C-Methylated Spermidine Derivatives: Convenient Syntheses and Antizyme-Related Effects
Abstract
1. Introduction
2. Results
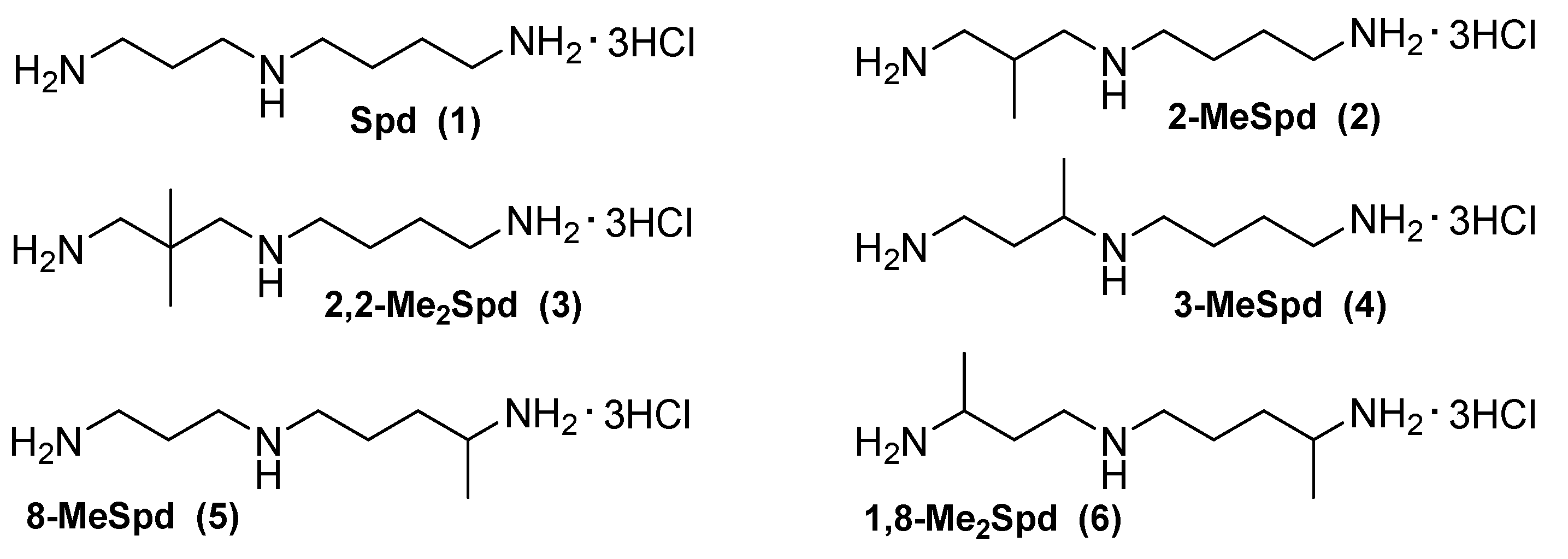
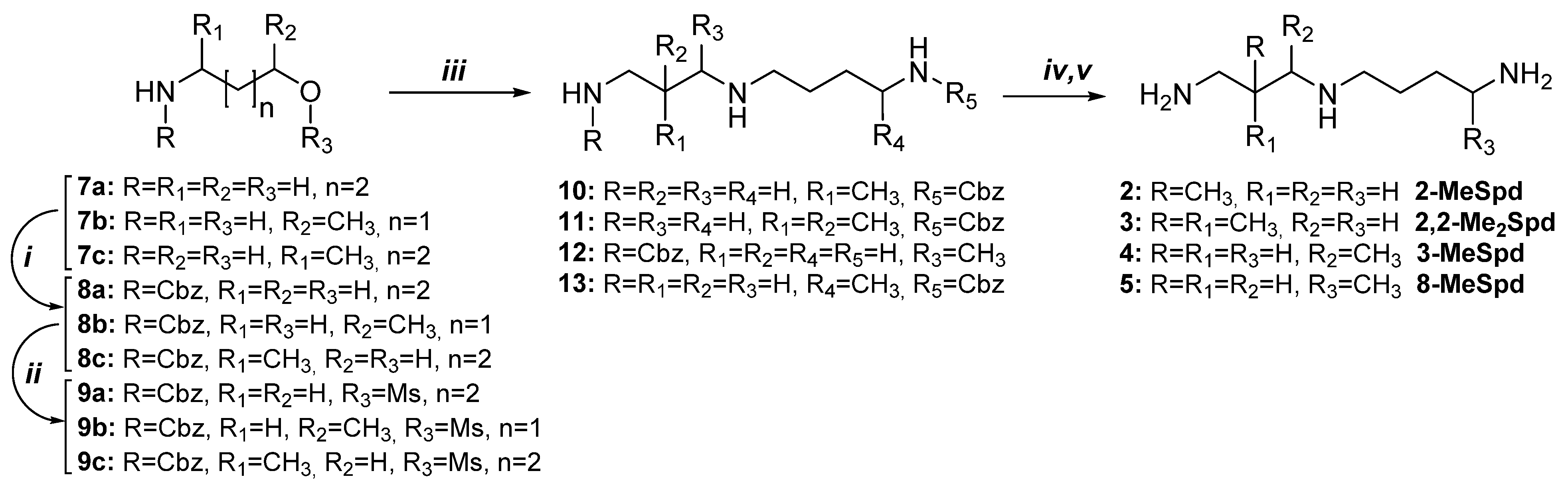
2.1. Syntheses of 2-MeSpd, 2,2-Me2Spd, 3-MeSpd, and 8-MeSpd
2.2. Synthesis of 1,8-Me2Spd
2.3. Spermine Binds to mOAZ1 and Induces Its Dimerization In Vitro
2.4. MeSpds Stabilize the Stem-Loop Region of mOAZ1 mRNA Differently
3. Discussion
3.1. Chemistry
3.2. Biochemistry
4. Materials and Methods
4.1. Materials
4.2. Syntheses of 2-MeSpd, 2,2-Me2Spd, 3-MeSpd and 8-MeSpd
4.2.1. General Method for Synthesis of N-Cbz-Protected C-Methylated Analogs of Spd (10–13)
4.2.2. General Method for the Syntheses of C-Methylated Spermidines Trihydrochloride (2–5)
4.3. Synthesis of 2,9-Diamino-4-azadecane (1,8-Me2Spd)
4.4. Plasmid Construction
4.5. The Expression and Purification of mOAZ1 Protein
4.6. Isothermal Titration Calorimetry (ITC)
4.7. The Stem-Loop Region of mOAZ1 mRNA Melting Point Experiments
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining mysteries of molecular biology: The role of polyamines in the cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E. Functions of polyamines in mammals. J. Biol. Chem. 2016, 291, 14904–14912. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, S.; Cleveland, J.L. Polyamine homeostasis in development and disease. Med. Sci. 2021, 9, 28. [Google Scholar] [CrossRef]
- Madeo, F.; Eisenberg, T.; Pietrocola, F.; Kroemer, G. Spermidine in health and disease. Science 2018, 359, eaan2788. [Google Scholar] [CrossRef] [PubMed]
- Casero, R.A., Jr.; Murray Stewart, T.; Pegg, A.E. Polyamine metabolism and cancer: Treatments, challenges and opportunities. Nat. Rev. Cancer 2018, 18, 681–695. [Google Scholar] [CrossRef]
- Park, M.H.; Wolff, E.C. Hypusine, a polyamine-derived amino acid critical for eukaryotic translation. J. Biol. Chem. 2018, 293, 18710–18718. [Google Scholar] [CrossRef]
- Chattopadhyay, M.K.; Park, M.H.; Tabor, H. Hypusine modification for growth is the major function of spermidine in Saccharomyces cerevisiae polyamine auxotrophs grown in limiting spermidine. Proc. Natl. Acad. Sci. USA 2008, 105, 6554–6559. [Google Scholar] [CrossRef]
- Phillips, M.A. Polyamines in protozoan pathogens. J. Biol. Chem. 2018, 293, 18746–18756. [Google Scholar] [CrossRef]
- Cason, A.L.; Ikeguchi, Y.; Skinner, C.; Wood, T.C.; Holden, K.R.; Lubs, H.A.; Martinez, F.; Simensen, R.J.; Stevenson, R.E.; Pegg, A.E.; et al. X-linked spermine synthase gene (SMS) defect: The first polyamine deficiency syndrome. Eur. J. Hum. Genet. 2003, 11, 937–944. [Google Scholar] [CrossRef]
- Murray-Stewart, T.; Dunworth, M.; Foley, J.R.; Schwartz, C.E.; Casero, R.A., Jr. Polyamine homeostasis in Snyder-Robinson syndrome. Med. Sci. 2018, 6, 112. [Google Scholar] [CrossRef]
- Casero, R.A., Jr.; Woster, P.M. Recent advances in the development of polyamine analogues as antitumor agents. J. Med. Chem. 2009, 52, 4551–4573. [Google Scholar] [CrossRef] [PubMed]
- Verlinden, B.K.; de Beer, M.; Pachaiyappan, B.; Besaans, E.; Andayi, W.A.; Reader, J.; Niemand, J.; van Biljon, R.; Guy, K.; Egan, T.; et al. Interrogating alkyl and arylalkylpolyamino (bis)urea and (bis)thiourea isosteres as potent antimalarial chemotypes against multiple lifecycle forms of Plasmodium falciparum parasites. Bioorg. Med. Chem. 2015, 23, 5131–5143. [Google Scholar] [CrossRef]
- Birkholtz, L.M.; Williams, M.; Niemand, J.; Louw, A.I.; Persson, L.; Heby, O. Polyamine homoeostasis as a drug target in pathogenic protozoa: Peculiarities and possibilities. Biochem. J. 2011, 438, 229–244. [Google Scholar] [CrossRef]
- Wang, B.; Pachaiyappan, B.; Gruber, J.D.; Schmidt, M.G.; Zhang, Y.M.; Woster, P.M. Antibacterial diamines targeting bacterial membranes. J. Med. Chem. 2016, 59, 3140–3151. [Google Scholar]
- Hobley, L.; Kim, S.H.; Maezato, Y.; Wyllie, S.; Fairlamb, A.H.; Stanley-Wall, N.R.; Michael, A.J. Norspermidine is not a self-produced trigger for biofilm disassembly. Cell 2014, 156, 844–854. [Google Scholar] [CrossRef]
- Sjögren, T.; Wassvik, C.M.; Snijder, A.; Aagaard, A.; Kumanomidou, T.; Barlind, L.; Kaminski, T.P.; Kashima, A.; Yokota, T.; Fjellström, O. The structure of murine N1-acetylspermine oxidase reveals molecular details of vertebrate polyamine catabolism. Biochemistry 2017, 56, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Diaz, E.; Adhikary, S.; Tepper, A.W.J.W.; Riley, D.; Ortiz-Meoz, R.; Krosky, D.; Buyck, C.; Lamenca, C.M.; Llaveria, J.; Fang, L.; et al. Structure of human spermine oxidase in complex with a highly selective allosteric inhibitor. Commun. Biol. 2022, 5, 787. [Google Scholar]
- Furbish, A.B.; Alford, A.S.; Burger, P.; Peterson, Y.K.; Murray-Stewart, T.; Casero, R.A., Jr.; Woster, P.M. Identification and characterization of novel small-molecule SMOX inhibitors. Med. Sci. 2022, 10, 47. [Google Scholar]
- Dobrovolskaite, A.; Gardner, R.A.; Delcros, J.G.; Phanstiel, O., 4th. Development of polyamine lassos as polyamine transport inhibitors. ACS Med. Chem. Lett. 2022, 13, 319–326. [Google Scholar] [CrossRef]
- Ayoola, M.B.; Shack, L.A.; Lee, J.H.; Lim, J.; Eoh, H.; Swiatlo, E.; Phanstiel, O., 4th; Nanduri, B. Difluoromethylornithine (DFMO) and AMXT 1501 inhibit capsule biosynthesis in pneumococci. Sci. Rep. 2022, 12, 11804. [Google Scholar] [CrossRef]
- Burns, M.R.; Graminski, G.F.; Weeks, R.S.; Chen, Y.; O’Brien, T.G. Lipophilic lysine-spermine conjugates are potent polyamine transport inhibitors for use in combination with a polyamine biosynthesis inhibitor. J. Med. Chem. 2009, 52, 1983–1993. [Google Scholar] [CrossRef]
- Petros, L.M.; Graminski, G.F.; Robinson, S.; Burns, M.R.; Kisiel, N.; Gesteland, R.F.; Atkins, J.F.; Kramer, D.L.; Howard, M.T.; Weeks, R.S. Polyamine analogs with xylene rings induce antizyme frameshifting, reduce ODC activity, and deplete cellular polyamines. J. Biochem. 2006, 140, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.L.; Leyser, A.; Holtorff, M.S.; Bates, J.S.; Frydman, B.; Valasinas, A.L.; Reddy, V.K.; Marton, L.J. Antizyme induction by polyamine analogues as a factor of cell growth inhibition. Biochem. J. 2002, 366, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Urban-Wójciuk, Z.; Graham, A.; Barker, K.; Kwok, C.; Sbirkov, Y.; Howell, L.; Campbell, J.; Woster, P.M.; Poon, E.; Petrie, K.; et al. The biguanide polyamine analog verlindamycin promotes differentiation in neuroblastoma via induction of antizyme. Cancer Gene Ther. 2022, 29, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Khomutov, M.A.; Mikhura, I.V.; Kochetkov, S.N.; Khomutov, A.R. C-Methylated analogs of spermine and spermidine: Synthesis and biological activity. Russ. J. Bioorg. Chem. 2019, 45, 463–487. [Google Scholar] [CrossRef]
- Keinänen, T.A.; Hyvönen, M.T.; Alhonen, L.; Vepsäläinen, J.; Khomutov, A.R. Selective regulation of polyamine metabolism with methylated polyamine analogues. Amino Acids 2014, 46, 605–620. [Google Scholar] [CrossRef]
- Räsänen, T.L.; Alhonen, L.; Sinervirta, R.; Keinänen, T.; Herzig, K.H.; Suppola, S.; Khomutov, A.R.; Vepsäläinen, J.; Jänne, J. A polyamine analogue prevents acute pancreatitis and restores early liver regeneration in transgenic rats with activated polyamine catabolism. J. Biol. Chem. 2002, 277, 39867–39872. [Google Scholar] [CrossRef]
- Fashe, T.M.; Keinanen, T.A.; Grigorenko, N.A.; Khomutov, A.R.; Janne, J.; Alhonen, L.; Pietila, M. Cutaneous application of α-methylspermidine activates the growth of resting hair follicles in mice. Amino Acids 2010, 38, 583–590. [Google Scholar] [CrossRef]
- Hyvönen, M.T.; Keinänen, T.A.; Khomutov, M.; Simonian, A.; Weisell, J.; Kochetkov, S.N.; Vepsäläinen, J.; Alhonen, L.; Khomutov, A.R. The use of novel C-methylated spermidine derivatives to investigate the regulation of polyamine metabolism. J. Med. Chem. 2011, 54, 4611–4618. [Google Scholar] [CrossRef]
- Khomutov, M.; Hyvönen, M.T.; Simonian, A.; Formanovsky, A.A.; Mikhura, I.V.; Chizhov, A.O.; Kochetkov, S.N.; Alhonen, L.; Vepsäläinen, J.; Keinänen, T.A.; et al. Unforeseen possibilities to investigate the regulation of polyamine metabolism revealed by novel C-methylated spermine derivatives. J. Med. Chem. 2019, 62, 11335–11347. [Google Scholar] [CrossRef]
- Hyvönen, M.T.; Smirnova, O.A.; Mitkevich, V.A.; Tunitskaya, V.L.; Khomutov, M.; Karpov, D.S.; Korolev, S.P.; Häkkinen, M.R.; Pietilä, M.; Gottikh, M.B.; et al. Role of polyamine-induced dimerization of antizyme in its cellular functions. Int. J. Mol. Sci. 2022, 23, 4614. [Google Scholar] [CrossRef]
- Stewart, T.M.; Khomutov, M.; Foley, J.R.; Guo, X.; Holbert, C.E.; Dunston, T.T.; Schwartz, C.E.; Gabrielson, K.; Khomutov, A.; Casero, R.A. (R,R)-1,12-Dimethylspermine can mitigate abnormal spermidine accumulation in Snyder-Robinson syndrome. J. Biol. Chem. 2020, 295, 3247–3256. [Google Scholar] [CrossRef] [PubMed]
- Hyvönen, M.T.; Khomutov, M.; Petit, M.; Weisell, J.; Kochetkov, S.N.; Alhonen, L.; Vepsäläinen, J.; Khomutov, A.R.; Keinänen, T.A. Enantiomers of 3-methylspermidine selectively modulate deoxyhypusine synthesis and reveal important determinants for spermidine transport. ACS Chem. Biol. 2015, 10, 1417–1424. [Google Scholar] [CrossRef] [PubMed]
- Järvinen, A.J.; Cerrada-Gimenez, M.; Grigorenko, N.A.; Khomutov, A.R.; Vepsäläinen, J.J.; Sinervirta, R.M.; Keinänen, T.A.; Alhonen, L.I.; Jänne, J.E. Alpha-methyl polyamines: Efficient synthesis and tolerance studies in vivo and in vitro. First evidence for dormant stereospecificity of polyamine oxidase. J. Med. Chem. 2006, 49, 399–406. [Google Scholar] [CrossRef]
- Atkins, J.F.; Loughran, G.; Bhatt, P.R.; Firth, A.E.; Baranov, P.V. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016, 44, 7007–7078. [Google Scholar] [CrossRef]
- Kuksa, V.; Buchan, R.; Lin, P.K.T. Synthesis of polyamines, their derivatives, analogues and conjugates. Synthesis 2000, 9, 1189–1207. [Google Scholar] [CrossRef]
- Nichugovskiy, A.; Tron, G.C.; Maslov, M. Recent advances in the synthesis of polyamine derivatives and their applications. Molecules 2021, 26, 6579. [Google Scholar] [CrossRef]
- Hahn, F.; Schepers, U. Solid phase chemistry for the directed synthesis of biologically active polyamine analogs, derivatives, and conjugates. Top. Curr. Chem. 2007, 278, 135–208. [Google Scholar]
- Nagarajan, S.; Ganem, B. Chemistry of naturally occurring polyamines. Nonmetabolizable derivatives of spermine and spermidine. J. Org. Chem. 1986, 51, 4856–4861. [Google Scholar] [CrossRef]
- Lakanen, J.R.; Coward, J.K.; Pegg, A.E. α-Methyl Polyamines: Metabolically stable spermidine and spermine mimics capable of supporting growth in cells depleted of polyamines. J. Med. Chem. 1992, 35, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Khomutov, A.R.; Weisell, J.; Khomutov, M.A.; Grigorenko, N.A.; Simonian, A.R.; Hakkinen, M.R.; Keinanen, T.A.; Hyvonen, M.T.; Alhonen, L.; Kochetkov, S.N.; et al. Methylated Polyamines as Research Tools. In Polyamines; Pegg, A.E., Casero, R.A., Eds.; Springer Science + Business Media. LLC.: Berlin/Heidelberg, Germany, 2011; Volume 720, pp. 449–461. [Google Scholar]
- Fukuyama, T.; Chung-Kuang, J.; Cheung, M. 2- and 4-Nitrobenzenesulfonamides: Exceptionally versatile means for preparation of secondary amines and protection of amines. Tetrahedron Lett. 1995, 36, 6373–6374. [Google Scholar] [CrossRef]
- Murakami, Y.; Matsufuji, S.; Kameji, T.; Hayashi, S.; Igarashi, K.; Tamura, T.; Tanaka, K.; Ichihara, A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 1992, 360, 597–599. [Google Scholar] [CrossRef]
- Suzuki, T.; He, Y.; Kashiwagi, K.; Murakami, Y.; Hayashi, S.; Igarashi, K. Antizyme protects against abnormal accumulation and toxicity of polyamines in ornithine decarboxylase-overproducing cells. Proc. Natl. Acad. Sci. USA 1994, 91, 8930–8934. [Google Scholar] [CrossRef] [PubMed]
- Matsufuji, S.; Kanamoto, R.; Murakami, Y.; Hayashi, S. Monoclonal antibody studies on the properties and regulation of murine ornithine decarboxylase antizymes. J. Biochem. 1990, 107, 87–91. [Google Scholar] [CrossRef]
- Matsufuji, S.; Matsufuji, T.; Miyazaki, Y.; Murakami, Y.; Atkins, J.F.; Gesteland, R.F.; Hayashi, S.-I. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell 1995, 80, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Rom, E.; Kahana, C. Polyamines regulate the expression of ornithine decarboxylase antizyme in vitro by inducing ribosomal frameshifting. Proc. Natl. Acad. Sci. USA 1994, 91, 3959–3963. [Google Scholar] [CrossRef]
- Ivanov, I.P.; Atkins, J.F. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: Close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007, 35, 1842–1858. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.P.; Matsufuji, S.; Murakami, Y.; Gesteland, R.F.; Atkins, J.F. Conservation of polyamine regulation by translational frameshifting from yeast to mammals. EMBO J. 2000, 19, 1907–1917. [Google Scholar] [CrossRef]
- Petros, L.M.; Howard, M.T.; Gesteland, R.F.; Atkins, J.F. Polyamine sensing during antizyme mRNA programmed frameshifting. Biochem. Biophys. Res. Commun. 2005, 338, 1478–1489. [Google Scholar] [CrossRef]
- Yordanova, M.M.; Wu, C.; Andreev, D.E.; Sachs, M.S.; Atkins, J.F. A nascent peptide signal responsive to endogenous levels of polyamines acts to stimulate regulatory frameshifting on antizyme mRNA. J. Biol. Chem. 2015, 290, 17863–17878. [Google Scholar] [CrossRef]
- Coffino, P. Regulation of cellular polyamines by antizyme. Nat. Rev. Mol. Cell Biol. 2001, 2, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Kurian, L.; Palanimurugan, R.; Godderz, D.; Dohmen, R.J. Polyamine sensing by nascent ornithine decarboxylase antizyme stimulates decoding of its mRNA. Nature 2011, 477, 490–494. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, D.W.; Carroll, D.; Martinez, N.; Hackert, M.L. Solution structure of a conserved domain of antizyme: A protein regulator of polyamines. Biochemistry 2005, 44, 11777–11785. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Chen, S.F.; Hsieh, J.Y.; Chou, F.; Wang, Y.H.; Lin, W.T.; Lee, P.Y.; Yu, Y.J.; Lin, L.Y.; Lin, T.S.; et al. Structural basis of antizyme-mediated regulation of polyamine homeostasis. Proc. Natl. Acad. Sci. USA 2015, 112, 11229–11234. [Google Scholar] [CrossRef]
- Hsieh, J.-Y.; Liu, Y.-C.; Cheng, I.-T.; Lee, C.-J.; Wang, Y.-H.; Fang, Y.-S.; Liu, Y.-L.; Liu, G.-Y.; Hung, H.-C. Critical factors in human antizymes that determine the differential binding, inhibition, and degradation of human ornithine decarboxylase. Biomolecules 2019, 9, 864. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-C.; Lee, C.-Y.; Lin, C.-L.; Chen, H.-Y.; Liu, G.-Y.; Hung, H.-C. Multifaceted interactions and regulation between antizyme and its interacting proteins cyclin D1, ornithine decarboxylase and antizyme inhibitor. Oncotarget 2015, 6, 23917–23929. [Google Scholar] [CrossRef]
- Kankare, K.; Uusi-Oukari, M.; Janne, O.A. Structure, organization and expression of the mouse ornithine decarboxylase antizyme gene. Biochem. J. 1997, 324, 807–813. [Google Scholar] [CrossRef]
- Tsedilin, A.M.; Fakhrutdinov, A.N.; Eremin, D.B.; Zalesskiy, S.S.; Chizhov, A.O.; Kolotyrkina, N.G.; Ananikov, V.P. How sensitive and accurate are routine NMR and MS measurements? Mend. Commun. 2015, 25, 454–456. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Korovina, A.N.; Tunitskaya, V.L.; Kostyuk, D.A.; Rechinsky, V.O.; Kukhanova, M.K.; Kochetkov, S.N. Development of the system ensuring a high-level expression of hepatitis C virus nonstructural NS5B and NS5A proteins. Protein Expr. Purif. 2006, 48, 14–23. [Google Scholar] [CrossRef]
- Petrushanko, I.Y.; Mitkevich, V.A.; Anashkina, A.A.; Klimanova, E.A.; Dergousova, E.A.; Lopina, O.D.; Makarov, A.A. Critical role of gamma-phosphate in structural transition of Na,K-ATPase upon ATP binding. Sci. Rep. 2014, 4, 5165. [Google Scholar] [CrossRef]
- Hou, M.H.; Lin, S.B.; Yuann, J.M.; Lin, W.C.; Wang, A.H.; Kan Ls, L. Effects of polyamines on the thermal stability and formation kinetics of DNA duplexes with abnormal structure. Nucleic Acids Res. 2001, 29, 5121–5128. [Google Scholar] [CrossRef] [PubMed]

| Stoichiometry mOAZ1/Spm | Ka b, M−1 | Kd c, µM | ΔH d, kcal/mole | TΔS e, kcal/mole |
|---|---|---|---|---|
| mOAZ1‒6XHis-tag * | ||||
| 2:1 | 9.2 × 105 | 1.1 | −3.7 | 4.5 |
| 6XHis-tag‒mOAZ1 | ||||
| No binding f | ||||
| mOAZ1 | ||||
| 2:1 | 2.7 × 106 | 0.37 | −3.6 | 5.5 |
| Polyamine Concentration, μM | Tm, °C | |||||
|---|---|---|---|---|---|---|
| Spd a | 1-MeSpd a | 2-MeSpd a | 3-MeSpd a | 8-MeSpd | 2,2-Me2Spd | |
| 0 | 65.5 ± 0.6 | |||||
| 20 | 73.7 ± 0.6 | 73.0 ± 0.4 | 70.5 ± 0.5 *** | 71.9 ± 0.5 * | 72.7 ± 0.4 | 71.6 ± 0.9 |
| 50 | 75.3 ± 0.4 | 74.0 ± 0.3 | 73.4 ± 0.4 * | 75.2 ± 0.4 | 76.2 ± 0.7 | 74.4 ± 0.6 |
| 200 | 79.7 ± 1.1 | 80.2 ± 0.6 | 77.5 ± 0.5 ** | 79.4 ± 0.5 | 81.3 ± 1.4 | 76.7 ± 0.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khomutov, M.A.; Salikhov, A.I.; Mitkevich, V.A.; Tunitskaya, V.L.; Smirnova, O.A.; Korolev, S.P.; Chizhov, A.O.; Gottikh, M.B.; Kochetkov, S.N.; Khomutov, A.R. C-Methylated Spermidine Derivatives: Convenient Syntheses and Antizyme-Related Effects. Biomolecules 2023, 13, 916. https://doi.org/10.3390/biom13060916
Khomutov MA, Salikhov AI, Mitkevich VA, Tunitskaya VL, Smirnova OA, Korolev SP, Chizhov AO, Gottikh MB, Kochetkov SN, Khomutov AR. C-Methylated Spermidine Derivatives: Convenient Syntheses and Antizyme-Related Effects. Biomolecules. 2023; 13(6):916. https://doi.org/10.3390/biom13060916
Chicago/Turabian StyleKhomutov, Maxim A., Arthur I. Salikhov, Vladimir A. Mitkevich, Vera L. Tunitskaya, Olga A. Smirnova, Sergey P. Korolev, Alexander O. Chizhov, Marina B. Gottikh, Sergey N. Kochetkov, and Alex R. Khomutov. 2023. "C-Methylated Spermidine Derivatives: Convenient Syntheses and Antizyme-Related Effects" Biomolecules 13, no. 6: 916. https://doi.org/10.3390/biom13060916
APA StyleKhomutov, M. A., Salikhov, A. I., Mitkevich, V. A., Tunitskaya, V. L., Smirnova, O. A., Korolev, S. P., Chizhov, A. O., Gottikh, M. B., Kochetkov, S. N., & Khomutov, A. R. (2023). C-Methylated Spermidine Derivatives: Convenient Syntheses and Antizyme-Related Effects. Biomolecules, 13(6), 916. https://doi.org/10.3390/biom13060916








