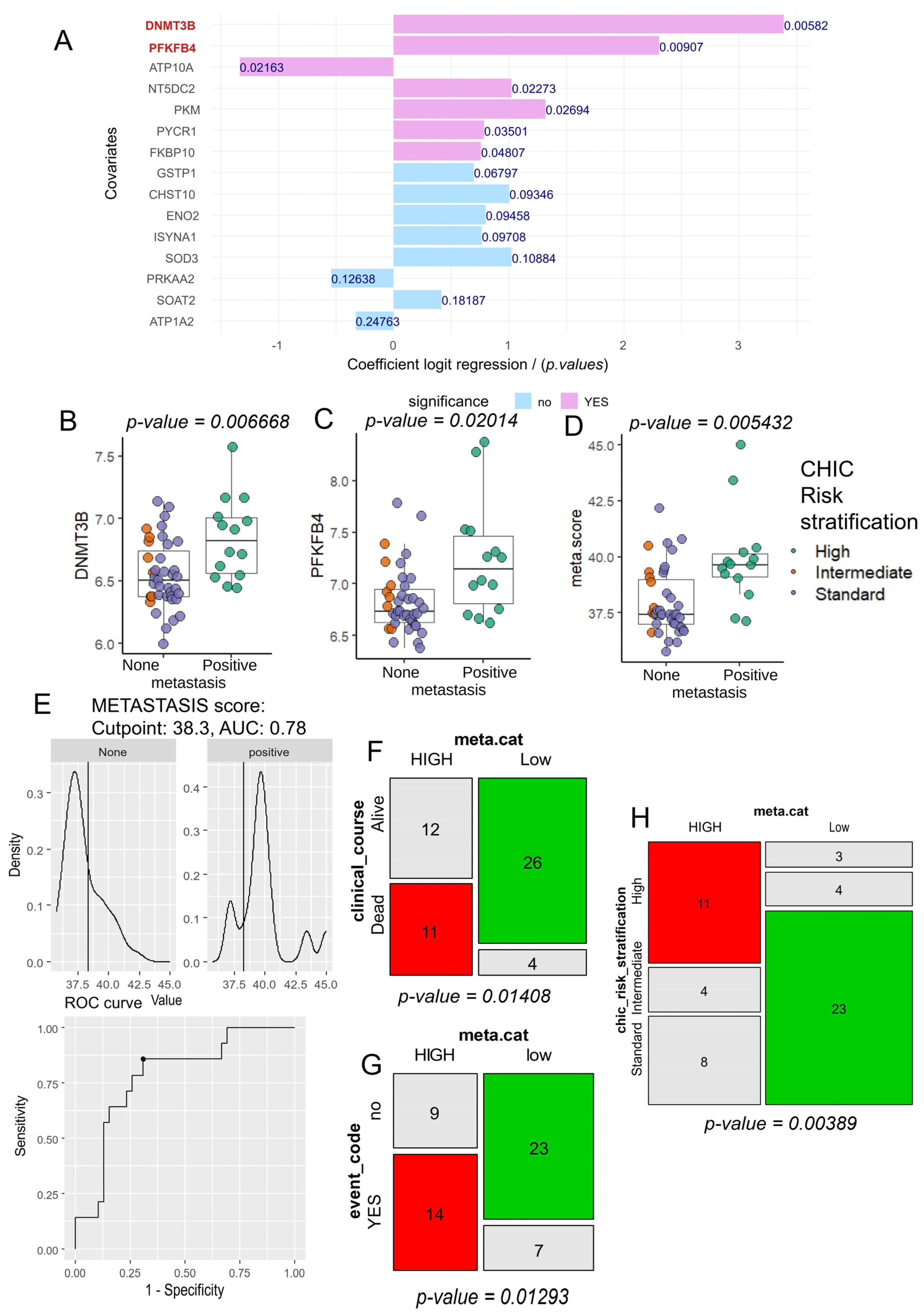
3.2. Computational Analysis of Metabolic Enzymes for Predicting Metastatic Potential in Hepatoblastoma
3.3. Tumor Co-Expression of DNMT3B and PFKFB4 as a Predictor of Metastasis and CHIC2 Risk Stratification in Hepatoblastoma
3.4. Improved Metastasis Risk Assessment with the Combined DNMT3B and PFKFB4 Metabolic Expression Score
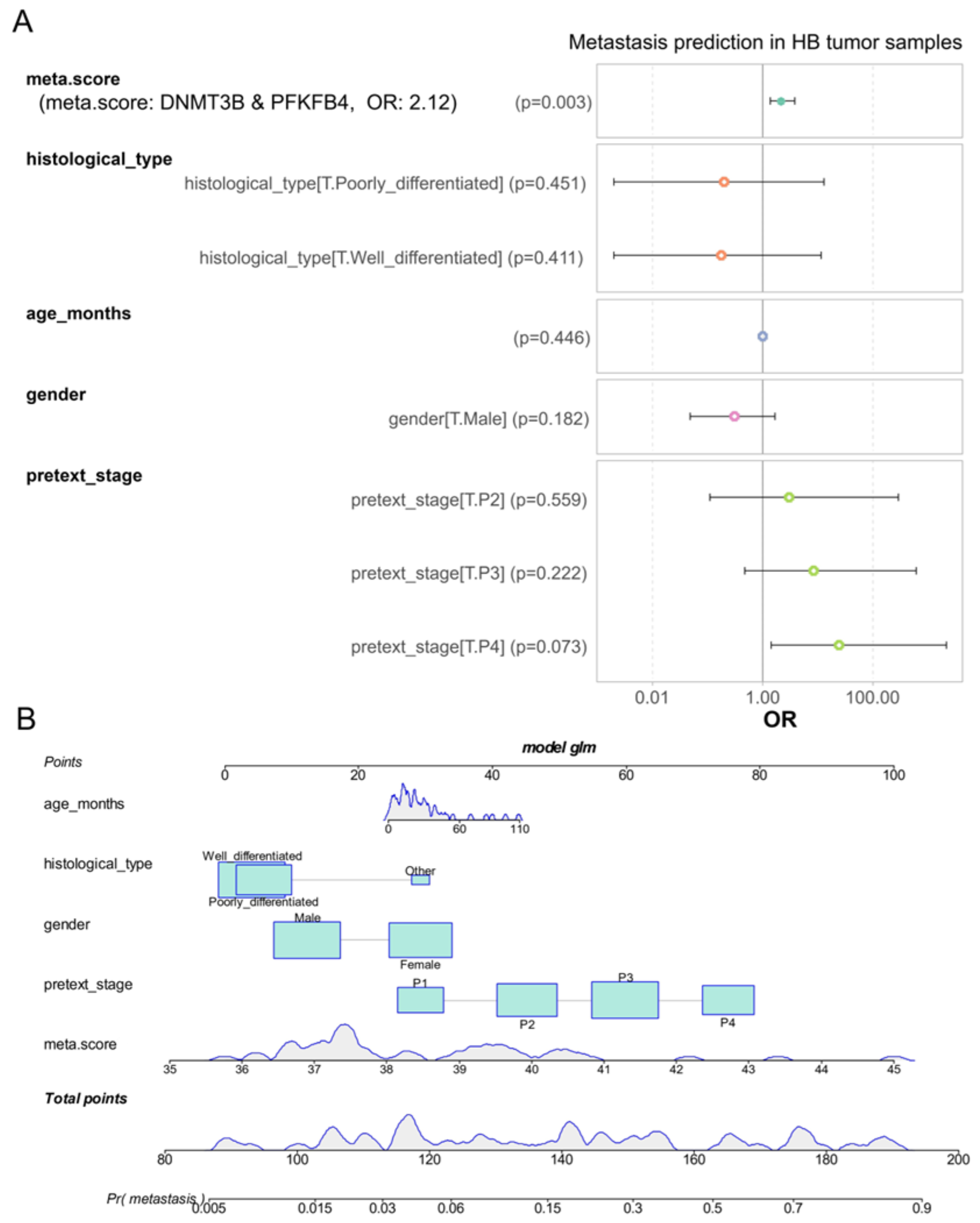
3.5. DNMT3B and PFKFB4 Metabolic Expression Score as an Independent Predictor of Metastasis in Hepatoblastoma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perilongo, G.; Shafford, E.; Plaschkes, J.; Liver Tumour Study Group of the International Society of Paediatric Oncology. SIOPEL Trials Using Preoperative Chemotherapy in Hepatoblastoma. Lancet Oncol. 2000, 1, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, F.; Matsunaga, T.; Iwafuchi, M.; Hayashi, Y.; Ohkawa, H.; Ohira, M.; Okamatsu, T.; Sugito, T.; Tsuchida, Y.; Toyosaka, A.; et al. Outcome of Hepatoblastoma Treated with the JPLT-1 (Japanese Study Group for Pediatric Liver Tumor) Protocol-1: A Report from the Japanese Study Group for Pediatric Liver Tumor. J. Pediatr. Surg. 2002, 37, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Meyers, R.L.; Maibach, R.; Hiyama, E.; Häberle, B.; Krailo, M.; Rangaswami, A.; Aronson, D.C.; Malogolowkin, M.H.; Perilongo, G.; von Schweinitz, D.; et al. Risk-Stratified Staging in Paediatric Hepatoblastoma: A Unified Analysis from the Children’s Hepatic Tumors International Collaboration. Lancet Oncol. 2017, 18, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Koh, K.N.; Park, M.; Kim, B.E.; Bae, K.W.; Kim, K.M.; Im, H.J.; Seo, J.J. Prognostic Implications of Serum Alpha-Fetoprotein Response during Treatment of Hepatoblastoma. Pediatr. Blood Cancer 2011, 57, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Meyers, R.L.; Rowland, J.R.; Krailo, M.; Chen, Z.; Katzenstein, H.M.; Malogolowkin, M.H. Predictive Power of Pretreatment Prognostic Factors in Children with Hepatoblastoma: A Report from the Children’s Oncology Group. Pediatr. Blood Cancer 2009, 53, 1016–1022. [Google Scholar] [CrossRef]
- Hata, Y. The Clinical Features and Prognosis of Hepatoblastoma: Follow-up Studies Done on Pediatric Tumors Enrolled in the Japanese Pediatric Tumor Registry between 1971 and 1980. Part I. Jpn. J. Surg. 1990, 20, 498–502. [Google Scholar] [CrossRef]
- Hishiki, T.; Matsunaga, T.; Sasaki, F.; Yano, M.; Ida, K.; Horie, H.; Kondo, S.; Watanabe, K.-I.; Oue, T.; Tajiri, T.; et al. Outcome of Hepatoblastomas Treated Using the Japanese Study Group for Pediatric Liver Tumor (JPLT) Protocol-2: Report from the JPLT. Pediatr. Surg. Int. 2011, 27, 1–8. [Google Scholar] [CrossRef]
- Czauderna, P.; Haeberle, B.; Hiyama, E.; Rangaswami, A.; Krailo, M.; Maibach, R.; Rinaldi, E.; Feng, Y.; Aronson, D.; Malogolowkin, M.; et al. The Children’s Hepatic Tumors International Collaboration (CHIC): Novel Global Rare Tumor Database Yields New Prognostic Factors in Hepatoblastoma and Becomes a Research Model. Eur. J. Cancer 2016, 52, 92–101. [Google Scholar] [CrossRef]
- Hiyama, E. Gene Expression Profiling in Hepatoblastoma Cases of the Japanese Study Group for Pediatric Liver Tumors-2 (JPLT-2) Trial; Science Repository OU: Tallinn, Estonia, 2019. [Google Scholar]
- Dai, Z.; Ramesh, V.; Locasale, J.W. The Evolving Metabolic Landscape of Chromatin Biology and Epigenetics. Nat. Rev. Genet. 2020, 21, 737–753. [Google Scholar] [CrossRef]
- Geiman, T.M.; Robertson, K.D. Chromatin Remodeling, Histone Modifications, and DNA Methylation—How Does It All Fit Together? J. Cell. Biochem. 2002, 87, 117–125. [Google Scholar] [CrossRef]
- Rodríguez-Paredes, M.; Esteller, M. Cancer Epigenetics Reaches Mainstream Oncology. Nat. Med. 2011, 17, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Liu, B.; Zheng, S.; Dong, K.; Dong, R. Genome-Wide Analysis of DNA Methylation in Hepatoblastoma Tissues. Oncol. Lett. 2016, 12, 1529–1534. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Meltzer, P.S. GEOquery: A Bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Monge, C.; Francés, R.; Marchio, A.; Pineau, P.; Desterke, C.; Matta-Garrido, J. Activated Metabolic Transcriptional Program in Tumor Cells from Hepatoblastoma. Int. J. Mol. Sci. Preprints 2024, 2024090699. [Google Scholar] [CrossRef]
- Corcoran, C.C.; Grady, C.R.; Pisitkun, T.; Parulekar, J.; Knepper, M.A. From 20th Century Metabolic Wall Charts to 21st Century Systems Biology: Database of Mammalian Metabolic Enzymes. Am. J. Physiol. Ren. Physiol. 2017, 312, F533–F542. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the Caret Package. J. Stat. Soft. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Tay, J.K.; Narasimhan, B.; Hastie, T. Elastic Net Regularization Paths for All Generalized Linear Models. J. Stat. Soft. 2023, 106. [Google Scholar] [CrossRef]
- Zeileis, A.; Meyer, D.; Hornik, K. Residual-Based Shadings for Visualizing (Conditional) Independence. J. Comput. Graph. Stat. 2007, 16, 507–525. [Google Scholar] [CrossRef]
- Cairo, S.; Armengol, C.; De Reyniès, A.; Wei, Y.; Thomas, E.; Renard, C.-A.; Goga, A.; Balakrishnan, A.; Semeraro, M.; Gresh, L.; et al. Hepatic Stem-like Phenotype and Interplay of Wnt/Beta-Catenin and Myc Signaling in Aggressive Childhood Liver Cancer. Cancer Cell 2008, 14, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. The DNA Methylation Paradox. Trends Genet. 1999, 15, 34–37. [Google Scholar] [CrossRef] [PubMed]
- Rivas, M.P.; Aguiar, T.F.M.; Fernandes, G.R.; Caires-Júnior, L.C.; Goulart, E.; Telles-Silva, K.A.; Cypriano, M.; De Toledo, S.R.C.; Rosenberg, C.; Carraro, D.M.; et al. TET Upregulation Leads to 5-Hydroxymethylation Enrichment in Hepatoblastoma. Front. Genet. 2019, 10, 553. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, M.Z.; Sabitaliyevich, U.Y.; Rabandiyarov, M.; Arystanbekuly, A.T. Role of DNA Methyltransferases (DNMTs) in Metastasis. Cell Mol. Biol. 2022, 68, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Oshima, G.; Poli, E.C.; Bolt, M.J.; Chlenski, A.; Forde, M.; Jutzy, J.M.S.; Biyani, N.; Posner, M.C.; Pitroda, S.P.; Weichselbaum, R.R.; et al. DNA Methylation Controls Metastasis-Suppressive 14q32-Encoded miRNAs. Cancer Res. 2019, 79, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ren, M.; Zhao, Y.; Lu, X.; Wang, M.; Hu, J.; Lu, G.; He, S. MicroRNA-26a Inhibits Proliferation and Metastasis of Human Hepatocellular Carcinoma by Regulating DNMT3B-MEG3 Axis. Oncol. Rep. 2017, 37, 3527–3535. [Google Scholar] [CrossRef]
- Rider, M.H.; Bertrand, L.; Vertommen, D.; Michels, P.A.; Rousseau, G.G.; Hue, L. 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase: Head-to-Head with a Bifunctional Enzyme That Controls Glycolysis. Biochem. J. 2004, 381, 561–579. [Google Scholar] [CrossRef]
- Chesney, J.; Clark, J.; Klarer, A.C.; Imbert-Fernandez, Y.; Lane, A.N.; Telang, S. Fructose-2,6-Bisphosphate Synthesis by 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase 4 (PFKFB4) Is Required for the Glycolytic Response to Hypoxia and Tumor Growth. Oncotarget 2014, 5, 6670–6686. [Google Scholar] [CrossRef]
- Gao, R.; Li, D.; Xun, J.; Zhou, W.; Li, J.; Wang, J.; Liu, C.; Li, X.; Shen, W.; Qiao, H.; et al. CD44ICD Promotes Breast Cancer Stemness via PFKFB4-Mediated Glucose Metabolism. Theranostics 2018, 8, 6248–6262. [Google Scholar] [CrossRef]
- Minchenko, O.H.; Ochiai, A.; Opentanova, I.L.; Ogura, T.; Minchenko, D.O.; Caro, J.; Komisarenko, S.V.; Esumi, H. Overexpression of 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase-4 in the Human Breast and Colon Malignant Tumors. Biochimie 2005, 87, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Minchenko, O.H.; Ogura, T.; Opentanova, I.L.; Minchenko, D.O.; Ochiai, A.; Caro, J.; Komisarenko, S.V.; Esumi, H. 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase Gene Family Overexpression in Human Lung Tumor. Ukr. Biokhim. Zh. 2005, 77, 46–50. [Google Scholar]
- Olaizola, P.; Banales, J.M. PFKFB4 Is a Metabolic Driver of HCC Progression and Chemoresistance Through ROS Mitigation. Cell. Mol. Gastroenterol. Hepatol. 2023, 15, 1527–1529. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.; Rosario, S.R.; Katsuta, E.; Sawant Dessai, A.; Paterson, E.J.; Novickis, A.T.; Cortes Gomez, E.; Zhu, B.; Liu, S.; Wang, H.; et al. Hypoxic Activation of PFKFB4 in Breast Tumor Microenvironment Shapes Metabolic and Cellular Plasticity to Accentuate Metastatic Competence. Cell Rep. 2022, 41, 111756. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, S.; Rajapakshe, K.; Zhu, B.; Nikolai, B.C.; Yi, P.; Putluri, N.; Choi, J.M.; Jung, S.Y.; Coarfa, C.; Westbrook, T.F.; et al. Metabolic Enzyme PFKFB4 Activates Transcriptional Coactivator SRC-3 to Drive Breast Cancer. Nature 2018, 556, 249–254. [Google Scholar] [CrossRef]
- Sittewelle, M.; Kappès, V.; Zhou, C.; Lécuyer, D.; Monsoro-Burq, A.H. PFKFB4 Interacts with ICMT and Activates RAS/AKT Signaling-Dependent Cell Migration in Melanoma. Life Sci. Alliance 2022, 5, e202201377. [Google Scholar] [CrossRef]
- Hsin, M.-C.; Hsieh, Y.-H.; Hsiao, Y.-H.; Chen, P.-N.; Wang, P.-H.; Yang, S.-F. Carbonic Anhydrase IX Promotes Human Cervical Cancer Cell Motility by Regulating PFKFB4 Expression. Cancers 2021, 13, 1174. [Google Scholar] [CrossRef]
- Zhu, X.; Xue, C.; Kang, X.; Jia, X.; Wang, L.; Younis, M.H.; Liu, D.; Huo, N.; Han, Y.; Chen, Z.; et al. DNMT3B-mediated FAM111B methylation promotes papillary thyroid tumor glycolysis, growth and metastasis. Int. J. Biol. Sci. 2022, 18, 4372–4387. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shen, L.; Du, J.; Xia, Y.; Tan, Z.; Fu, Y.; Yang, Q.; Li, X.; Tang, G.; Jiang, Y.; Wang, J.; et al. Genome-wide landscape of DNA methylomes and their relationship with mRNA and miRNA transcriptomes in oxidative and glycolytic skeletal muscles. Sci. Rep. 2016, 6, 32186. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mei, C.; Sun, L.; Liu, Y.; Yang, Y.; Cai, X.; Liu, M.; Yao, W.; Wang, C.; Li, X.; Wang, L.; et al. Transcriptional and post-transcriptional control of DNA methyltransferase 3B is regulated by phosphatidylinositol 3 kinase/Akt pathway in human hepatocellular carcinoma cell lines. J. Cell Biochem. 2010, 111, 158–167. [Google Scholar] [CrossRef] [PubMed]

| Variable | Level | Low (n = 30) | HIGH (n = 23) | Total (n = 53) | p-Value |
|---|---|---|---|---|---|
| age_months | Mean (sd) | 24 (22.8) | 31.4 (25.7) | 27.2 (24.1) | 0.26766 |
| CHIC_risk_stratification | Standard | 23 (76.7) | 8 (34.8) | 31 (58.5) | |
| High | 3 (10.0) | 11 (47.8) | 14 (26.4) | ||
| Intermediate | 4 (13.3) | 4 (17.4) | 8 (15.1) | 0.00389 | |
| clinical_course | Alive | 26 (86.7) | 12 (52.2) | 38 (71.7) | |
| Dead | 4 (13.3) | 11 (47.8) | 15 (28.3) | 0.01408 | |
| Clinical event during follow up | No | 23 (76.7) | 9 (39.1) | 32 (60.4) | |
| YES | 7 (23.3) | 14 (60.9) | 21 (39.6) | 0.01293 | |
| histological_type | Well diff. | 17 (56.7) | 13 (56.5) | 30 (56.6) | |
| Other | 1 (3.3) | 1 (4.3) | 2 (3.8) | ||
| Poorly diff. | 12 (40.0) | 9 (39.1) | 21 (39.6) | 0.98116 | |
| Sex | Female | 14 (46.7) | 11 (47.8) | 25 (47.2) | |
| Male | 16 (53.3) | 12 (52.2) | 28 (52.8) | 1.00000 | |
| PRETEXT stage | P3 | 10 (33.3) | 8 (34.8) | 18 (34.0) | |
| P2 | 10 (33.3) | 5 (21.7) | 15 (28.3) | ||
| P4 | 4 (13.3) | 7 (30.4) | 11 (20.8) | ||
| P1 | 6 (20.0) | 3 (13.0) | 9 (17.0) | 0.41827 |
| Predictors | Beta Coefficients | Odds Ratios | p-Values |
|---|---|---|---|
| DNMT3B | 3.389 | 29.638 | 5.82 × 10−3 |
| PFKFB4 | 2.310 | 10.071 | 9.07× 10−3 |
| NT5DC2 | 1.030 | 2.801 | 2.27× 10−2 |
| PKM | 1.321 | 3.745 | 2.69 × 10−2 |
| PYCR1 | 0.792 | 2.208 | 3.50 × 10−2 |
| FKBP10 | 0.764 | 2.146 | 4.81 × 10−2 |
| GSTP1 | 0.702 | 2.017 | 6.80 × 10−2 |
| CHST10 | 1.009 | 2.742 | 9.35 × 10−2 |
| ENO2 | 0.802 | 2.230 | 9.46 × 10−2 |
| ISYNA1 | 0.772 | 2.163 | 9.71 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desterke, C.; Francés, R.; Monge, C.; Marchio, A.; Pineau, P.; Mata-Garrido, J. Single-Cell RNA-Seq Analysis Links DNMT3B and PFKFB4 Transcriptional Profiles with Metastatic Traits in Hepatoblastoma. Biomolecules 2024, 14, 1394. https://doi.org/10.3390/biom14111394
Desterke C, Francés R, Monge C, Marchio A, Pineau P, Mata-Garrido J. Single-Cell RNA-Seq Analysis Links DNMT3B and PFKFB4 Transcriptional Profiles with Metastatic Traits in Hepatoblastoma. Biomolecules. 2024; 14(11):1394. https://doi.org/10.3390/biom14111394
Chicago/Turabian StyleDesterke, Christophe, Raquel Francés, Claudia Monge, Agnès Marchio, Pascal Pineau, and Jorge Mata-Garrido. 2024. "Single-Cell RNA-Seq Analysis Links DNMT3B and PFKFB4 Transcriptional Profiles with Metastatic Traits in Hepatoblastoma" Biomolecules 14, no. 11: 1394. https://doi.org/10.3390/biom14111394
APA StyleDesterke, C., Francés, R., Monge, C., Marchio, A., Pineau, P., & Mata-Garrido, J. (2024). Single-Cell RNA-Seq Analysis Links DNMT3B and PFKFB4 Transcriptional Profiles with Metastatic Traits in Hepatoblastoma. Biomolecules, 14(11), 1394. https://doi.org/10.3390/biom14111394







