Cytochalasans and Their Impact on Actin Filament Remodeling
Abstract
1. Introduction
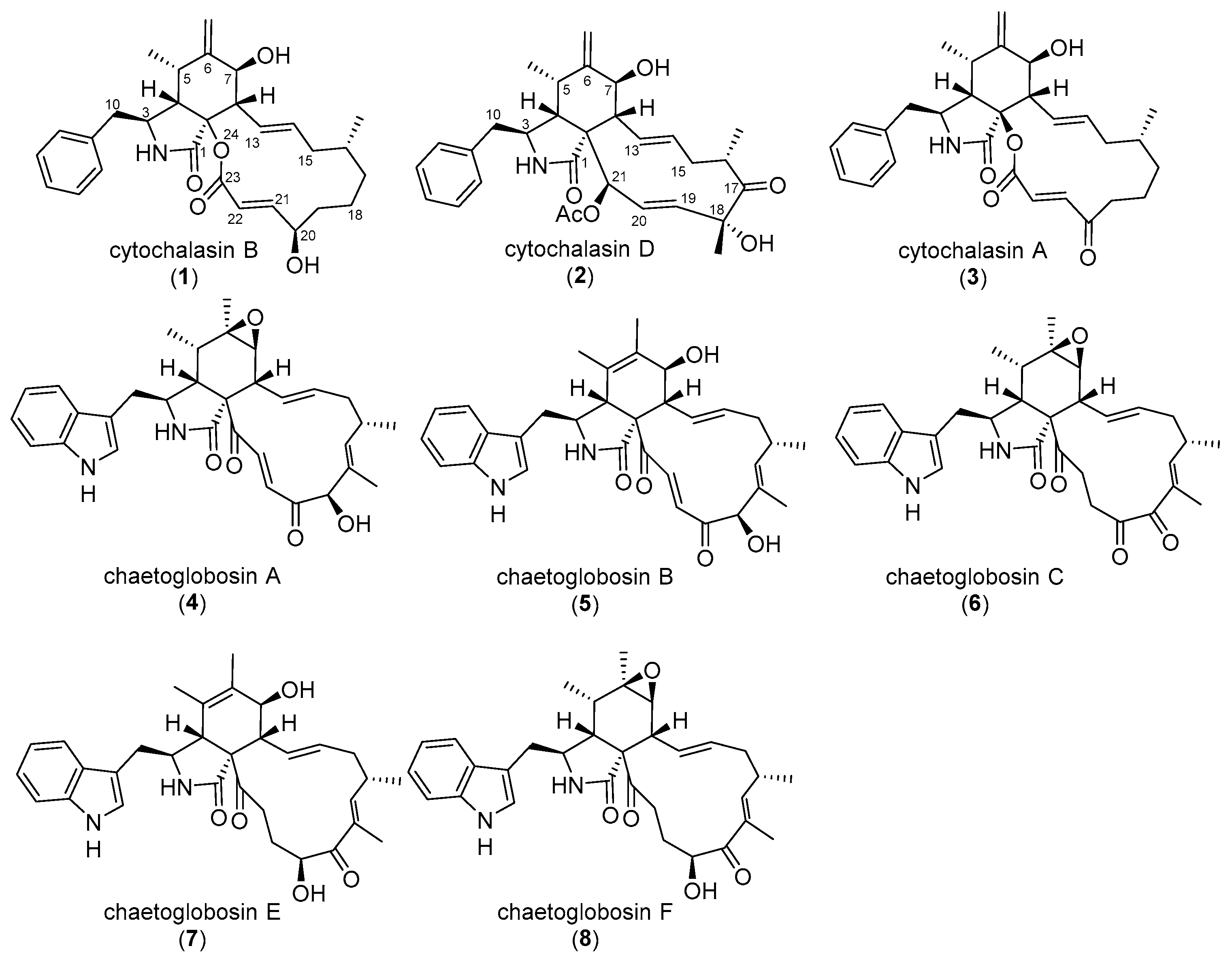
2. Reports of Cytochalasans Impacting on the Morphology of Cellular Model Systems
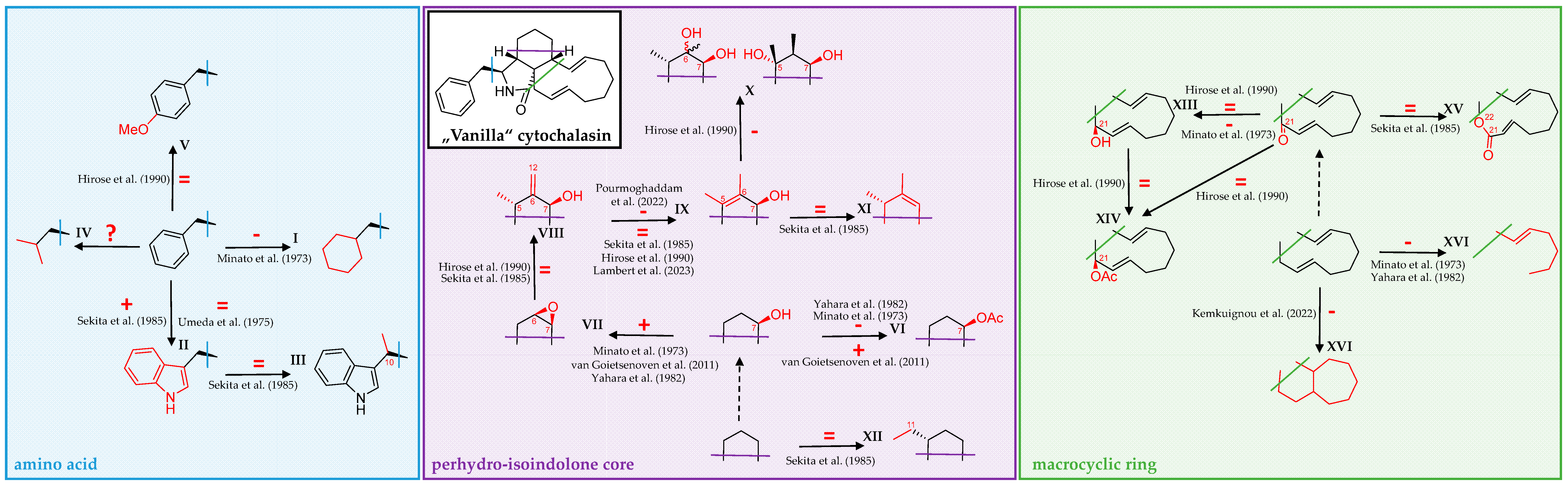
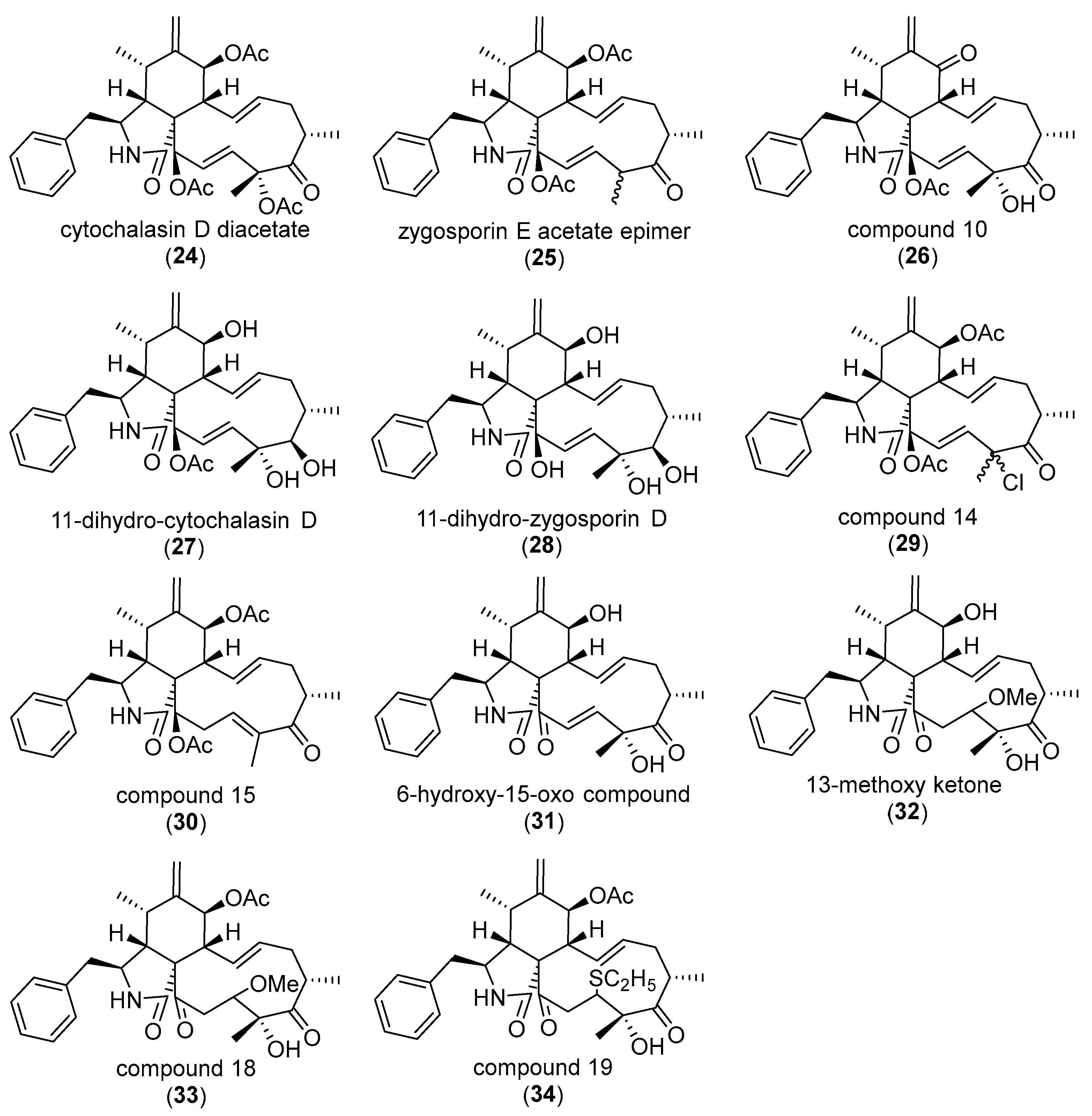
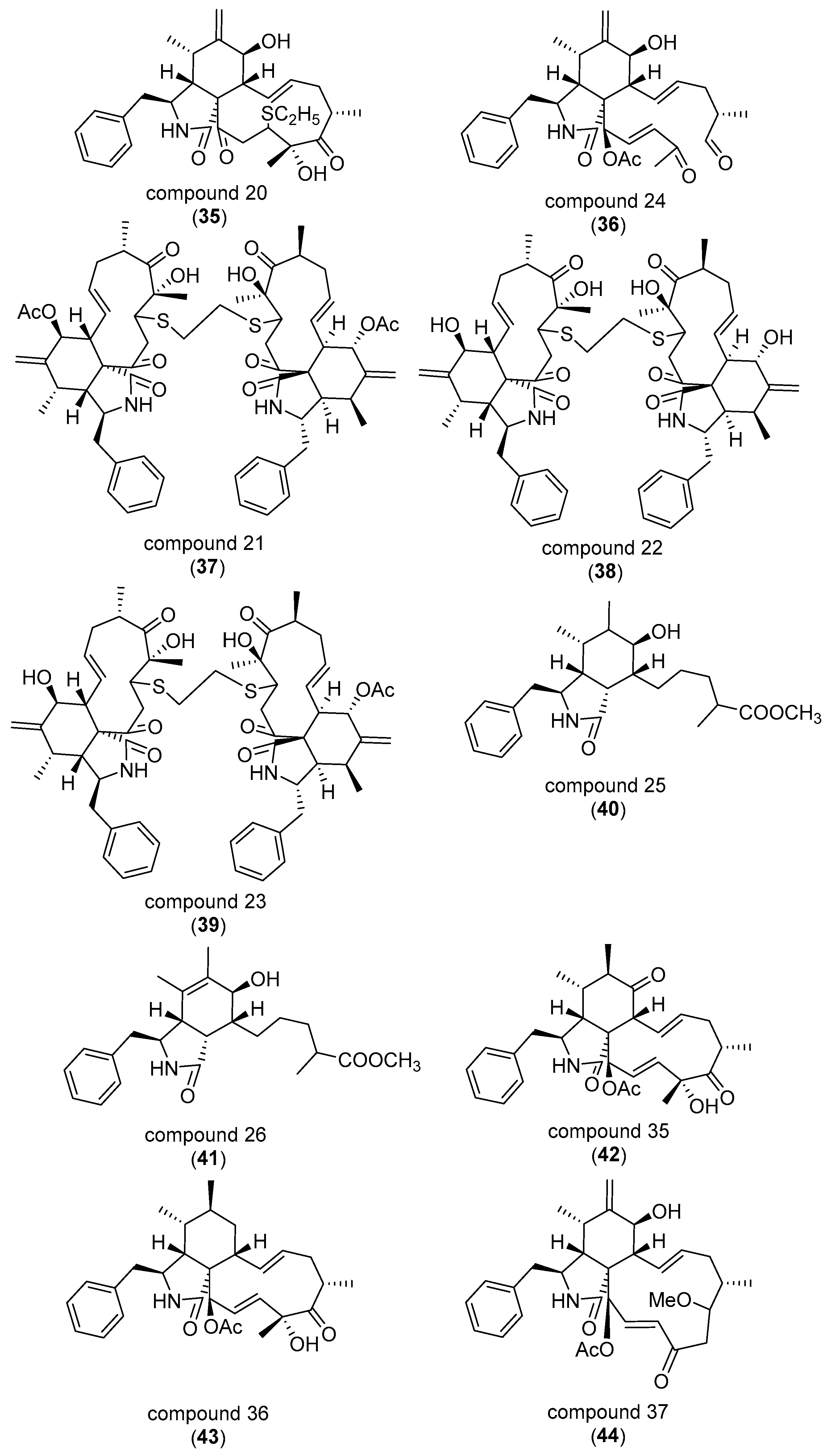
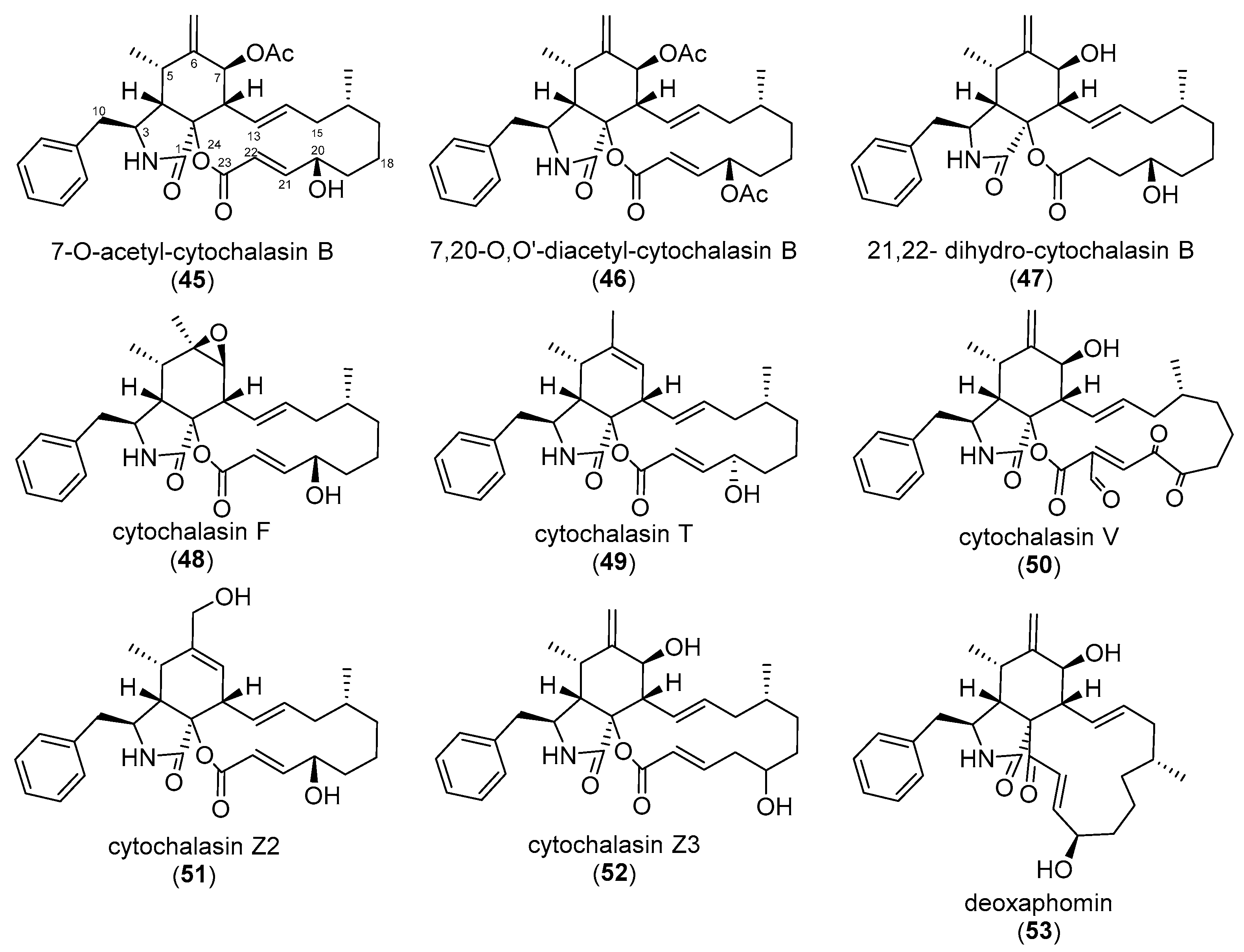
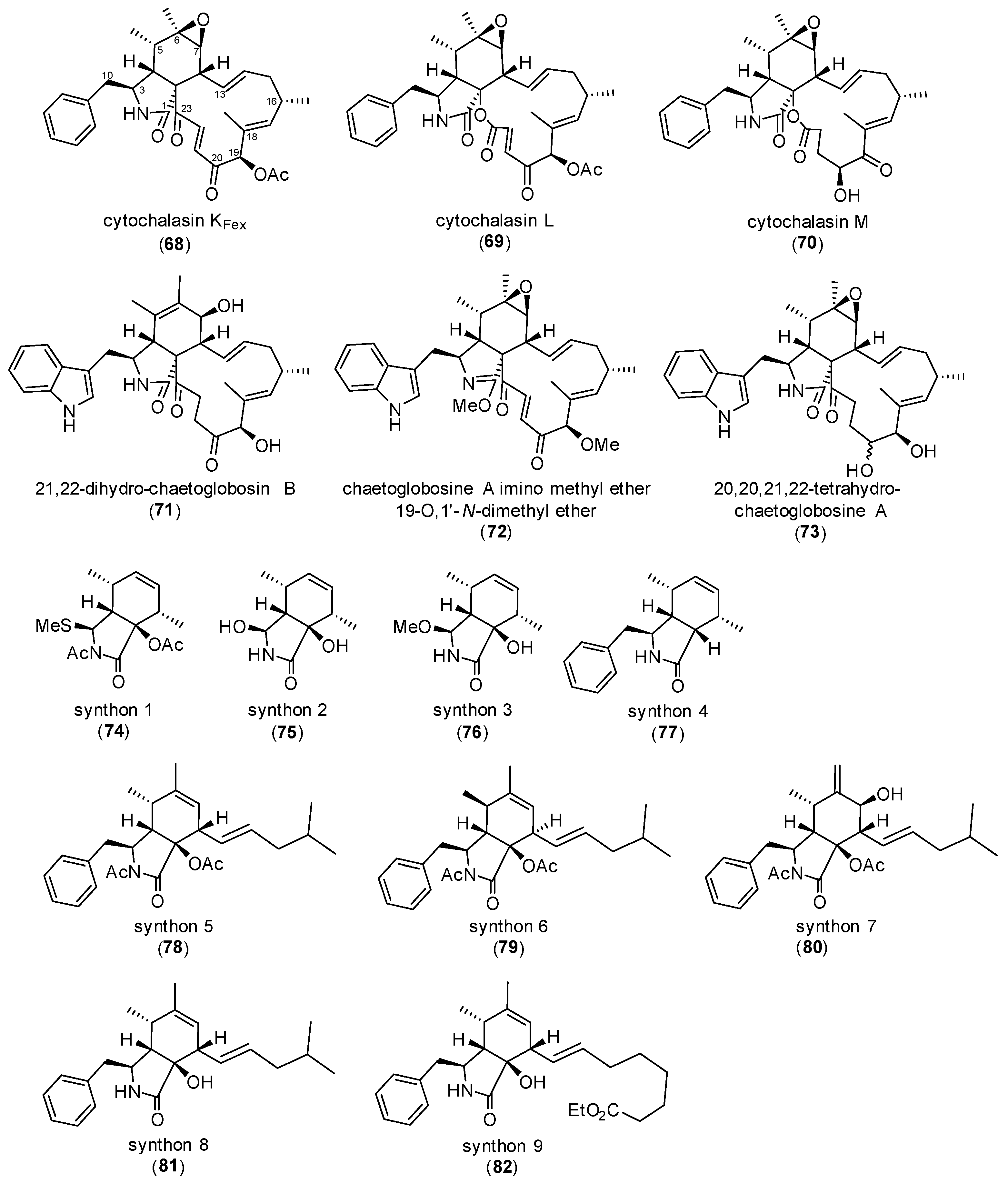
3. Correlating Cytochalasan Chemodiversity with the Activity Spectrum towards the Actin Cytoskeleton and Cell Morphology
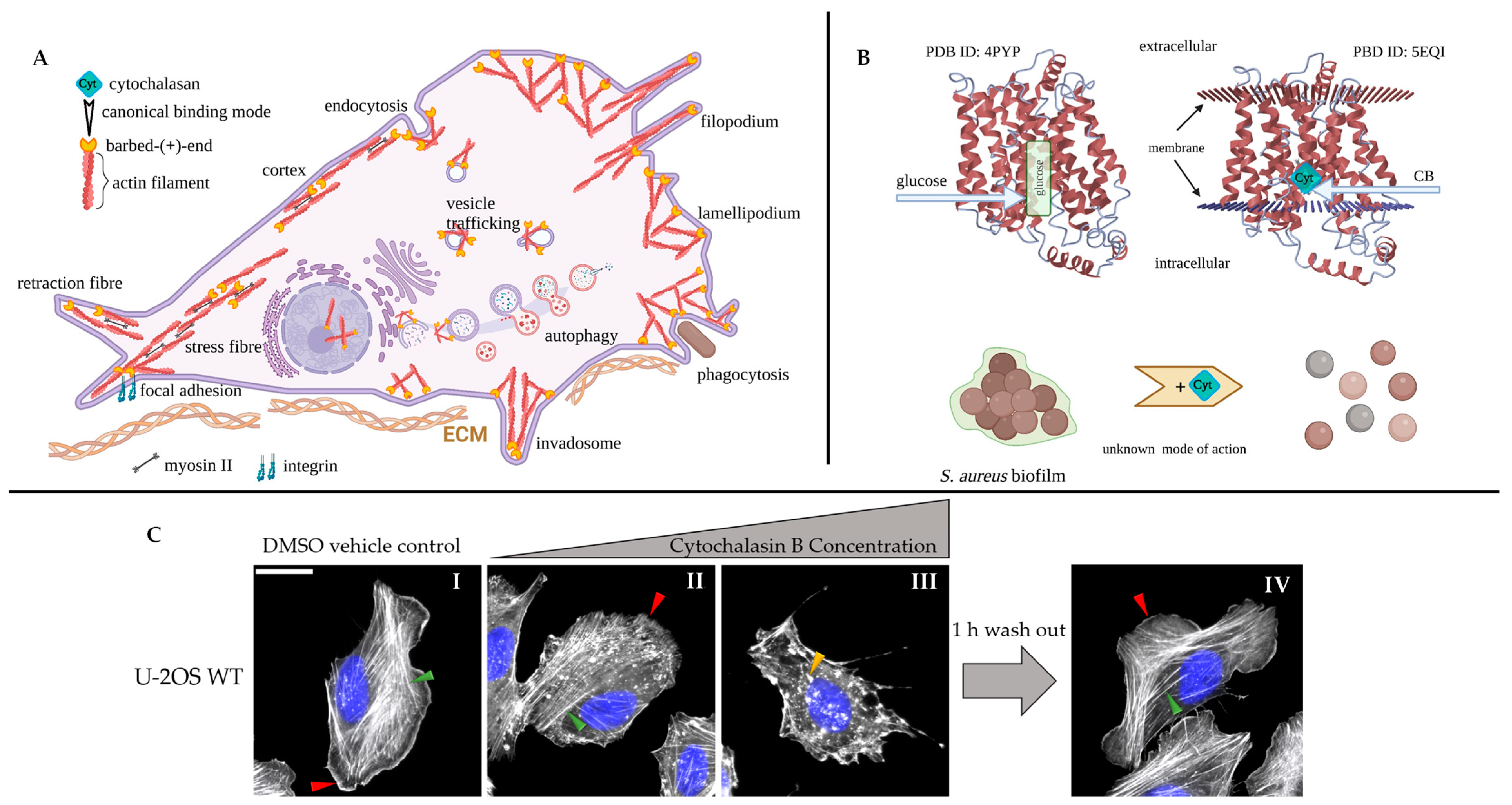
4. Cytochalasins Interfere with Cytoskeletal Dynamics
5. Cytochalasins and Reported Effects on Actin-Binding Proteins and Actin Architecture
6. Potential Non-Barbed End Binding Activities of Cytochalasans in Actin-Dependent Model Systems
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brakhage, A.A. Regulation and role of fungal secondary metabolites. Annu. Rev. Genet. 2016, 50, 371–392. [Google Scholar] [CrossRef]
- Skellam, E. The biosynthesis of cytochalasans. Nat. Prod. Rep. 2017, 34, 1252–1263. [Google Scholar] [CrossRef] [PubMed]
- Chepkirui, C.; Stadler, M. The genus Diaporthe: A rich source of diverse and bioactive metabolites. Mycol. Prog. 2017, 16, 477–494. [Google Scholar] [CrossRef]
- Becker, K.; Stadler, M. Recent progress in biodiversity research on the Xylariales and their secondary metabolism. J. Antibiot. 2021, 74, 1–23. [Google Scholar] [CrossRef]
- Charria-Girón, E.; Surup, F.; Marin-Felix, Y. Diversity of biologically active secondary metabolites in the ascomycete order Sordariales. Mycol. Prog. 2022, 21, 43. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, C.; Tong, Q.; Zhou, Y.; Ye, Y.; Gu, L.; Zhang, Y. Progress in the chemistry of cytochalasans. Prog. Chem. Org. Nat. Prod. 2021, 114, 1–134. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, D.C.; Armstrong, J.J.; Speake, R.N.; Turner, W.B. The cytochalasins, a new class of biologically active mould metabolites. Chem. Commun. 1967, 26–27. [Google Scholar] [CrossRef]
- Schroeder, T.E. The contractile ring. I. Fine structure of dividing mammalian (HeLa) cells and the effects of cytochalasin B. Z. Zellforsch. Mikrosk. Anat. 1970, 109, 431–449. [Google Scholar] [CrossRef]
- Spudich, J.A.; Lin, S. Cytochalasin B, its interaction with actin and actomyosin from muscle (cell movement-microfilaments-rabbit striated muscle). Proc. Natl. Acad. Sci. USA 1972, 69, 442–446. [Google Scholar] [CrossRef]
- Peterson, J.R.; Mitchison, T.J. Small molecules, big impact: A history of chemical inhibitors and the cytoskeleton. Chem. Biol. 2002, 9, 1275–1285. [Google Scholar] [CrossRef]
- Sampath, P.; Pollard, T.D. Effects of cytochalasin, phalloidin, and pH on the elongation of actin filaments. Biochemistry 1991, 30, 1973–1980. [Google Scholar] [CrossRef]
- Brown, S.S.; Spudich, J.A. Cytochalasin inhibits the rate of elongation of actin filament fragments. J. Cell Biol. 1979, 83, 657–662. [Google Scholar] [CrossRef]
- Scherlach, K.; Boettger, D.; Remme, N.; Hertweck, C. The chemistry and biology of cytochalasans. Nat. Prod. Rep. 2010, 27, 869–886. [Google Scholar] [CrossRef]
- Yuyama, K.T.; Wendt, L.; Surup, F.; Kretz, R.; Chepkirui, C.; Wittstein, K.; Boonlarppradab, C.; Wongkanoun, S.; Luangsa-ard, J.; Stadler, M.; et al. Cytochalasans act as inhibitors of biofilm formation of Staphylococcus aureus. Biomolecules 2018, 8, 129. [Google Scholar] [CrossRef]
- Kapoor, K.; Finer-Moore, J.S.; Pedersen, B.P.; Caboni, L.; Waight, A.; Hillig, R.C.; Bringmann, P.; Heisler, I.; Müller, T.; Siebeneicher, H.; et al. Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides. Proc. Natl. Acad. Sci. USA 2016, 113, 4711–4716. [Google Scholar] [CrossRef]
- Reckzeh, E.S.; Waldmann, H. Small-molecule inhibition of glucose transporters GLUT-1–4. ChemBioChem 2020, 21, 45–52. [Google Scholar] [CrossRef]
- Wang, L.; Chen, K.; He, J.; Kenny, J.; Yuan, Y.; Chen, J.; Liu, Q.; Tan, R.; Zhao, J.; Xu, J. Cytochalasin Z11 inhibits RANKL-induced osteoclastogenesis via suppressing NFATc1 activation. RSC Adv. 2019, 9, 38438–38446. [Google Scholar] [CrossRef]
- Sen, B.; Xie, Z.; Uzer, G.; Thompson, W.; Styner, M.; Wu, X.; Rubin, J. Intranuclear actin regulates osteogenesis. Stem Cells 2015, 33, 3065–3076. [Google Scholar] [CrossRef] [PubMed]
- Xiu, Z.; Liu, J.; Wu, X.; Li, X.; Li, S.; Wu, X.; Lv, X.; Ye, H.; Tang, X. Cytochalasin H isolated from mangrove-derived endophytic fungus inhibits epithelial-mesenchymal transition and cancer stemness via YAP/TAZ signaling pathway in non-small cell lung cancer cells. J. Cancer 2021, 12, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Török, D.; Patel, N.; JeBailey, L.; Thong, F.S.L.; Randhawa, V.K.; Klip, A.; Rudich, A. Insulin but not PDGF relies on actin remodeling and on VAMP2 for GLUT4 translocation in myoblasts. J. Cell Sci. 2004, 117, 5447–5455. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.H.; Park, J.A.; Kim, K.R.; Lee, G.I.; Lee, Y.T.; Choe, H.; Ko, S.H.; Kim, M.H.; Seo, Y.H.; Kwak, Y.G. Direct block of cloned hKv1.5 channel by cytochalasins, actin-disrupting agents. Am. J. Physiol.-Cell Physiol. 2005, 289, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Reifenberger, M.S.; Yu, L.; Bao, H.-F.; Duke, B.J.; Liu, B.-C.; Ma, H.-P.; Alli, A.A.; Eaton, D.C.; Alli, A.A. Cytochalasin E alters the cytoskeleton and decreases ENaC activity in Xenopus 2F3 cells. Am. J. Physiol. Renal Physiol. 2014, 307, F86–F95. [Google Scholar] [CrossRef]
- Rueckschloss, U.; Isenberg, G. Cytochalasin D reduces Ca2+ currents via cofilin-activated depolymerization of F-actin in guinea-pig cardiomyocytes. J. Physiol. 2001, 537, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Nazari, H.; Khaleghian, A.; Takahashi, A.; Harada, N.; Webster, N.J.G.; Nakano, M.; Kishi, K.; Ebina, Y.; Nakaya, Y. Cortactin, an actin binding protein, regulates GLUT4 translocation via actin filament remodeling. Biochemistry 2011, 76, 1262–1269. [Google Scholar] [CrossRef][Green Version]
- Bockholt, S.M.; Burridge, K. Cell spreading on extracellular matrix proteins induces tyrosine phosphorylation of tensin. J. Biol. Chem. 1993, 268, 14565–14567. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, P.F.; Paulsen, L.A.; Fondy, C.; Lipski, K.M.; Loucy, K.J.; Fondy, T.P. Effects of cytochalasin B in culture and in vivo on murine Madison 109 lung carcinoma and on B16 melanoma. Cancer Res. 1990, 50, 1431–1439. [Google Scholar]
- Trendowski, M. Exploiting the cytoskeletal filaments of neoplastic cells to potentiate a novel therapeutic approach. Biochim. Biophys. Acta-Rev. Cancer 2014, 1846, 599–616. [Google Scholar] [CrossRef]
- Trendowski, M. Using cytochalasins to improve current chemotherapeutic approaches. Anticancer Agents Med. Chem. 2015, 15, 327–335. [Google Scholar] [CrossRef]
- Trendowski, M.; Christen, T.D.; Acquafondata, C.; Fondy, T.P. Effects of cytochalasin congeners, microtubule-directed agents, and doxorubicin alone or in combination against human ovarian carcinoma cell lines in vitro. BMC Cancer 2015, 15, 632. [Google Scholar] [CrossRef]
- Lingham, R.B.; Hsu, A.; Silverman, K.C.; Bills, G.F.; Dombrowski, A.; Goldman, M.E.; Darke, P.L.; Huang, L.; Koch, G.; Ondeyka, J.G. L-696,474, a novel cytochalasin as an inhibitor of HIV-1 protease. III. Biological activity. J. Antibiot. 1992, 45, 686–691. [Google Scholar] [CrossRef]
- Jayasuriya, H.; Herath, K.B.; Ondeyka, J.G.; Polishook, J.D.; Bills, G.F.; Dombrowski, A.W.; Springer, M.S.; Siciliano, S.; Malkowitz, L.; Sanchez, M.; et al. Isolation and structure of antagonists of chemokine receptor (CCR5). J. Nat. Prod. 2004, 67, 1036–1038. [Google Scholar] [CrossRef]
- Rochfort, S.; Ford, J.; Ovenden, S.; Wan, S.S.; George, S.; Wildman, H.; Tait, R.M.; Meurer-Grimes, B.; Cox, S.; Coates, J.; et al. A novel aspochalasin with HIV-1 integrase inhibitory activity from Aspergillus flavipes. J. Antibiot. 2005, 58, 279–283. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, R.; Liu, S.; Chen, X.; Liu, X.; Che, Y. Alachalasins A–G, new cytochalasins from the fungus Stachybotrys charatum. Bioorg. Med. Chem. 2008, 16, 2627–2634. [Google Scholar] [CrossRef]
- Wang, H.; Yang, P.; Liu, K.; Guo, F.; Zhang, Y.; Zhang, G.; Jiang, C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008, 18, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Valli, M.; Souza, J.M.; Chelucci, R.C.; Biasetto, C.R.; Araujo, A.R.; Bolzani, V.d.S.; Andricopulo, A.D. Identification of natural cytochalasins as leads for neglected tropical diseases drug discovery. PLoS ONE 2022, 17, e0275002. [Google Scholar] [CrossRef]
- Bräse, S.; Gläser, F.; Kramer, C.S.; Lindner, S.; Linsenmeier, A.M.; Masters, K.-S.; Meister, A.C.; Ruff, B.M.; Zhong, S.; Bräse, S.; et al. Cytochalasans. In The Chemistry of Mycotoxins; Springer: Vienna, Austria, 2013; pp. 207–223. [Google Scholar]
- Blanchoin, L.; Boujemaa-Paterski, R.; Sykes, C.; Plastino, J. Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 2014, 94, 235–263. [Google Scholar] [CrossRef]
- Lappalainen, P.; Kotila, T.; Jégou, A.; Romet-Lemonne, G. Biochemical and mechanical regulation of actin dynamics. Nat. Rev. Mol. Cell Biol. 2022, 23, 836–852. [Google Scholar] [CrossRef]
- Rothweiler, W.; Tamm, C. Isolation and structure of Phomin. Experientia 1966, 22, 750–752. [Google Scholar] [CrossRef]
- Carter, S.B. Effects of cytochalasins on mammalian cells. Nature 1967, 213, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Ridler, M.A.; Smith, G.F. The response of human cultured lymphocytes to cytochalasin B. J. Cell Sci. 1968, 3, 595–602. [Google Scholar] [CrossRef]
- Krishan, A.; Ray-Chaudhuri, R. Asynchrony of nuclear development in cytochalasin-induced multinucleate cells. J. Cell Biol. 1969, 43, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Löw, I.; Dancker, P.; Wieland, T. Stabilization of F-actin by phalloidin reversal of the destabilizing effect of cytochalasin B. FEBS Lett. 1975, 54, 263–265. [Google Scholar] [CrossRef]
- Ohmori, H.; Toyama, S.; Toyama, S. Direct proof that the primary site of action of cytochalasin on cell motility processes is actin. J. Cell Biol. 1992, 116, 933–941. [Google Scholar] [CrossRef]
- Spooner, B.S.; Yamada, K.M.; Wessells, N.K. Microfilaments and cell locomotion. J. Cell Biol. 1971, 49, 595–613. [Google Scholar] [CrossRef] [PubMed]
- Wessells, N.K.; Spooner, B.S.; Ash, J.F.; Bradley, M.O.; Luduena, M.A.; Taylor, E.L.; Wrenn, J.T.; Yamada, K.M. Microfilaments in cellular and developmental processes. Science (80-) 1971, 171, 135–143. [Google Scholar] [CrossRef]
- Weber, K.; Rathke, P.C.; Osborn, M.; Franke, W.W. Distribution of actin and tubulin in cells and in glycerinated cell models after treatment with cytochalasin B (CB). Exp. Cell Res. 1976, 102, 285–297. [Google Scholar] [CrossRef]
- Brenner, S.L.; Korn, E.D. The effects of cytochalasins on actin polymerization and actin ATPase provide insights into the mechanism of polymerization. J. Biol. Chem. 1980, 255, 841–844. [Google Scholar] [CrossRef] [PubMed]
- MacLean-Fletcher, S.; Pollard, T.D. Mechanism of action of cytochalasin B on actin. Cell 1980, 20, 329–341. [Google Scholar] [CrossRef]
- Lin, D.C.; Tobin, K.D.; Grumet, M.; Lin, S. Cytochalasins inhibit nuclei-induced actin polymerization by blocking filament elongation. J. Cell Biol. 1980, 84, 455–460. [Google Scholar] [CrossRef]
- Cooper, J.A.; Bryan, J.; Schwab, B.; Frieden, C.; Loftus, D.J.; Elson, E.L. Microinjection of gelsolin into living cells. J. Cell Biol. 1987, 104, 491–501. [Google Scholar] [CrossRef]
- Katagiri, K.; Matsuura, S. Antitumor activity of cytochalasin D. J. Antibiot. 1971, 24, 722–723. [Google Scholar] [CrossRef]
- Miranda, A.F.; Godman, G.C.; Deitch, A.D.; Tanenbaum, S.W. Action of cytochalasin D on cells of established lines. I. Early events. J. Cell Biol. 1974, 61, 481–500. [Google Scholar] [CrossRef]
- Cooper, J.A. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 1987, 105, 1473–1478. [Google Scholar] [CrossRef] [PubMed]
- Umeda, M.; Ohtsubo, K.; Saito, M.; Sekita, S.; Yoshihira, K. Cytotoxicity of new cytochalasans from Chaetomium globosum. Experientia 1975, 31, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Minato, H.; Katayama, T.; Matsumoto, M.; Katagiri, K.; Matsuura, S. Structure-activity relationships among zygosporin derivatives. Chem. Pharm. Bull. 1973, 21, 2268–2277. [Google Scholar] [CrossRef]
- Sekita, S.; Yoshihira, K.; Natori, S.; Harada, F.; Iida, K.; Yahara, I. Structure-activity relationship of thirty-nine cytochalasans observed in the effects on cellular structures and cellular events and on actin polymerization in vitro. J. Pharmacobiodyn. 1985, 8, 906–916. [Google Scholar] [CrossRef] [PubMed]
- Van Goietsenoven, G.; Mathieu, V.; Andolfi, A.; Cimmino, A.; Lefranc, F.; Kiss, R.; Evidente, A. In vitro growth inhibitory effects of cytochalasins and derivatives in cancer cells. Planta Med. 2011, 77, 711–717. [Google Scholar] [CrossRef]
- Li, H.; Xiao, J.; Gao, Y.-Q.Q.; Tang, J.J.; Zhang, A.-L.L.; Gao, J.-M.M. Chaetoglobosins from Chaetomium globosum, an endophytic fungus in Ginkgo biloba, and their phytotoxic and cytotoxic activities. J. Agric. Food Chem. 2014, 62, 3734–3741. [Google Scholar] [CrossRef]
- Beno, M.A.; Cox, R.H.; Wells, J.M.; Cole, R.J.; Kirksey, J.W.; Christoph, G.G. Structure of a new [11]cytochalasin, cytochalasin H or kodo-cytochalasin-1. J. Am. Chem. Soc. 1977, 99, 4123–4130. [Google Scholar] [CrossRef]
- Miao, S.; Liu, M.; Qi, S.; Wu, Y.; Sun, K.; Zhang, Z.; Zhu, K.; Cai, G.; Gong, K. Cytochalasins from coastal saline soil-derived fungus Aspergillus flavipes RD-13 and their cytotoxicities. J. Antibiot. 2022, 75, 410–414. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Rich, S.A.; Estes, J.E. Detection of conformational changes in actin by proteolytic digestion: Evidence for a new monomeric species. J. Mol. Biol. 1976, 104, 777–792. [Google Scholar] [CrossRef] [PubMed]
- Lai, F.P.L.; Szczodrak, M.; Block, J.; Faix, J.; Breitsprecher, D.; Mannherz, H.G.; Stradal, T.E.B.; Dunn, G.A.; Small, J.V.; Rottner, K. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 2008, 27, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Matsunaga, D.; Deguchi, S. Long-term fluorescence recovery after photobleaching (FRAP). Methods Mol. Biol. 2023, 2600, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Campellone, K.G.; Welch, M.D. A nucleator arms race: Cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 2010, 11, 237–251. [Google Scholar] [CrossRef]
- Löw, I.; Jahn, W.; Wieland, T.; Sekita, S.; Yoshihira, K.; Natori, S. Interaction between rabbit muscle actin and several chaetoglobosins or cytochalasins. Anal. Biochem. 1979, 95, 14–18. [Google Scholar] [CrossRef]
- Rottner, K.; Schaks, M. Assembling actin filaments for protrusion. Curr. Opin. Cell Biol. 2019, 56, 53–63. [Google Scholar] [CrossRef]
- Yahara, I.; Harada, F.; Sekita, S.; Yoshihira, K.; Natori, S. Correlation between effects of 24 different cytochalasins on cellular structures and cellular events and those on actin in vitro. J. Cell Biol. 1982, 92, 69–78. [Google Scholar] [CrossRef]
- Domnina, L.V.; Gelfand, V.I.; Ivanova, O.Y.; Leonova, E.V.; Pletjushkina, O.Y.; Vasiliev, J.M.; Gelfand, I.M. Effects of small doses of cytochalasins on fibroblasts: Preferential changes of active edges and focal contacts. Proc. Natl. Acad. Sci. USA 1982, 79, 7754–7757. [Google Scholar] [CrossRef]
- Sanger, J.W. The use of cytochalasin B to distinguish myoblasts from fibroblasts in cultures of developing chick striated muscle. Proc. Natl. Acad. Sci. USA 1974, 71, 3621–3625. [Google Scholar] [CrossRef]
- Small, J.V.; Rottner, K.; Kaverina, I. Functional design in the actin cytoskeleton. Curr. Opin. Cell Biol. 1999, 11, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Kaverina, I.; Krylyshkina, O.; Gimona, M.; Beningo, K.; Wang, Y.L.; Small, J.V. Enforced polarisation and locomotion of fibroblasts lacking microtubules. Curr. Biol. 2000, 10, 739–742. [Google Scholar] [CrossRef]
- Revach, O.Y.; Grosheva, I.; Geiger, B. Biomechanical regulation of focal adhesion and invadopodia formation. J. Cell Sci. 2020, 133, jcs244848. [Google Scholar] [CrossRef] [PubMed]
- Zapotoczny, B.; Szafranska, K.; Lekka, M.; Ahluwalia, B.S.; McCourt, P. Tuning of liver sieve: The interplay between actin and myosin regulatory light chain regulates fenestration size and number in murine liver sinusoidal endothelial cells. Int. J. Mol. Sci. 2022, 23, 9850. [Google Scholar] [CrossRef]
- Lambert, C.; Pourmoghaddam, M.J.; Cedeño-Sanchez, M.; Surup, F.; Khodaparast, S.A.; Krisai-Greilhuber, I.; Voglmayr, H.; Stradal, T.E.B.B.; Stadler, M. Resolution of the Hypoxylon fuscum complex (Hypoxylaceae, Xylariales) and discovery and biological characterization of two of its prominent secondary metabolites. J. Fungi 2021, 7, 131. [Google Scholar] [CrossRef] [PubMed]
- Garcia, K.Y.M.; Quimque, M.T.J.; Lambert, C.; Schmidt, K.; Primahana, G.; Stradal, T.E.B.; Ratzenböck, A.; Dahse, H.M.; Phukhamsakda, C.; Stadler, M.; et al. Antiproliferative and cytotoxic cytochalasins from Sparticola triseptata inhibit actin polymerization and aggregation. J. Fungi 2022, 8, 560. [Google Scholar] [CrossRef]
- Kretz, R.; Wendt, L.; Wongkanoun, S.; Luangsa-ard, J.J.; Surup, F.; Helaly, S.E.; Noumeur, S.R.; Stadler, M.; Stradal, T.E.B. The effect of cytochalasans on the actin cytoskeleton of eukaryotic cells and preliminary structure–activity relationships. Biomolecules 2019, 9, 73. [Google Scholar] [CrossRef]
- Pourmoghaddam, M.J.; Ekiz, G.; Lambert, C.; Surup, F.; Primahana, G.; Wittstein, K.; Khodaparast, S.A.; Voglmayr, H.; Krisai-Greilhuber, I.; Stradal, T.E.B.; et al. Studies on the secondary metabolism of Rosellinia and Dematophora strains (Xylariaceae) from Iran. Mycol. Prog. 2022, 21, 65. [Google Scholar] [CrossRef]
- Rampal, A.L.; Pinkofsky, H.B.; Jung, C.Y. Structure of cytochalasins and cytochalasin B binding sites in human erythrocyte membranes. Biochemistry 1980, 19, 679–683. [Google Scholar] [CrossRef]
- Griffin, J.F.; Rampal, A.L.; Jung, C.Y. Inhibition of glucose transport in human erythrocytes by cytochalasins: A model based on diffraction studies. Proc. Natl. Acad. Sci. USA 1982, 79, 3759–3763. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y.W.; Tokuyasu, K.T.; Singer, S.J. The capping of lymphocytes and other cells, studied by an improved method for immunofluorescence staining of frozen sections. J. Cell. Physiol. 1978, 95, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Kouyama, T.; Mihashi, K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur. J. Biochem. 1981, 114, 33–38. [Google Scholar] [CrossRef]
- Cooper, J.A.; Walker, S.B.; Pollard, T.D. Pyrene actin: Documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 1983, 4, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Izawa, Y.; Koyama, K.; Natori, S.; Iida, K.; Yahara, I.; Shimaoka, S.; Maruyama, K. The effects of new cytochalasins from Phomopsis sp. and the derivatives on cellular structure and actin polymerization. Chem. Pharm. Bull. 1990, 38, 971–974. [Google Scholar] [CrossRef]
- Nukina, M. Pyrichalasin H, a new phytotoxic metabolite belonging to the cytochalasans from Pyricularia grisea (Cooke) Saccardo. Agric. Biol. Chem. 1987, 51, 2625–2628. [Google Scholar] [CrossRef]
- Maruyama, K.; Oosawa, M.; Tashiro, A.; Suzuki, T.; Tanikawa, M.; Kikuchi, M.; Sekita, S.; Natori, S. Effects of chaetoglobosin J on the G-F transformation of actin. Biochim. Biophys. Acta-Protein Struct. Mol. Enzymol. 1986, 874, 137–143. [Google Scholar] [CrossRef]
- Goddette, D.W.; Frieden, C. The kinetics of cytochalasin D binding to monomeric actin. J. Biol. Chem. 1986, 261, 15970–15973. [Google Scholar] [CrossRef]
- Brenner, S.L.; Korn, E.D. Substoichiometric concentrations of cytochalasin D inhibit actin polymerization. Additional evidence for an F-actin treadmill. J. Biol. Chem. 1979, 254, 9982–9985. [Google Scholar] [CrossRef]
- Blikstad, I.; Markey, F.; Carlsson, L.; Persson, T.; Lindberg, U. Selective assay of monomeric and filamentous actin in cell extracts, using inhibition of deoxyribonuclease I. Cell 1978, 15, 935–943. [Google Scholar] [CrossRef]
- Hitchcock, S.E. Actin deoxyroboncuclease I interaction. Depolymerization and nucleotide exchange. J. Biol. Chem. 1980, 255, 5668–5673. [Google Scholar] [CrossRef]
- Thohinung, S.; Kanokmedhakul, S.; Kanokmedhakul, K.; Kukongviriyapan, V.; Tusskorn, O.; Soytong, K. Cytotoxic 10-(indol-3-yl)-[13]cytochalasans from the fungus Chaetomium elatum ChE01. Arch. Pharm. Res. 2010, 33, 1135–1141. [Google Scholar] [CrossRef]
- Zhu, X.; Zhou, D.; Liang, F.; Wu, Z.; She, Z.; Li, C. Penochalasin K, a new unusual chaetoglobosin from the mangrove endophytic fungus Penicillium chrysogenum V11 and its effective semi-synthesis. Fitoterapia 2017, 123, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, S.; Helaly, S.; Schroers, H.-J.; Stadler, M.; Richert-Poeggeler, K.R.; Dababat, A.A.; Maier, W. Ijuhya vitellina sp. nov., a novel source for chaetoglobosin A, is a destructive parasite of the cereal cyst nematode Heterodera filipjevi. PLoS ONE 2017, 12, e0180032. [Google Scholar] [CrossRef] [PubMed]
- Ruan, B.H.; Yu, Z.F.; Yang, X.Q.; Yang, Y.B.; Hu, M.; Zhang, Z.X.; Zhou, Q.Y.; Zhou, H.; Ding, Z.T. New bioactive compounds from aquatic endophyte Chaetomium globosum. Nat. Prod. Res. 2018, 32, 1050–1055. [Google Scholar] [CrossRef] [PubMed]
- Lambert, C.; Shao, L.; Zeng, H.; Surup, F.; Saetang, P.; Aime, M.C.; Husbands, D.R.; Rottner, K.; Stradal, T.E.B.; Stadler, M. Cytochalasans produced by Xylaria karyophthora and their biological activities. Mycologia 2023, 115, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Kemkuignou, B.M.; Lambert, C.; Schmidt, K.; Schweizer, L.; Anoumedem, E.G.M.; Kouam, S.F.; Stadler, M.; Stradal, T.; Marin-Felix, Y. Unreported cytochalasins from an acid-mediated transformation of cytochalasin J isolated from Diaporthe cf. ueckeri. Fitoterapia 2023, 166, 105434. [Google Scholar] [CrossRef] [PubMed]
- Kemkuignou, B.M.; Schweizer, L.; Lambert, C.; Gisèle, E.; Anoumedem, M.; Kouam, S.F.; Stadler, M.; Marin-Felix, Y.; Felix, Y.M.; Anoumedem, E.G.M.; et al. New polyketides from the liquid culture of Diaporthe breyniae sp. nov. (Diaporthales, Diaporthaceae). MycoKeys 2022, 90, 85–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Ge, H.; Xie, D.; Chen, R.; Zou, J.; Tao, X.; Dai, J. Periconiasins A–C, new cytotoxic cytochalasans with an unprecedented 9/6/5 tricyclic ring system from endophytic fungus Periconia sp. Org. Lett. 2013, 15, 1674–1677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tao, X.; Chen, R.; Liu, J.; Li, L.; Fang, X.; Yu, L.; Dai, J. Pericoannosin A, a polyketide synthase-nonribosomal peptide synthetase hybrid metabolite with new carbon skeleton from the endophytic fungus Periconia sp. Org. Lett. 2015, 17, 4304–4307. [Google Scholar] [CrossRef]
- Bear, J.E.; Svitkina, T.M.; Krause, M.; Schafer, D.A.; Loureiro, J.J.; Strasser, G.A.; Maly, I.V.; Chaga, O.Y.; Cooper, J.A.; Borisy, G.G.; et al. Antagonism between Ena/VASP Proteins and actin filament capping regulates fibroblast motility. Cell 2002, 109, 509–521. [Google Scholar] [CrossRef]
- Pruyne, D.; Evangelista, M.; Yang, C.; Bi, E.; Zigmond, S.; Bretscher, A.; Boone, C. Role of formins in actin assembly: Nucleation and barbed-end association. Science 2002, 297, 612–615. [Google Scholar] [CrossRef] [PubMed]
- Mehidi, A.; Kage, F.; Karatas, Z.; Cercy, M.; Schaks, M.; Polesskaya, A.; Sainlos, M.; Gautreau, A.M.; Rossier, O.; Rottner, K.; et al. Forces generated by lamellipodial actin filament elongation regulate the WAVE complex during cell migration. Nat. Cell Biol. 2021, 23, 1148–1162. [Google Scholar] [CrossRef]
- Steffan, A.-M.; Gendrault, J.-L.; Kirn, A. Increase in the number of fenestrae in mouse endothelial liver cells by altering the cytoskeleton with cytochalasin B. Hepatology 1987, 7, 1230–1238. [Google Scholar] [CrossRef]
- Rottner, K.; Faix, J.; Bogdan, S.; Linder, S.; Kerkhoff, E. Actin assembly mechanisms at a glance. J. Cell Sci. 2017, 130, 3427–3435. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J. Life at the leading edge. Cell 2011, 145, 1012–1022. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, S.; Pernier, J.; Carlier, M.F. Regulators of actin filament barbed ends at a glance. J. Cell Sci. 2016, 129, 1085–1091. [Google Scholar] [CrossRef] [PubMed]
- Small, J.V. The actin cytoskeleton. Electron Microsc. Rev. 1988, 1, 155–174. [Google Scholar] [CrossRef]
- Small, J.V.; Stradal, T.; Vignal, E.; Rottner, K. The lamellipodium: Where motility begins. Trends Cell Biol. 2002, 12, 112–120. [Google Scholar] [CrossRef]
- Damiano-Guercio, J.; Kurzawa, L.; Mueller, J.; Dimchev, G.; Schaks, M.; Nemethova, M.; Pokrant, T.; Brühmann, S.; Linkner, J.; Blanchoin, L.; et al. Loss of Ena/VASP interferes with lamellipodium architecture, motility and integrin-dependent adhesion. Elife 2020, 9, e55351. [Google Scholar] [CrossRef] [PubMed]
- Steffen, A.; Faix, J.; Resch, G.P.; Linkner, J.; Wehland, J.; Small, J.V.; Rottner, K.; Stradal, T.E.B. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol. Biol. Cell 2006, 17, 2581–2591. [Google Scholar] [CrossRef]
- Dimchev, V.; Lahmann, I.; Koestler, S.A.; Kage, F.; Dimchev, G.; Steffen, A.; Stradal, T.E.B.; Vauti, F.; Arnold, H.-H.; Rottner, K. Induced Arp2/3 complex depletion increases FMNL2/3 formin expression and filopodia formation. Front. Cell Dev. Biol. 2021, 9, 634708. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Blanchoin, L.; Mullins, R.D. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 545–576. [Google Scholar] [CrossRef]
- Vinzenz, M.; Nemethova, M.; Schur, F.; Mueller, J.; Narita, A.; Urban, E.; Winkler, C.; Schmeiser, C.; Koestler, S.A.; Rottner, K.; et al. Actin branching in the initiation and maintenance of lamellipodia. J. Cell Sci. 2012, 125, 2775–2785. [Google Scholar] [CrossRef]
- Koestler, S.A.; Steffen, A.; Nemethova, M.; Winterhoff, M.; Luo, N.; Holleboom, J.M.; Krupp, J.; Jacob, S.; Vinzenz, M.; Schur, F.; et al. Arp2/3 complex is essential for actin network treadmilling as well as for targeting of capping protein and cofilin. Mol. Biol. Cell 2013, 24, 2861–2875. [Google Scholar] [CrossRef]
- Kovar, D.R.; Harris, E.S.; Mahaffy, R.; Higgs, H.N.; Pollard, T.D. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 2006, 124, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Ferron, F.; Rebowski, G.; Lee, S.H.; Dominguez, R. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 2007, 26, 4597–4606. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.D.; Mullins, R.D. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J. Cell Biol. 2010, 191, 571–584. [Google Scholar] [CrossRef]
- Barzik, M.; Kotova, T.I.; Higgs, H.N.; Hazelwood, L.; Hanein, D.; Gertler, F.B.; Schafer, D.A. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J. Biol. Chem. 2005, 280, 28653–28662. [Google Scholar] [CrossRef]
- Pasic, L.; Kotova, T.; Schafer, D.A. Ena/VASP proteins capture actin filament barbed ends. J. Biol. Chem. 2008, 283, 9814–9819. [Google Scholar] [CrossRef]
- Breitsprecher, D.; Kiesewetter, A.K.; Linkner, J.; Urbanke, C.; Resch, G.P.; Small, J.V.; Faix, J. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 2008, 27, 2943–2954. [Google Scholar] [CrossRef]
- Hansen, S.D.; Mullins, R.D. Lamellipodin promotes actin assembly by clustering Ena/VASP proteins and tethering them to actin filaments. Elife 2015, 4, e06585. [Google Scholar] [CrossRef]
- Pantaloni, D.; Boujemaa, R.; Didry, D.; Gounon, P.; Carlier, M.F. The Arp2/3 complex branches filament barbed ends: Functional antagonism with capping proteins. Nat. Cell Biol. 2000, 2, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Akin, O.; Mullins, R.D. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell 2008, 133, 841–851. [Google Scholar] [CrossRef]
- Funk, J.; Merino, F.; Schaks, M.; Rottner, K.; Raunser, S.; Bieling, P. A barbed end interference mechanism reveals how capping protein promotes nucleation in branched actin networks. Nat. Commun. 2021, 12, 5329. [Google Scholar] [CrossRef] [PubMed]
- Wioland, H.; Guichard, B.; Senju, Y.; Myram, S.; Lappalainen, P.; Jégou, A.; Romet-Lemonne, G. ADF/Cofilin accelerates actin dynamics by severing filaments and promoting their depolymerization at both ends. Curr. Biol. 2017, 27, 1956–1967.e7. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, S.; Carlier, M.F. Enhanced depolymerization of actin filaments by ADF/Cofilin and monomer funneling by capping protein cooperate to accelerate barbed-end growth. Curr. Biol. 2017, 27, 1990–1998.e5. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.K.; Charras, G.T.; Mitchison, T.J.; Brieher, W.M.; Kueh, H.Y.; Charras, G.T.; Mitchison, T.J.; Brieher, W.M. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J. Cell Biol. 2008, 182, 341–353. [Google Scholar] [CrossRef]
- Rottner, K.; Stradal, T.E.B.; Chen, B. WAVE regulatory complex. Curr. Biol. 2021, 31, R512–R517. [Google Scholar] [CrossRef]
- Pollard, T.D. Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 2019, 8, a018226. [Google Scholar] [CrossRef]
- Gao, J.; Nakamura, F. Actin-associated proteins and small molecules targeting the actin cytoskeleton. Int. J. Mol. Sci. 2022, 23, 2118. [Google Scholar] [CrossRef] [PubMed]
- Bonder, E.M.; Mooseker, M.S. Cytochalasin B slows but does not prevent monomer addition at the barbed end of the actin filament. J. Cell Biol. 1986, 102, 282–288. [Google Scholar] [CrossRef]
- Goddette, D.W.; Frieden, C. Actin polymerization. The mechanism of action of cytochalasin D. J. Biol. Chem. 1986, 261, 15974–15980. [Google Scholar] [CrossRef] [PubMed]
- Suarez, C.; Roland, J.; Boujemaa-Paterski, R.; Kang, H.; McCullough, B.R.; Reymann, A.C.; Guérin, C.; Martiel, J.L.; De La Cruz, E.M.; Blanchoin, L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr. Biol. 2011, 21, 862–868. [Google Scholar] [CrossRef]
- Schliwa, M. Action of cytochalasin D on cytoskeletal networks. J. Cell Biol. 1982, 92, 79–91. [Google Scholar] [CrossRef]
- Hartwig, J.H.; Stossel, T.P. Cytochalasin B and the structure of actin gels. J. Mol. Biol. 1979, 134, 539–553. [Google Scholar] [CrossRef]
- Wakatsuki, T.; Schwab, B.; Thompson, N.C.; Elson, E.L. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J. Cell Sci. 2001, 114, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Theodoropoulos, P.A.; Gravanis, A.; Tsapara, A.; Margioris, A.N.; Papadogiorgaki, E.; Galanopoulos, V.; Stournaras, C. Cytochalasin B may shorten actin filaments by a mechanism independent of barbed end capping. Biochem. Pharmacol. 1994, 47, 1875–1881. [Google Scholar] [CrossRef] [PubMed]
- Nair, U.B.; Joel, P.B.; Wan, Q.; Lowey, S.; Rould, M.A.; Trybus, K.M. Crystal structures of monomeric actin bound to cytochalasin D. J. Mol. Biol. 2008, 384, 848–864. [Google Scholar] [CrossRef]
- McLaughlin, P.J.; Gooch, J.T.; Mannherz, H.G.; Weeds, A.G. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature 1993, 364, 685–692. [Google Scholar] [CrossRef]
- Hertzog, M.; van Heijenoort, C.; Didry, D.; Gaudier, M.; Coutant, J.; Gigant, B.; Didelot, G.; Préat, T.; Knossow, M.; Guittet, E.; et al. The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell 2004, 117, 611–623. [Google Scholar] [CrossRef]
- Otterbein, L.R.; Cosio, C.; Graceffa, P.; Dominguez, R. Crystal structures of the vitamin D-binding protein and its complex with actin: Structural basis of the actin-scavenger system. Proc. Natl. Acad. Sci. USA 2002, 99, 8003–8008. [Google Scholar] [CrossRef]
- Head, J.F.; Swamy, N.; Ray, R. Crystal structure of the complex between actin and human vitamin D-binding protein at 2.5 A resolution. Biochemistry 2002, 41, 9015–9020. [Google Scholar] [CrossRef]
- Otomo, T.; Tomchick, D.R.; Otomo, C.; Panchal, S.C.; Machius, M.; Rosen, M.K. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 2005, 433, 488–494. [Google Scholar] [CrossRef]
- Chereau, D.; Kerff, F.; Graceffa, P.; Grabarek, Z.; Langsetmo, K.; Dominguez, R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. USA 2005, 102, 16644–16649. [Google Scholar] [CrossRef]
- Dominguez, R. The WH2 domain and actin nucleation: Necessary but insufficient. Trends Biochem. Sci. 2016, 41, 478–490. [Google Scholar] [CrossRef]
- Burnette, D.T.; Schaefer, A.W.; Ji, L.; Danuser, G.; Forscher, P. Filopodial actin bundles are not necessary for microtubule advance into the peripheral domain of Aplysia neuronal growth cones. Nat. Cell Biol. 2007, 9, 1360–1369. [Google Scholar] [CrossRef]
- Faix, J.; Rottner, K. Ena/VASP proteins in cell edge protrusion, migration and adhesion. J. Cell Sci. 2022, 135, jcs259226. [Google Scholar] [CrossRef] [PubMed]
- Neel, N.F.; Barzik, M.; Raman, D.; Sobolik-Delmaire, T.; Sai, J.; Ham, A.J.; Mernaugh, R.L.; Gertler, F.B.; Richmond, A. VASP is a CXCR2-interacting protein that regulates CXCR2-mediated polarization and chemotaxis. J. Cell Sci. 2009, 122, 1882–1894. [Google Scholar] [CrossRef]
- Krause, M.; Leslie, J.D.; Stewart, M.; Lafuente, E.M.; Valderrama, F.; Jagannathan, R.; Strasser, G.A.; Rubinson, D.A.; Liu, H.; Way, M.; et al. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial Dynamics. Dev. Cell 2004, 7, 571–583. [Google Scholar] [CrossRef]
- Lacayo, C.I.; Pincus, Z.; VanDuijn, M.M.; Wilson, C.A.; Fletcher, D.A.; Gertler, F.B.; Mogilner, A.; Theriot, J.A. Emergence of large-scale cell morphology and movement from local actin filament growth dynamics. PLoS Biol. 2007, 5, 2035–2052. [Google Scholar] [CrossRef]
- Scott, J.A.; Shewan, A.M.; den Elzen, N.R.; Loureiro, J.J.; Gertler, F.B.; Yap, A.S. Ena/VASP proteins can regulate distinct modes of actin organization at cadherin-adhesive contacts. Mol. Biol. Cell 2006, 17, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Schnoor, M.; Stradal, T.E.; Rottner, K. Cortactin: Cell functions of a multifaceted actin-binding protein. Trends Cell Biol. 2018, 28, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Small, J.V.; Rottner, K.; Carlier, M.F. Actin-Based Motility; Carlier, M.-F., Ed.; Springer: Dordrecht, The Netherlands, 2010; ISBN 978-90-481-9300-4. [Google Scholar]
- Flanagan, M.D.; Lin, S. Cytochalasins block actin filament elongation by binding to high affinity sites associated with F-actin. J. Biol. Chem. 1980, 255, 835–838. [Google Scholar] [CrossRef] [PubMed]
- Perrin, B.J.; Ervasti, J.M. The actin gene family: Function follows isoform. Cytoskeleton 2010, 67, 630–634. [Google Scholar] [CrossRef]
- Vedula, P.; Kurosaka, S.; Leu, N.A.; Wolf, Y.I.; Shabalina, S.A.; Wang, J.; Sterling, S.; Dong, D.W.; Kashina, A. Diverse functions of homologous actin isoforms are defined by their nucleotide, rather than their amino acid sequence. Elife 2017, 6, e31661. [Google Scholar] [CrossRef]
- Chen, A.; Arora, P.D.; McCulloch, C.A.; Wilde, A. Cytokinesis requires localized β-actin filament production by an actin isoform specific nucleator. Nat. Commun. 2017, 8, 1530. [Google Scholar] [CrossRef]
- Takenawa, T.; Suetsugu, S. The WASP-WAVE protein network: Connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2007, 8, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Dugina, V.; Zwaenepoel, I.; Gabbiani, G.; Clément, S.; Chaponnier, C. Beta and gamma-cytoplasmic actins display distinct distribution and functional diversity. J. Cell Sci. 2009, 122, 2980–2988. [Google Scholar] [CrossRef]
- Toyama, S.; Toyama, S. A variant form of β-actin in a mutant of KB cells resistant to cytochalasin B. Cell 1984, 37, 609–614. [Google Scholar] [CrossRef]
- Toyama, S.; Toyama, S. Functional alterations in beta’-actin from a KB cell mutant resistant to cytochalasin B. J. Cell Biol. 1988, 107, 1499–1504. [Google Scholar] [CrossRef]
- Wu, C.; Asokan, S.B.; Berginski, M.E.; Haynes, E.M.; Sharpless, N.E.; Griffith, J.D.; Gomez, S.M.; Bear, J.E. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 2012, 148, 973–987. [Google Scholar] [CrossRef]
- Suraneni, P.; Rubinstein, B.; Unruh, J.R.; Durnin, M.; Hanein, D.; Li, R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 2012, 197, 239–251. [Google Scholar] [CrossRef]
- Forscher, P.; Smith, S.J. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J. Cell Biol. 1988, 107, 1505–1516. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.I.; Wang, Y.L.; Small, J.V. Coordination of protrusion and translocation of the keratocyte involves rolling of the cell body. J. Cell Biol. 1996, 134, 1209–1218. [Google Scholar] [CrossRef]
- Weisswange, I.; Newsome, T.P.; Schleich, S.; Way, M. The rate of N-WASP exchange limits the extent of ARP2/3-complex-dependent actin-based motility. Nature 2009, 458, 87–91. [Google Scholar] [CrossRef]
- Rotty, J.D.; Wu, C.; Haynes, E.M.; Suarez, C.; Winkelman, J.D.; Johnson, H.E.; Haugh, J.M.; Kovar, D.R.; Bear, J.E. Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev. Cell 2015, 32, 54–67. [Google Scholar] [CrossRef]
- Weiner, O.D.; Marganski, W.A.; Wu, L.F.; Altschuler, S.J.; Kirschner, M.W. An actin-based wave generator organizes cell motility. PLoS Biol. 2007, 5, 2053–2063. [Google Scholar] [CrossRef]
- Shuster, C.B.; Lin, A.Y.; Nayak, R.; Herman, I.M. βCAP73: A novel β actin-specific binding protein. Cell Motil. Cytoskeleton 1996, 35, 175–187. [Google Scholar] [CrossRef]
- Nishida, E.; Ohta, Y.; Sakai, H. The regulation of actin polymerization by the 88K protein/actin complex and cytochalasin B. J. Biochem. 1983, 94, 1671–1683. [Google Scholar] [PubMed]
- Lo, S.H.; Janmey, P.A.; Hartwig, J.H.; Chen, L.B. Interactions of tensin with actin and identification of its three distinct actin-binding domains. J. Cell Biol. 1994, 125, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Schafer, D.A.; Welch, M.D.; Machesky, L.M.; Bridgman, P.C.; Meyer, S.M.; Cooper, J.A. Visualization and molecular analysis of actin assembly in living cells. J. Cell Biol. 1998, 143, 1919–1930. [Google Scholar] [CrossRef] [PubMed]
- Shuster, C.B.; Herman, I.M. Indirect association of ezrin with F-actin: Isoform specificity and calcium sensitivity. J. Cell Biol. 1995, 128, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Dabrowska, R.; Próchniewicz, E.; Drabikowski, W. The effect of cytochalasin and glutaraldehyde on F-actin filaments containing muscle and non-muscle tropomyosin. J. Muscle Res. Cell Motil. 1983, 4, 83–93. [Google Scholar] [CrossRef]
- Zheng, B.; Wen, J. hhLIM is a novel F-actin binding protein involved in actin cytoskeleton remodeling. FEBS J. 2008, 275, 1568–1578. [Google Scholar] [CrossRef]
- Dimchev, G.; Amiri, B.; Humphries, A.C.; Dimchev, V.; Stradal, T.E.B.B.; Faix, J.; Way, M.; Falcke, M.; Rottner, K.; Schaks, M.; et al. Lamellipodin tunes cell migration by stabilizing protrusions and promoting adhesion formation. J. Cell Sci. 2020, 133, jcs239020. [Google Scholar] [CrossRef]
- Block, J.; Breitsprecher, D.; Kühn, S.; Winterhoff, M.; Kage, F.; Geffers, R.; Duwe, P.; Rohn, J.L.; Baum, B.; Brakebusch, C.; et al. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr. Biol. 2012, 22, 1005–1012. [Google Scholar] [CrossRef]
- Tellam, R.; Frieden, C. Cytochalasin D and platelet gelsolin accelerate actin polymer formation. A model for regulation of the extent of actin polymer formation in vivo. Biochemistry 1982, 21, 3207–3214. [Google Scholar] [CrossRef]
- Okada, K.; Ravi, H.; Smith, E.M.; Goode, B.L. Aip1 and Cofilin promote rapid turnover of yeast actin patches and cables: A coordinated mechanism for severing and capping filaments. Mol. Biol. Cell 2006, 17, 2855–2868. [Google Scholar] [CrossRef]
- Brieher, W.M.; Hao, Y.K.; Ballif, B.A.; Mitchison, T.J. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J. Cell Biol. 2006, 175, 315–324. [Google Scholar] [CrossRef]
- McGough, A.; Pope, B.; Chiu, W.; Weeds, A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J. Cell Biol. 1997, 138, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Andrianantoandro, E.; Pollard, T.D. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/Cofilin. Mol. Cell 2006, 24, 13–23. [Google Scholar] [CrossRef]
- Shoji, K.; Ohashi, K.; Sampei, K.; Oikawa, M.; Mizuno, K. Cytochalasin D acts as an inhibitor of the actin-cofilin interaction. Biochem. Biophys. Res. Commun. 2012, 424, 52–57. [Google Scholar] [CrossRef]
- Liem, R.K.H. Cytoskeletal integrators: The spectrin superfamily. Cold Spring Harb. Perspect. Biol. 2016, 8, a018259. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, C.Y.; Zhao, Q.; Li, D.H. Spectrin: Structure, function and disease. Sci. China Life Sci. 2013, 56, 1076–1085. [Google Scholar] [CrossRef]
- Dutta, S.; Bose, D.; Ghosh, S.; Chakrabarti, A. Spectrin: An alternate target for cytoskeletal drugs. J. Biomol. Struct. Dyn. 2022, 41, 6534–6545. [Google Scholar] [CrossRef]
- Stephens, D.J.; Banting, G. In vivo dynamics of the F-actin-binding protein neurabin-II. Biochem. J. 2000, 345 Pt 2, 185–194. [Google Scholar] [CrossRef]
- Xiao, H.; Han, B.; Lodyga, M.; Bai, X.-H.; Wang, Y.; Liu, M. The actin-binding domain of actin filament-associated protein (AFAP) is involved in the regulation of cytoskeletal structure. Cell. Mol. Life Sci. 2012, 69, 1137–1151. [Google Scholar] [CrossRef]
- Wang, K.; Spector, A. α-Crystallin stabilizes actin filaments and prevents cytochalasin-induced depolymerization in a phosphorylation-dependent manner. Eur. J. Biochem. 1996, 242, 56–66. [Google Scholar] [CrossRef]
- Selden, L.A. The effect of profilin on the interaction between actin and cytochalasin D. Biophys. J. 2001, 1, 433. [Google Scholar]
- Foissner, I.; Wasteneys, G.O. Wide-ranging effects of eight cytochalasins and latrunculin A and B on intracellular motility and actin filament reorganization in characean internodal cells. Plant Cell Physiol. 2007, 48, 585–597. [Google Scholar] [CrossRef]
- Burant, C.F.; Bell, G.I. Mammalian facilitative glucose transporters: Evidence for similar substrate recognition sites in functionally monomeric proteins. Biochemistry 1992, 31, 10414–10420. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Manolescu, A.; Ritzel, M.; Yao, S.; Slugoski, M.; Young, J.D.; Chen, X.-Z.; Cheeseman, C.I. Cloning and functional characterization of the human GLUT7 isoform SLC2A7 from the small intestine. Am. J. Physiol. Liver Physiol. 2004, 287, G236–G242. [Google Scholar] [CrossRef]
- Schwingshackl, A.; Roan, E.; Teng, B.; Waters, C.M. TREK-1 regulates cytokine secretion from cultured human alveolar epithelial cells independently of cytoskeletal rearrangements. PLoS ONE 2015, 10, e0126781. [Google Scholar] [CrossRef]
- Morris, A.; Tannenbaum, J. Cytochalasin D does not produce net depolymerization of actin filaments in HEp-2 cells. Nature 1980, 287, 637–639. [Google Scholar] [CrossRef] [PubMed]
- Verkhovsky, A.B.; Svitkina, T.M.; Borisy, G.G. Polarity sorting of actin filaments in cytochalasin-treated fibroblasts. J. Cell Sci. 1997, 110, 1693–1704. [Google Scholar] [CrossRef]
- Davis, C.M.; Gruebele, M. Cytoskeletal drugs modulate off-target protein folding landscapes inside cells. Biochemistry 2020, 59, 2650–2659. [Google Scholar] [CrossRef]
- Uezato, T.; Fujita, M. Cytochalasin-B-binding proteins related to glucose transport across the basolateral membrane of the intestinal epithelial cell. J. Cell Sci. 1986, 85, 177–185. [Google Scholar] [CrossRef]
- Kletzien, R.F.; Perdue, J.F.; Springer, A. Cytochalasin A and B. Inhibition of sugar uptake in cultured cells. J. Biol. Chem. 1972, 247, 2964–2966. [Google Scholar] [CrossRef]
- Wang, F.; Arauz-Lara, B.J.J.L.; Ware, B.R. Actin critical concentration optimizes at intermediate [cytochalasin B]/[actin] ratios. Biochem. Biophys. Res. Commun. 1990, 171, 543–547. [Google Scholar] [CrossRef]
- Wang, C.; Lambert, C.; Hauser, M.; Deuschmann, A.; Zeilinger, C.; Rottner, K.; Stradal, T.E.B.; Stadler, M.; Skellam, E.J.; Cox, R.J. Diversely functionalised cytochalasins through mutasynthesis and semi-synthesis. Chem.-A Eur. J. 2020, 26, 13578–13583. [Google Scholar] [CrossRef]




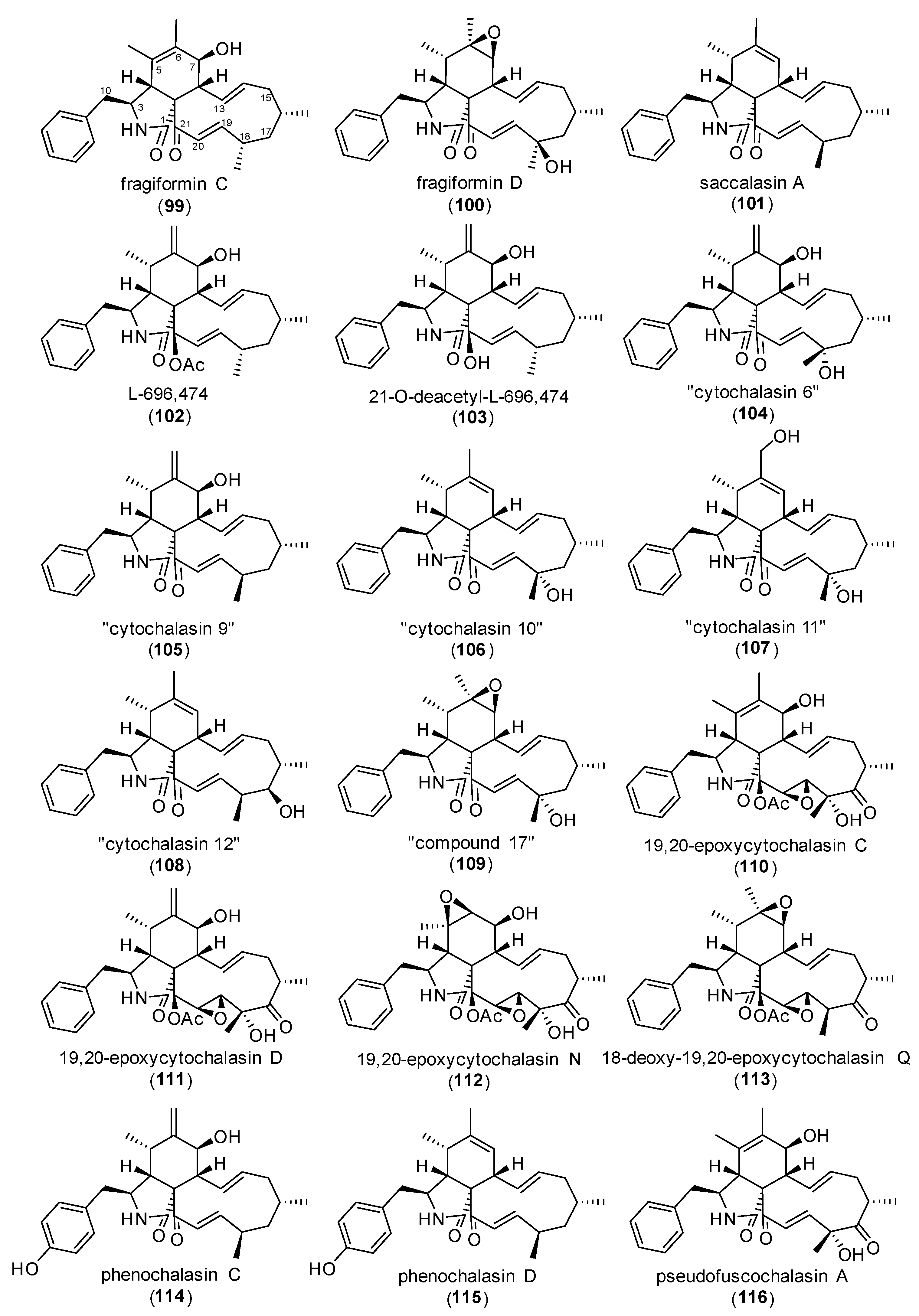
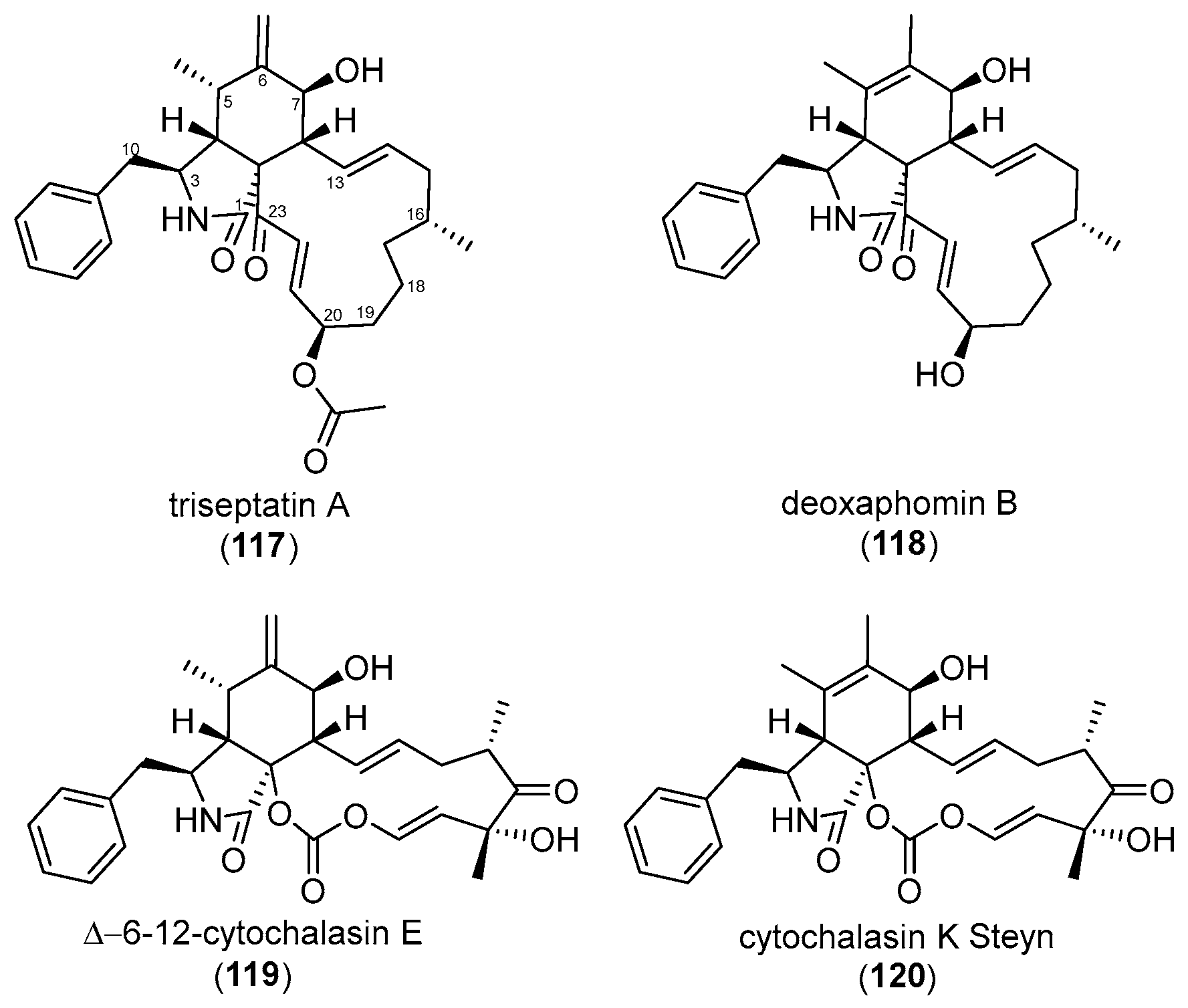

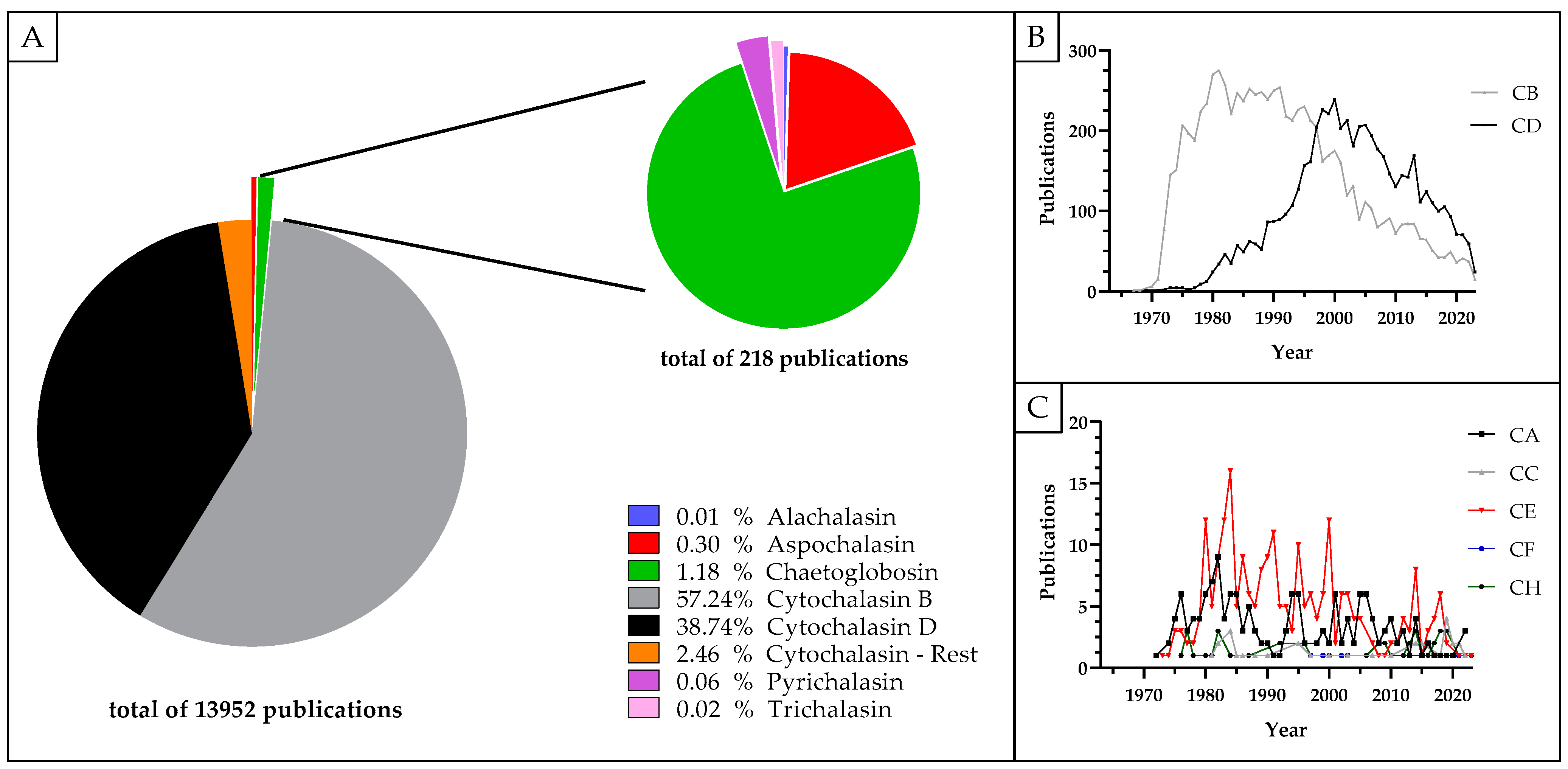
| Protein | Cytochalasan | Dosage | Treatment Duration | Experiment | Observed Effect | Source |
|---|---|---|---|---|---|---|
| Arp2/3 complex | ||||||
| Arp3 | CD (2) | 0.1 µM | 30 min | Immunofluorescence | Localization unaffected | [160] |
| p34Arc (Arp2/3) | CD | 0.1 µM | 30 min | Immunofluorescence | Localization unaffected | [160] |
| Capping proteins | ||||||
| 88 K actin binding protein (capping protein) | CB (1) | 0.1–2.5 µM | Viscometry, light scattering, A237 measurement | No inhibition by CB, but 88K inhibits CB effect | [171] | |
| CP-β2 (mouse capping protein) | CD | 1 µM | 2 min | Live cell imaging of GFP-tagged CP-β2 | No effect on CP-β2 distribution | [173] |
| Gelsolin | CB | 2 µM | Pyrene assay | Negligible effect on gelsolin-bound filaments | [102] | |
| Tensin (crosslinking and capping protein) | CD | 1.5 µg/mL | 30 min | Immunoprecipitation | Tyrosine phosphorylation inhibited | [25] |
| βcap73 (capping protein) | CD | 10 nM- 15 µg/mL | Actin affinity matrices | Binding to β-actin largely inhibited | [170] | |
| Ena/VASP family proteins | ||||||
| Mena | CD | 25–150 nM | 5–30 min | Immunofluorescence, live cell imaging | Delocalization | [101,150,152] |
| VASP | CB | 52 µM (in needle) | 2 min | Live cell imaging | Accumulation | [154] |
| VASP | CD | 25 nM–1 µM | 2 min–2 h | Immunofluorescence, live cell imaging | Delocalization | [101,122,149,150,151,152] |
| Ezrin/radixin/moesin (ERM) family proteins | ||||||
| Ezrin | CE (55) | 1.5 µM | 60 min | Gradient centrifugation of treated cell lysate | Protein abundance unaffected | [22] |
| Ezrin | CD | 2 µM- 15 µg/mL | Actin affinity matrices | CD competes with binding on actin affinity matrices | [170,174] | |
| Radixin | CE | 1.5 µM | 60 min | Gradient centrifugation of treated cell lysate | Protein abundance unaffected | [22] |
| Fodrin | CE | 1.5 µM | 60 min | Gradient centrifugation of treated cell lysate | Protein abundance decreased | [22] |
| Moesin | CE | 1.5 µM | 60 min | Gradient centrifugation of treated cell lysate | Protein abundance unaffected | [22] |
| Proteins examined in context of L. monocytogenesactin comet tails | ||||||
| Cofilin, Coronin, Aip1 | CD | Concentration titration 0–3 µM | Single filament imaging | Activity on F-actin disassembly inhibited | [128] | |
| Myosins | ||||||
| Heavy meromyosin | CB | ~100–420 µM | ATPase activity assay | ATPase activity inhibited | [9] | |
| Heavy meromyosin (HMM) | ChJ (54) | 0.71 µM | 5 min | Electron microscopy of HMM-decorated F-actin | Binding to F-actin unaffected, but suppresses ChJ-mediated depolymerization | [87] |
| Myosin II | CB | 3–57 µM | 20 min | Viscometry | Competition of myosin II and CB for actin binding | [9] |
| Myosin II | ChJ | 2 mol per mol actin | 5 min | ATPase activity assay | ATPase activity reduced | [87] |
| Myosin II | CB | 350 nM | Live cell imaging | Localization unaffected | [147] | |
| Other ABPs | ||||||
| AFAP (actin filament-associated protein) | CD | Gradient centrifugation | Increased in cytosolic fraction and reduced in cytoskeletal fraction | [189] | ||
| Bn1pFH1FH2 | CB | 2 µM | Pyrene assay | Partial inhibition of filament elongation, but not nucleation | [102] | |
| Cofilin | CD | 0.3–10 µM | 10–120 min | In vitro sedimentation and BiFC assay | Binding to G-and F-actin inhibited | [184] |
| Cofilin | CD | 5–10 µM | 10 min | Western blots of whole cell lysates and comparison of phosphorylation status | Activated via dephosphorylation | [23] |
| Cortactin | CD | 25 nM | 30 min | Immunofluorescence | Localization unaffected | [152] |
| α-Crystallin | CD | 1 or 2 mol per mol actin | 35 min–20 h | Pyrene assay | CD-mediated depolymerization alleviated by α-crystallin | [190] |
| hhLIM (novel human heart LIM protein) | CB | ? | 10–120 min, 30 min | Live cell imaging, Immunoprecipitation | CB-mediated inhibition of polymerization and induced depolymerization alleviated by hhLIM | [176] |
| Lpd | CD | 150 nM | 30 min | Immunofluorescence | Localization unaffected | [150] |
| Lpd850–1250aa (ABD of Lpd) | CD | 100 nM | ~2 min | Live cell imaging | Delocalization | [122] |
| Neurabin-II (spinophilin) | CD | 10 nM–2 µM | 20 min–2 h | Immunofluorescence and live cell imaging | No effect on association with F-actin | [188] |
| N-WASP | CD | 10–200 nM | 30 min | Immunofluorescence | Localization unaffected | [101] |
| Profilin | CD | ? | ? | ? | Competitive binding of profilin and CD to Mg-ATP-actin | [191], Discussed by [192] |
| Spectrin | CE | 1.5 µM | 60 min | Gradient centrifugation of treated cell lysate | Protein abundance unaffected | [22] |
| Spectrin | CB | 2 µM | Pyrene assay | Lack of effect on inhibition of F-actin polymerization | [103] | |
| Spectrin | CB | 2–60 µM | Steady-state titration | In vitro fluorescence quenching, in silico computational modeling and docking | CB binding with moderate affinity | [187] |
| ABD of spectrin | CB | 2–60 µM | Steady-state titration | In vitro fluorescence quenching, in silico computational modeling and docking | 10× increase of affinity to CB as compared to spectrin | [187] |
| Tropomyosin | CD, CB | 2–20 µM, 100× higher | Viscometry | Protection of F-actin against CB and CD upon saturated binding | [175] | |
| Proteins examined in context ofVaccinia virus actin tails | ||||||
| ARPC5 | CD | 1 µM | Immediate, 30 min | Live cell imaging with FRAP, Immunofluorescence | Reduction of turnover at vaccinia virus actin tails | [167] |
| GRB2 [154]) | CD | 1 µM | Immediate, 30 min | Live cell imaging with FRAP, Immunofluorescence | Unaffected turnover at vaccinia virus actin tails | [167] |
| Nck | CD | 1 µM | Immediate, 30 min | Live cell imaging with FRAP, Immunofluorescence | Unaffected turnover at vaccinia virus actin tails | [167] |
| N-WASP | CD | 1 µM | 30 min | Live cell imaging | Reduction of turnover at vaccinia virus actin tails | [167] |
| WIP | CD | 1 µM | Immediate, 30 min | Live cell imaging with FRAP, Immunofluorescence | Reduction of turnover at vaccinia virus actin tails | [167] |
| WAVE and WAVE regulatory complex (WRC) | ||||||
| Abi-1 | CB | Live cell imaging | Accumulation | Unpublished [154] | ||
| Sra1 | CD | 0.1–1 µM | ~1–20 min | Live cell imaging | Reduction of turnover at and translocation along leading edge | [103] |
| Non-actin-related targets | ||||||
| Glucose transport through erythrocyte membranes | CA (3), CB, CB-7-monoacetate, 24-deoxaphomin, proxiphomin, protophomin, ChgB (5), ChgE (7), ChgF (8) | Inhibition of glucose transport | [80] | |||
| hGLUT1 | CB | 1 mM to 10 mg/mL of purified protein | Co-crystallization | CB binding to the central cavity of hGLUT1 | [15] | |
| GLUT1-4 | CB | Inhibition of glucose transport | [193,194] | |||
| hKv1.5 channel | CA (3) CB, CD, CJ (58) | Reduced activity upon CA and CB, less prominent effect upon CD and CJ | [21] | |||
| Further actin-associated effects | ||||||
| ENaC | CE CD | 1.5 µM | 15–60 min | Single channel patch clamp experiments, immunoprecipitation | Decrease of amiloride-sensitive transepithelial current, reduced interaction with MARCKS | [22] |
| GLUT4 | CD | Prevention of insulin induced translocation | [24] | |||
| L-type Ca2+ channel | CD | 5–10 µM | 5–10 min | Measurement of whole-cell currents | Phalloidin-preventable reduction of Ca2+ currents in guinea pig cardiomyocytes | [23] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lambert, C.; Schmidt, K.; Karger, M.; Stadler, M.; Stradal, T.E.B.; Rottner, K. Cytochalasans and Their Impact on Actin Filament Remodeling. Biomolecules 2023, 13, 1247. https://doi.org/10.3390/biom13081247
Lambert C, Schmidt K, Karger M, Stadler M, Stradal TEB, Rottner K. Cytochalasans and Their Impact on Actin Filament Remodeling. Biomolecules. 2023; 13(8):1247. https://doi.org/10.3390/biom13081247
Chicago/Turabian StyleLambert, Christopher, Katharina Schmidt, Marius Karger, Marc Stadler, Theresia E. B. Stradal, and Klemens Rottner. 2023. "Cytochalasans and Their Impact on Actin Filament Remodeling" Biomolecules 13, no. 8: 1247. https://doi.org/10.3390/biom13081247
APA StyleLambert, C., Schmidt, K., Karger, M., Stadler, M., Stradal, T. E. B., & Rottner, K. (2023). Cytochalasans and Their Impact on Actin Filament Remodeling. Biomolecules, 13(8), 1247. https://doi.org/10.3390/biom13081247











