Genome-Wide Acetylation Modification of H3K27ac in Bovine Rumen Cell Following Butyrate Exposure
Abstract
1. Introduction
2. Materials and Methods
2.1. Rumen Epithelial Primary Cells and Butyrate Treatment
2.2. Chromatin Immunoprecipitation and Sequencing
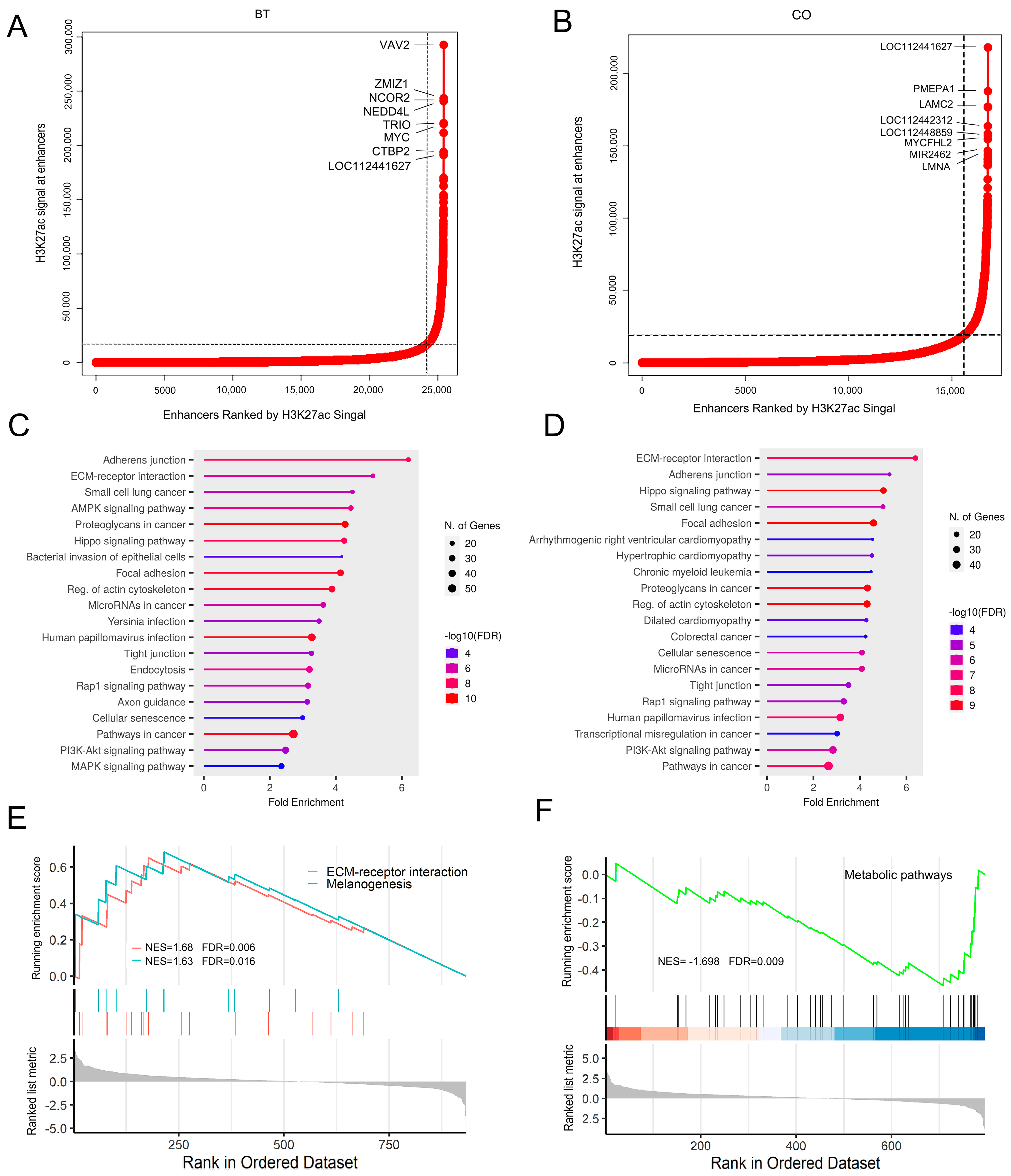
2.3. Differential Enrichment Regions and Super-Enhancer Identification
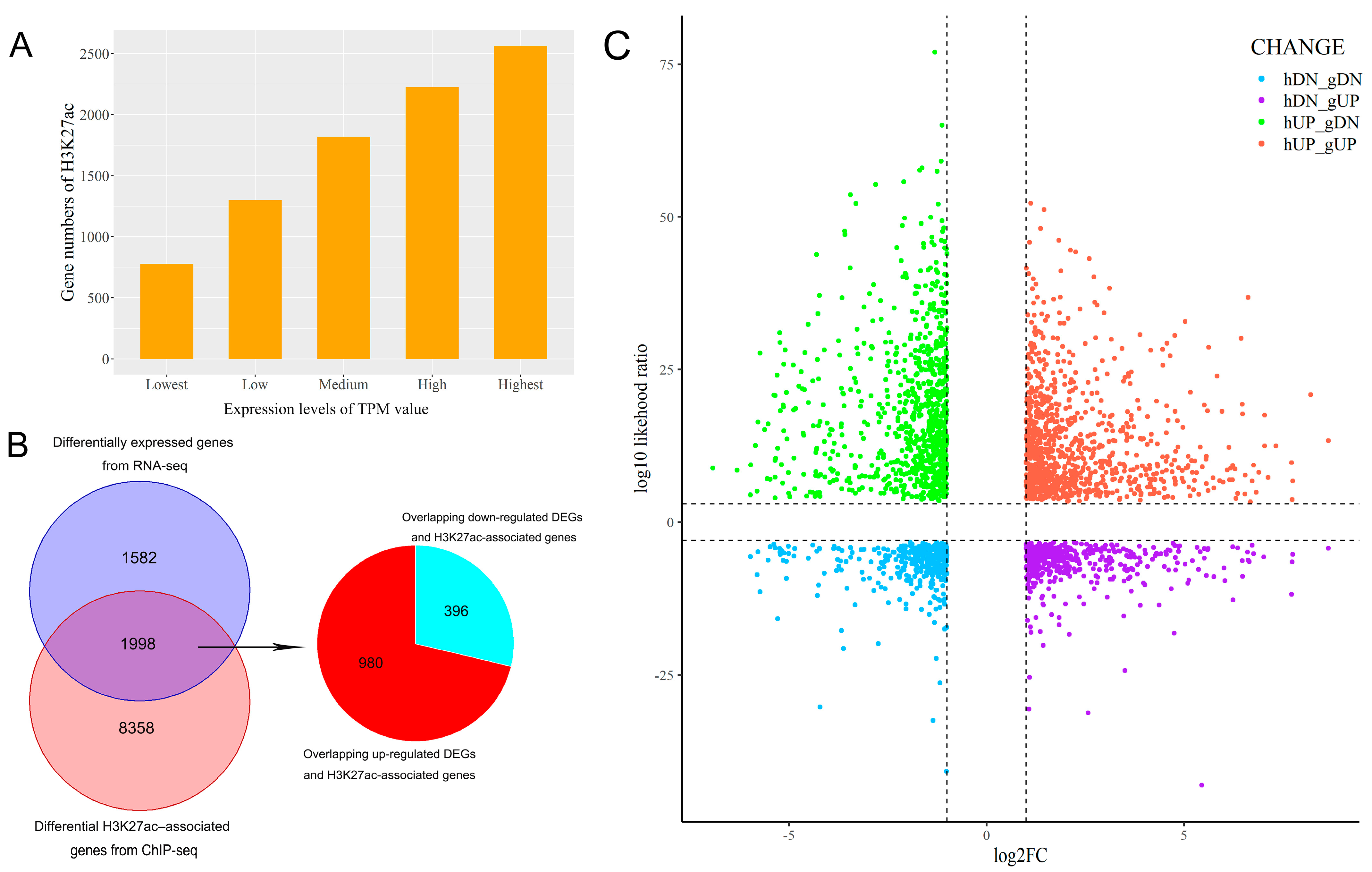
2.4. RNA-Seq Analysis
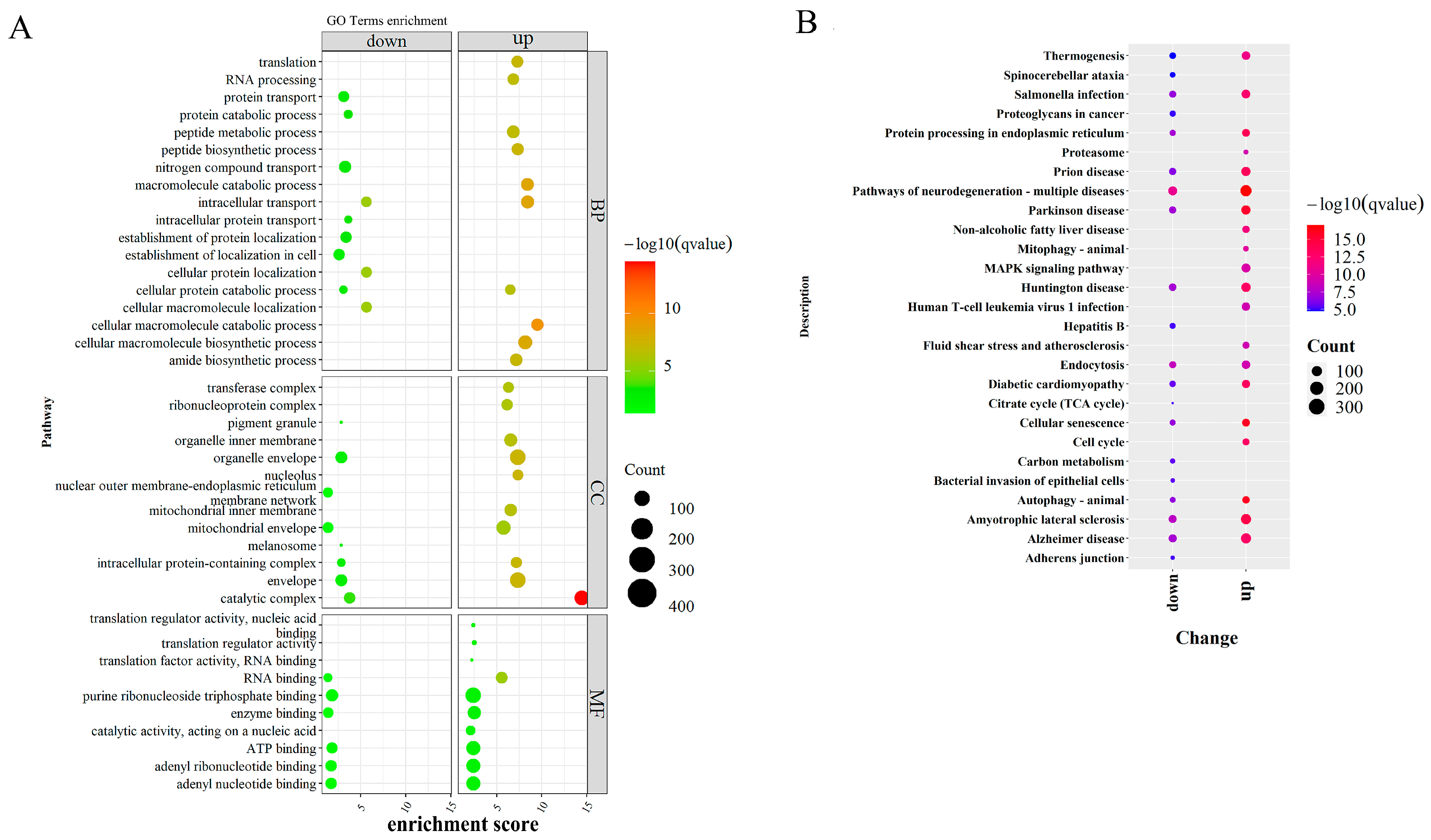
2.5. Functional Enrichment and Pathway Analysis
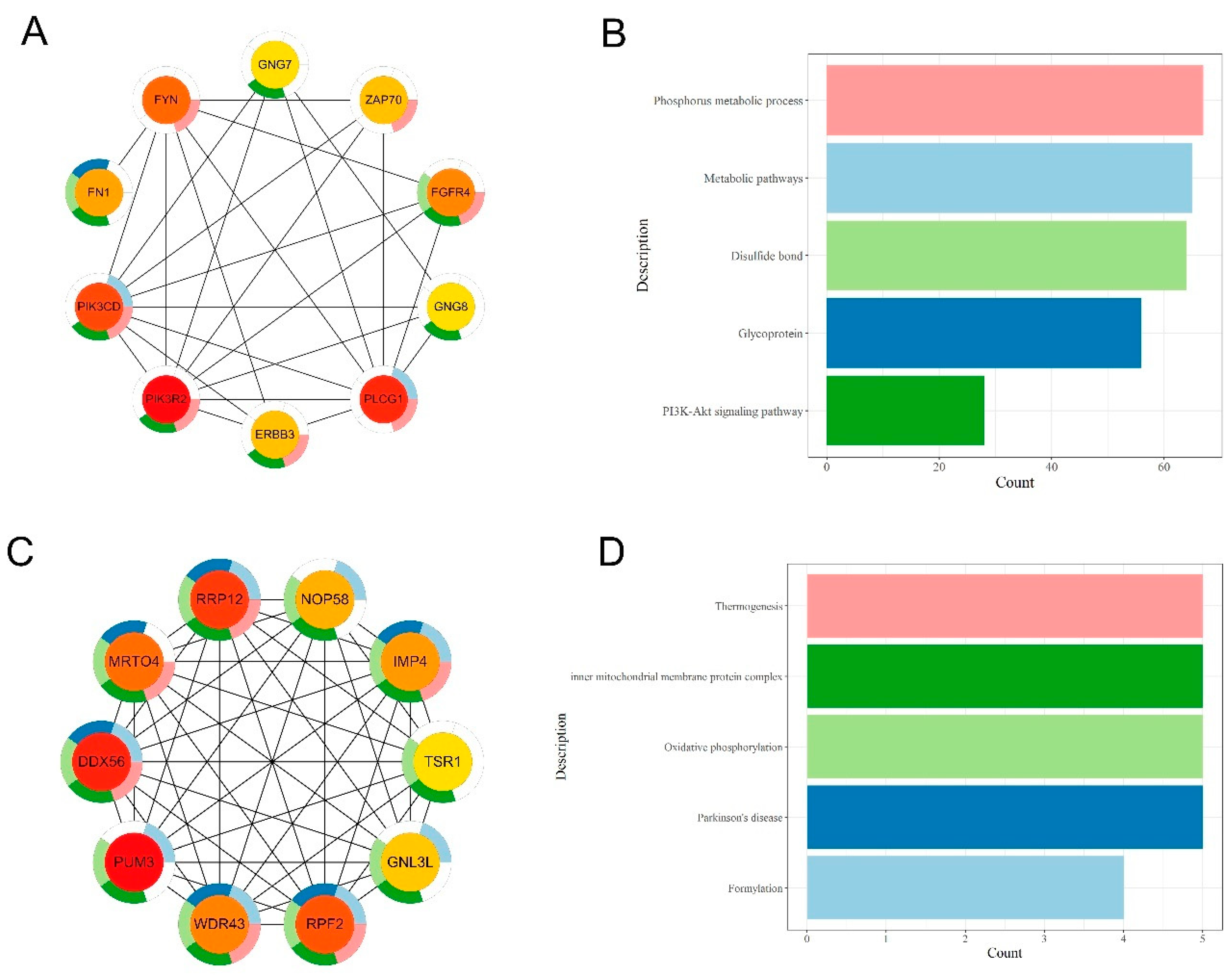
2.6. Protein-Protein Interaction Analysis
3. Results
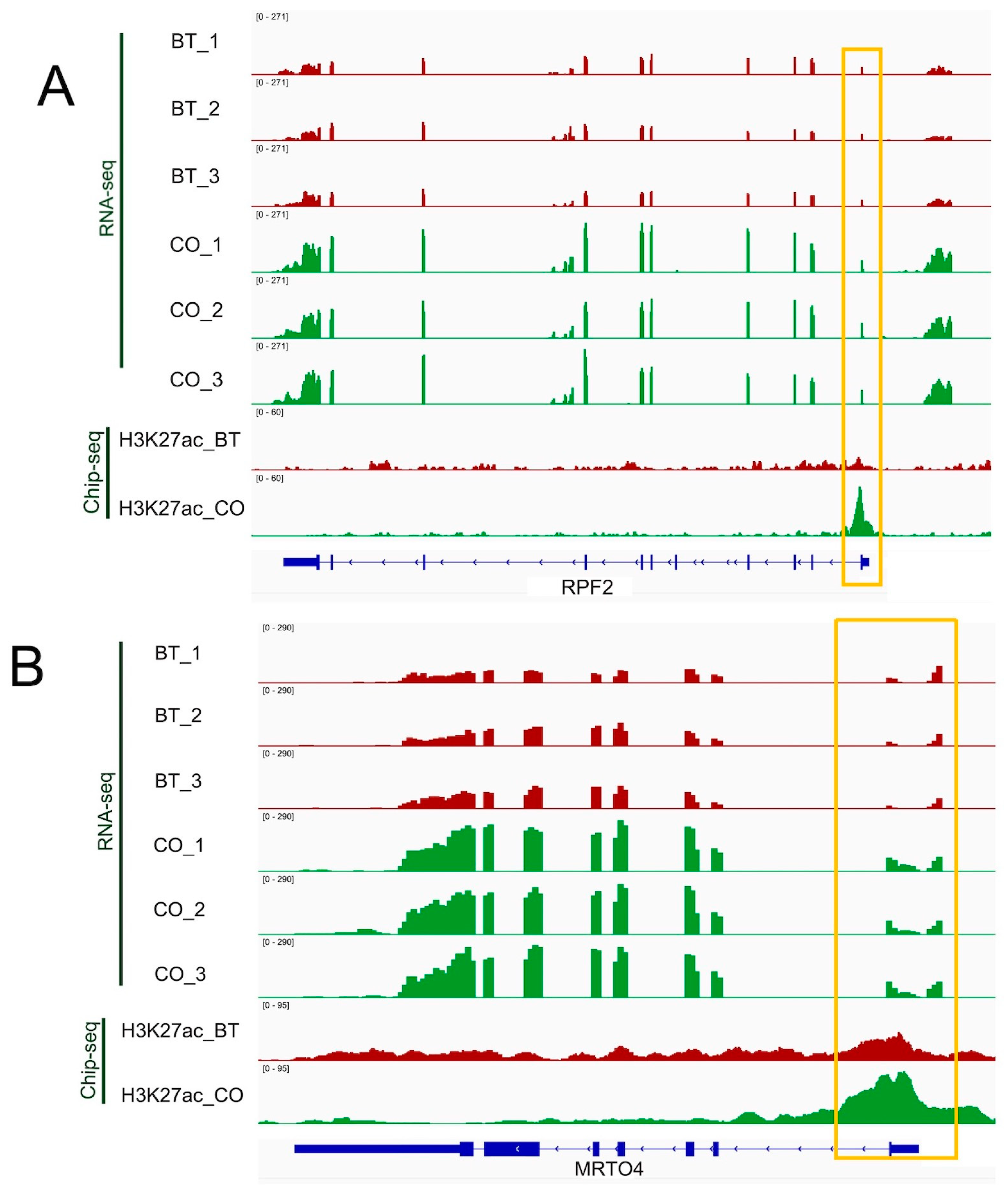
3.1. Genome-Wide Changes of H3K27ac Profiles in Butyrate-Induced REPC
3.2. H3K27ac Modified Regions That Regulate Gene Profiles in Butyrate-Induced REPC
3.3. Genome-Wide H3K27ac-Based Super-Enhancer Detection between Groups
3.4. Integrative Analysis of Butyrate-Associated Transcriptome and H3K27ac Modification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| REPC | rumen epithelial primary cell |
| SCFAs | short-chain fatty acids |
| HDAC | histone deacetylase |
| HDACi | histone deacetylase inhibitor |
| H3K27ac | histone H3 lysine 27 acetylation |
| IGV | integrative genomics viewer |
| DEG | differentially expressed gene |
| GSEA | gene set enrichment analysis |
| PPI | protein–protein interaction |
| TSS | transcription start site |
| TES | transcription end site |
| SE | super-enhancer |
| HAT | histone acetyltransferase |
| DHG | differentially H3K27ac-modified gene |
| TPM | transcripts per million |
| hUP-gUP | H3K27ac-gain and up-regulated DEG expression |
| hDN-gDN | H3K27ac-loss and down-regulated DEG expression |
| MAPK | mitogen-activated protein kinase |
References
- Shen, H.; Xu, Z.; Shen, Z.; Lu, Z. The Regulation of Ruminal Short-Chain Fatty Acids on the Functions of Rumen Barriers. Front. Physiol. 2019, 10, 1305. [Google Scholar] [CrossRef]
- Greco, G.; Hagen, F.; Meissner, S.; Shen, Z.; Lu, Z.; Amasheh, S.; Aschenbach, J.R. Effect of individual SCFA on the epithelial barrier of sheep rumen under physiological and acidotic luminal pH conditions. J. Anim. Sci. 2018, 96, 126–142. [Google Scholar] [CrossRef]
- Dai, L.N.; Yan, J.K.; Xiao, Y.T.; Wen, J.; Zhang, T.; Zhou, K.J.; Wang, Y.; Cai, W. Butyrate stimulates the growth of human intestinal smooth muscle cells by activation of yes-associated protein. J. Cell. Physiol. 2018, 233, 3119–3128. [Google Scholar] [CrossRef]
- Rashid, S.; Salim, A.; Qazi, R.; Malick, T.S.; Haneef, K. Sodium Butyrate Induces Hepatic Differentiation of Mesenchymal Stem Cells in 3D Collagen Scaffolds. Appl. Biochem. Biotechnol. 2022, 194, 3721–3732. [Google Scholar] [CrossRef]
- Pant, K.; Yadav, A.K.; Gupta, P.; Islam, R.; Saraya, A.; Venugopal, S.K. Butyrate induces ROS-mediated apoptosis by modulating miR-22/SIRT-1 pathway in hepatic cancer cells. Redox Biol. 2017, 12, 340–349. [Google Scholar] [CrossRef]
- Li, R.W.; Li, C. Butyrate induces profound changes in gene expression related to multiple signal pathways in bovine kidney epithelial cells. BMC Genom. 2006, 7, 234. [Google Scholar] [CrossRef]
- Lin, S.; Fang, L.; Kang, X.; Liu, S.; Liu, M.; Connor, E.E.; Baldwin, R.L.; Liu, G.; Li, C.J. Establishment and transcriptomic analyses of a cattle rumen epithelial primary cells (REPC) culture by bulk and single-cell RNA sequencing to elucidate interactions of butyrate and rumen development. Heliyon 2020, 6, e04112. [Google Scholar] [CrossRef]
- Prause, M.; Pedersen, S.S.; Tsonkova, V.; Qiao, M.; Billestrup, N. Butyrate Protects Pancreatic Beta Cells from Cytokine-Induced Dysfunction. Int. J. Mol. Sci. 2021, 22, 10427. [Google Scholar] [CrossRef]
- Chamberlain, D.; Thomas, P.; Anderson, F.J. Volatile fatty acid proportions and lactic acid metabolism in the rumen in sheep and cattle receiving silage diets. J. Agric. Sci. 1983, 101, 47–58. [Google Scholar] [CrossRef]
- Li, C.; Chen, X.; Zhang, B.; Liu, L.; Li, F. Sodium butyrate improved intestinal barrier in rabbits. Ital. J. Anim. Sci. 2020, 19, 1482–1492. [Google Scholar] [CrossRef]
- Song, B.; Li, H.; Wu, Y.; Zhen, W.; Wang, Z.; Xia, Z.; Guo, Y. Effect of microencapsulated sodium butyrate dietary supplementation on growth performance and intestinal barrier function of broiler chickens infected with necrotic enteritis. Anim. Feed Sci. Technol. 2017, 232, 6–15. [Google Scholar] [CrossRef]
- Naghizadeh, M.; Dalgaard, T.S.; Klaver, L.; Engberg, R.M. Effects of encapsulated butyrate and salinomycin on gut leakage and intestinal inflammation in broilers. Br. Poult. Sci. 2022, 63, 499–509. [Google Scholar] [CrossRef]
- Liu, H.; Zhao, J.; Zhang, W.; Nie, C. Impacts of sodium butyrate on intestinal mucosal barrier and intestinal microbial community in a weaned piglet model. Front. Microbiol. 2022, 13, 1041885. [Google Scholar] [CrossRef]
- Li, C.; Chen, J.; Zhao, M.; Liu, M.; Yue, Z.; Liu, L.; Li, F. Effect of sodium butyrate on slaughter performance, serum indexes and intestinal barrier of rabbits. J. Anim. Physiol. Anim. Nutr. 2022, 106, 156–166. [Google Scholar] [CrossRef]
- Morel, P.; Chidgey, K.; Jenkinson, C.; Lizarraga, I.; Schreurs, N. Effect of benzoic acid, sodium butyrate and sodium butyrate coated with benzoic acid on growth performance, digestibility, intestinal morphology and meat quality in grower-finisher pigs. Livest. Sci. 2019, 226, 107–113. [Google Scholar] [CrossRef]
- Chen, X.; Kong, Q.; Zhao, X.; Zhao, C.; Hao, P.; Irshad, I.; Lei, H.; Kulyar, M.F.; Bhutta, Z.A.; Ashfaq, H.; et al. Sodium acetate/sodium butyrate alleviates lipopolysaccharide-induced diarrhea in mice via regulating the gut microbiota, inflammatory cytokines, antioxidant levels, and NLRP3/Caspase-1 signaling. Front. Microbiol. 2022, 13, 1036042. [Google Scholar] [CrossRef]
- Baldwin, R.L.; Li, R.W.; Jia, Y.; Li, C.J. Transcriptomic Impacts of Rumen Epithelium Induced by Butyrate Infusion in Dairy Cattle in Dry Period. Gene Regul. Syst. Biol. 2018, 12, 1177625018774798. [Google Scholar] [CrossRef]
- Liu, J.; Chang, G.; Huang, J.; Ma, N.; Wang, Y.; Roy, A.C.; Shen, X. Sodium butyrate pretreatment mitigates lipopolysaccharide-induced inflammation through the TLR4/NF-κB signaling pathway in bovine embryo trachea cells. Anim. Biotechnol. 2022, 33, 1571–1581. [Google Scholar] [CrossRef]
- Boschiero, C.; Gao, Y.; Baldwin, R.L.; Ma, L.; Li, C.J.; Liu, G.E. Butyrate Induces Modifications of the CTCF-Binding Landscape in Cattle Cells. Biomolecules 2022, 12, 1177. [Google Scholar] [CrossRef]
- Fang, L.; Liu, S.; Liu, M.; Kang, X.; Lin, S.; Li, B.; Connor, E.E.; Baldwin, R.L.; Tenesa, A.; Ma, L. Functional annotation of the cattle genome through systematic discovery and characterization of chromatin states and butyrate-induced variations. BMC Biol. 2019, 17, 68. [Google Scholar] [CrossRef]
- Kang, X.; Liu, S.; Fang, L.; Lin, S.; Liu, M.; Baldwin, R.L.; Liu, G.E.; Li, C.-J. Data of epigenomic profiling of histone marks and CTCF binding sites in bovine rumen epithelial primary cells before and after butyrate treatment. Data Brief 2020, 28, 104983. [Google Scholar] [CrossRef]
- Wang, L.; Shannar, A.A.F.; Wu, R.; Chou, P.; Sarwar, M.S.; Kuo, H.C.; Peter, R.M.; Wang, Y.; Su, X.; Kong, A.N. Butyrate Drives Metabolic Rewiring and Epigenetic Reprogramming in Human Colon Cancer Cells. Mol. Nutr. Food Res. 2022, 66, e2200028. [Google Scholar] [CrossRef]
- Sharma, P.; Yadav, A.S.; Selokar, N.L.; Kumar, D.; Dhaka, S.S.; Yadav, P.S. Epigenetic status of buffalo fibroblasts treated with sodium butyrate a chromatin remodeling agent. Tissue Cell 2018, 50, 51–58. [Google Scholar] [CrossRef]
- Slaughter, M.J.; Shanle, E.K.; Khan, A.; Chua, K.F.; Hong, T.; Boxer, L.D.; Allis, C.D.; Josefowicz, S.Z.; Garcia, B.A.; Rothbart, S.B.; et al. HDAC inhibition results in widespread alteration of the histone acetylation landscape and BRD4 targeting to gene bodies. Cell Rep. 2021, 34, 108638. [Google Scholar] [CrossRef]
- Shahbazian, M.D.; Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100. [Google Scholar] [CrossRef]
- Zhu, I.; Song, W.; Ovcharenko, I.; Landsman, D. A model of active transcription hubs that unifies the roles of active promoters and enhancers. Nucleic Acids Res. 2021, 49, 4493–4505. [Google Scholar] [CrossRef]
- Shlyueva, D.; Stampfel, G.; Stark, A. Transcriptional enhancers: From properties to genome-wide predictions. Nat. Rev. Genet. 2014, 15, 272–286. [Google Scholar] [CrossRef]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef]
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-Andre, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-enhancers in the control of cell identity and disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef]
- Neville, J.J.; Orlando, J.; Mann, K.; McCloskey, B.; Antoniou, M.N. Ubiquitous Chromatin-opening Elements (UCOEs): Applications in biomanufacturing and gene therapy. Biotechnol. Adv. 2017, 35, 557–564. [Google Scholar] [CrossRef]
- Li, C.J.; Elsasser, T.H. Butyrate-induced apoptosis and cell cycle arrest in bovine kidney epithelial cells: Involvement of caspase and proteasome pathways. J. Anim. Sci. 2005, 83, 89–97. [Google Scholar] [CrossRef]
- Shin, J.H.; Li, R.W.; Gao, Y.; Baldwin, R.; Li, C.J. Genome-wide ChIP-seq mapping and analysis reveal butyrate-induced acetylation of H3K9 and H3K27 correlated with transcription activity in bovine cells. Funct. Integr. Genom. 2012, 12, 119–130. [Google Scholar] [CrossRef]
- De Sena Brandine, G.; Smith, A.D. Falco: High-speed FastQC emulation for quality control of sequencing data. F1000Research 2019, 8, 1874. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Ramirez, F.; Ryan, D.P.; Gruning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dundar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef]
- Zhu, L.J.; Gazin, C.; Lawson, N.D.; Pages, H.; Lin, S.M.; Lapointe, D.S.; Green, M.R. ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinform. 2010, 11, 237. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Cowan, J.E.; Malin, J.; Zhao, Y.; Seedhom, M.O.; Harly, C.; Ohigashi, I.; Kelly, M.; Takahama, Y.; Yewdell, J.W.; Cam, M. Myc controls a distinct transcriptional program in fetal thymic epithelial cells that determines thymus growth. Nat. Commun. 2019, 10, 5498. [Google Scholar] [CrossRef]
- Kimura, T.; Kawabe, H.; Jiang, C.; Zhang, W.; Xiang, Y.-Y.; Lu, C.; Salter, M.W.; Brose, N.; Lu, W.-Y.; Rotin, D. Deletion of the ubiquitin ligase Nedd4L in lung epithelia causes cystic fibrosis-like disease. Proc. Natl. Acad. Sci. USA 2011, 108, 3216–3221. [Google Scholar] [CrossRef]
- Zhang, J.; Deqing, Z.; Zhang, X.; Ta, N.; Gesang, J.; Luosang, C.; Pingcuo, B. Different feeding strategies can affect growth performance and rumen functions in Gangba sheep as revealed by integrated transcriptome and microbiome analyses. Front. Microbiol. 2022, 13, 908326. [Google Scholar]
- Levental, K.R.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.T.; Fong, S.F.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 2009, 139, 891–906. [Google Scholar] [CrossRef]
- Marinova, Z.; Leng, Y.; Leeds, P.; Chuang, D.M. Histone deacetylase inhibition alters histone methylation associated with heat shock protein 70 promoter modifications in astrocytes and neurons. Neuropharmacology 2011, 60, 1109–1115. [Google Scholar] [CrossRef]
- Kavurma, M.M.; Figg, N.; Bennett, M.R.; Mercer, J.; Khachigian, L.M.; Littlewood, T.D. Oxidative stress regulates IGF1R expression in vascular smooth-muscle cells via p53 and HDAC recruitment. Biochem. J. 2007, 407, 79–87. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Qian, Y.; He, X.; Mo, C.; Yang, X.; Xiao, Q. Sodium butyrate attenuates rotenone-induced toxicity by activation of autophagy through epigenetically regulating PGC-1α expression in PC12 cells. Brain Res. 2022, 1776, 147749. [Google Scholar] [CrossRef] [PubMed]
- Rada-Iglesias, A.; Enroth, S.; Ameur, A.; Koch, C.M.; Clelland, G.K.; Respuela-Alonso, P.; Wilcox, S.; Dovey, O.M.; Ellis, P.D.; Langford, C.F.; et al. Butyrate mediates decrease of histone acetylation centered on transcription start sites and down-regulation of associated genes. Genome Res. 2007, 17, 708–719. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, Y.; Sun, Y.; Dong, X.; Wang, Z.; Zhang, Z.; Xiao, Y.; Dong, G. Bacterial endotoxin decreased histone H3 acetylation of bovine mammary epithelial cells and the adverse effect was suppressed by sodium butyrate. BMC Vet. Res. 2019, 15, 267. [Google Scholar] [CrossRef] [PubMed]
- Davie, J.R. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 2003, 133 (Suppl. S7), 2485S–2493S. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Lou, X.; Hwang, P.; Rajashankar, K.R.; Wang, X.; Gustafsson, J.-Å.; Fletterick, R.J.; Jacobson, R.H.; Webb, P. Structural and functional insight into TAF1–TAF7, a subcomplex of transcription factor II D. Proc. Natl. Acad. Sci. USA 2014, 111, 9103–9108. [Google Scholar] [CrossRef]
- Irvin, J.D.; Pugh, B.F. Genome-wide transcriptional dependence on TAF1 functional domains. J. Biol. Chem. 2006, 281, 6404–6412. [Google Scholar] [CrossRef]
- Choi, J.K.; Kim, Y.J. Epigenetic regulation and the variability of gene expression. Nat. Genet. 2008, 40, 141–147. [Google Scholar] [CrossRef]
- Kloet, S.L.; Whiting, J.L.; Gafken, P.; Ranish, J.; Wang, E.H. Phosphorylation-dependent regulation of cyclin D1 and cyclin A gene transcription by TFIID subunits TAF1 and TAF7. Mol. Cell. Biol. 2012, 32, 3358–3369. [Google Scholar] [CrossRef]
- Dunphy, E.L.; Johnson, T.; Auerbach, S.S.; Wang, E.H. Requirement for TAF(II)250 acetyltransferase activity in cell cycle progression. Mol. Cell. Biol. 2000, 20, 1134–1139. [Google Scholar] [CrossRef]
- O’Brien, T.; Tjian, R. Different functional domains of TAFII250 modulate expression of distinct subsets of mammalian genes. Proc. Natl. Acad. Sci. USA 2000, 97, 2456–2461. [Google Scholar] [CrossRef]
- Gonçalves, P.; Gregório, I.; Catarino, T.A.; Martel, F. The effect of oxidative stress upon the intestinal epithelial uptake of butyrate. Eur. J. Pharmacol. 2013, 699, 88–100. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, A.M.; Somasekharan, S.P.; Wang, Y.; Cheng, H.; Negri, G.L.; Pan, M.; Wang, X.Q.; Delaidelli, A.; Rafn, B.; Cran, J.; et al. Class I HDAC inhibitors enhance YB-1 acetylation and oxidative stress to block sarcoma metastasis. EMBO Rep. 2019, 20, e48375. [Google Scholar] [CrossRef] [PubMed]
- Grilli, E.; Tugnoli, B.; Foerster, C.; Piva, A. Butyrate modulates inflammatory cytokines and tight junctions components along the gut of weaned pigs. J. Anim. Sci. 2016, 94 (Suppl. S3), 433–436. [Google Scholar] [CrossRef]
- Nielsen, D.S.G.; Jensen, B.B.; Theil, P.K.; Nielsen, T.S.; Knudsen, K.E.B.; Purup, S. Effect of butyrate and fermentation products on epithelial integrity in a mucus-secreting human colon cell line. J. Funct. Foods 2018, 40, 9–17. [Google Scholar] [CrossRef]
- Li, X.; Wang, C.; Zhu, J.; Lin, Q.; Yu, M.; Wen, J.; Feng, J.; Hu, C. Sodium butyrate ameliorates oxidative stress-induced intestinal epithelium barrier injury and mitochondrial damage through AMPK-mitophagy pathway. Oxidative Med. Cell. Longev. 2022, 2022, 3745135. [Google Scholar] [CrossRef]
- Khan, S.; Jena, G. Protective role of sodium butyrate, a HDAC inhibitor on beta-cell proliferation, function and glucose homeostasis through modulation of p38/ERK MAPK and apoptotic pathways: Study in juvenile diabetic rat. Chem.-Biol. Interact. 2014, 213, 1–12. [Google Scholar] [CrossRef]
- Sur, I.; Taipale, J. The role of enhancers in cancer. Nat. Rev. Cancer 2016, 16, 483–493. [Google Scholar] [CrossRef]
- Qiu, H.; Zhu, B.; Ni, S. Identification of genes associated with primary open-angle glaucoma by bioinformatics approach. Int. Ophthalmol. 2018, 38, 19–28. [Google Scholar] [CrossRef]
- Malhi, M.; Gui, H.; Yao, L.; Aschenbach, J.R.; Gabel, G.; Shen, Z. Increased papillae growth and enhanced short-chain fatty acid absorption in the rumen of goats are associated with transient increases in cyclin D1 expression after ruminal butyrate infusion. J. Dairy Sci. 2013, 96, 7603–7616. [Google Scholar] [CrossRef]
- Liu, L.; Sun, D.; Mao, S.; Zhu, W.; Liu, J. Infusion of sodium butyrate promotes rumen papillae growth and enhances expression of genes related to rumen epithelial VFA uptake and metabolism in neonatal twin lambs. J. Anim. Sci. 2019, 97, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Gorka, P.; Kowalski, Z.M.; Zabielski, R.; Guilloteau, P. Invited review: Use of butyrate to promote gastrointestinal tract development in calves. J. Dairy Sci. 2018, 101, 4785–4800. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, H.; Zhao, J.; Peng, P.; Shao, M.; Wu, H.; Wang, X.; Chen, L.; Zhang, Q.; Ruan, Y.; et al. High Intratumoral Expression of Tetranectin Associates with Poor Prognosis of Patients with Gastric Cancer after Gastrectomy. J. Cancer 2017, 8, 3623–3630. [Google Scholar] [CrossRef]
- Luo, C.; Wang, F.; Qin, S.; Chen, Q.; Wang, Q.K. Coronary artery disease susceptibility gene ADTRP regulates cell cycle progression, proliferation, and apoptosis by global gene expression regulation. Physiol. Genom. 2016, 48, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Defour, M.; van Weeghel, M.; Hermans, J.; Kersten, S. Hepatic ADTRP overexpression does not influence lipid and glucose metabolism. Am. J. Physiol.-Cell Physiol. 2021, 321, C585–C595. [Google Scholar] [CrossRef]
- Albelazi, M.S.; Martin, P.R.; Mohammed, S.; Mutti, L.; Parsons, J.L.; Elder, R.H. The Biochemical Role of the Human NEIL1 and NEIL3 DNA Glycosylases on Model DNA Replication Forks. Genes 2019, 10, 315. [Google Scholar] [CrossRef]
- Meng, F.; Zhang, S.; Song, R.; Liu, Y.; Wang, J.; Liang, Y.; Wang, J.; Han, J.; Song, X.; Lu, Z. NCAPG2 overexpression promotes hepatocellular carcinoma proliferation and metastasis through activating the STAT3 and NF-κB/miR-188-3p pathways. EBioMedicine 2019, 44, 237–249. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, L.; Liu, Y.; Zhang, W. NCAPG2 contributes to the progression of malignant melanoma through regulating proliferation and metastasis. Biochem. Cell Biol. 2022, 100, 473–484. [Google Scholar] [CrossRef]
- Shanware, N.P.; Bray, K.; Abraham, R.T. The PI3K, metabolic, and autophagy networks: Interactive partners in cellular health and disease. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 89–106. [Google Scholar] [CrossRef]
- Coleman-Derr, D.; Zilberman, D. Deposition of histone variant H2A. Z within gene bodies regulates responsive genes. PLoS Genet. 2012, 8, e1002988. [Google Scholar]
- Zerzaihi, O.; Chriett, S.; Vidal, H.; Pirola, L. Insulin-dependent transcriptional control in L6 rat myotubes is associated with modulation of histone acetylation and accumulation of the histone variant H2A. Z in the proximity of the transcriptional start site. Biochem. Cell Biol. 2014, 92, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Duteil, D.; Tosic, M.; Lausecker, F.; Nenseth, H.Z.; Muller, J.M.; Urban, S.; Willmann, D.; Petroll, K.; Messaddeq, N.; Arrigoni, L.; et al. Lsd1 Ablation Triggers Metabolic Reprogramming of Brown Adipose Tissue. Cell Rep. 2016, 17, 1008–1021. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, C.D.; Li, H.F.; Tian, M.L.; Pan, J.Q.; Shu, G.; Jiang, Q.Y.; Yin, Y.L.; Zhang, L. LSD1 mediates microbial metabolite butyrate-induced thermogenesis in brown and white adipose tissue. Metabolism 2020, 102, 154011. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | Gene Symbol | H3K27ac | DEGs | Function Description |
|---|---|---|---|---|
| ENSBTAG00000019533 | ADTRP | UP | UP | a serine hydrolase enzyme |
| ENSBTAG00000018331 | CLEC3B | UP | UP | a protein-coding gene; encodes tetranectin |
| ENSBTAG00000048075 | IGSF23 | UP | UP | Immunoglobulin superfamily member 23 |
| ENSBTAG00000016229 | KLF9 | UP | UP | a glucocorticoid-inducible factor |
| ENSBTAG00000013499 | LHX9 | UP | UP | LIM/homeobox protein; involved in gonadal development |
| ENSBTAG00000040111 | OTOP3 | UP | UP | Otopetrin 3 |
| ENSBTAG00000046544 | SLC7A10 | UP | UP | neutral amino acid transporter |
| ENSBTAG00000013176 | SMOC2 | UP | UP | secreted protein acidic |
| ENSBTAG00000006419 | TNNT1 | UP | UP | Troponin T, slow skeletal muscle |
| ENSBTAG00000005892 | ZAP70 | UP | UP | Tyrosine protein kinase |
| ENSBTAG00000000590 | POLE | DOWN | DOWN | DNA polymerase |
| ENSBTAG00000001938 | CKS2 | DOWN | DOWN | CDC28 protein kinase regulatory subunit |
| ENSBTAG00000005269 | CCNB2 | DOWN | DOWN | cyclin B2 |
| ENSBTAG00000005825 | NEIL3 | DOWN | DOWN | DNA glycosylase |
| ENSBTAG00000007749 | TONSL | DOWN | DOWN | DNA repair protein |
| ENSBTAG00000007783 | MYBL2 | DOWN | DOWN | a member of the MYB family,regulates cell proliferation |
| ENSBTAG00000008758 | KIF20A | DOWN | DOWN | kinesin family member 20A |
| ENSBTAG00000010766 | OIP5 | DOWN | DOWN | Opa interacting protein 5 |
| ENSBTAG00000016131 | NCAPG2 | DOWN | DOWN | non-SMC condensin II complex subunit G2 |
| ENSBTAG00000016619 | MIS18A | DOWN | DOWN | MIS18 kinetochore protein A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, X.; Li, C.; Liu, S.; Baldwin, R.L., VI; Liu, G.E.; Li, C.-J. Genome-Wide Acetylation Modification of H3K27ac in Bovine Rumen Cell Following Butyrate Exposure. Biomolecules 2023, 13, 1137. https://doi.org/10.3390/biom13071137
Kang X, Li C, Liu S, Baldwin RL VI, Liu GE, Li C-J. Genome-Wide Acetylation Modification of H3K27ac in Bovine Rumen Cell Following Butyrate Exposure. Biomolecules. 2023; 13(7):1137. https://doi.org/10.3390/biom13071137
Chicago/Turabian StyleKang, Xiaolong, Chenglong Li, Shuli Liu, Ransom L. Baldwin, VI, George E. Liu, and Cong-Jun Li. 2023. "Genome-Wide Acetylation Modification of H3K27ac in Bovine Rumen Cell Following Butyrate Exposure" Biomolecules 13, no. 7: 1137. https://doi.org/10.3390/biom13071137
APA StyleKang, X., Li, C., Liu, S., Baldwin, R. L., VI, Liu, G. E., & Li, C.-J. (2023). Genome-Wide Acetylation Modification of H3K27ac in Bovine Rumen Cell Following Butyrate Exposure. Biomolecules, 13(7), 1137. https://doi.org/10.3390/biom13071137






