mef2ca and mef2cb Double Mutant Zebrafish Show Altered Craniofacial Phenotype and Motor Behaviour
Abstract
1. Introduction
2. Material and Methods
2.1. Zebrafish Lines and Maintenance
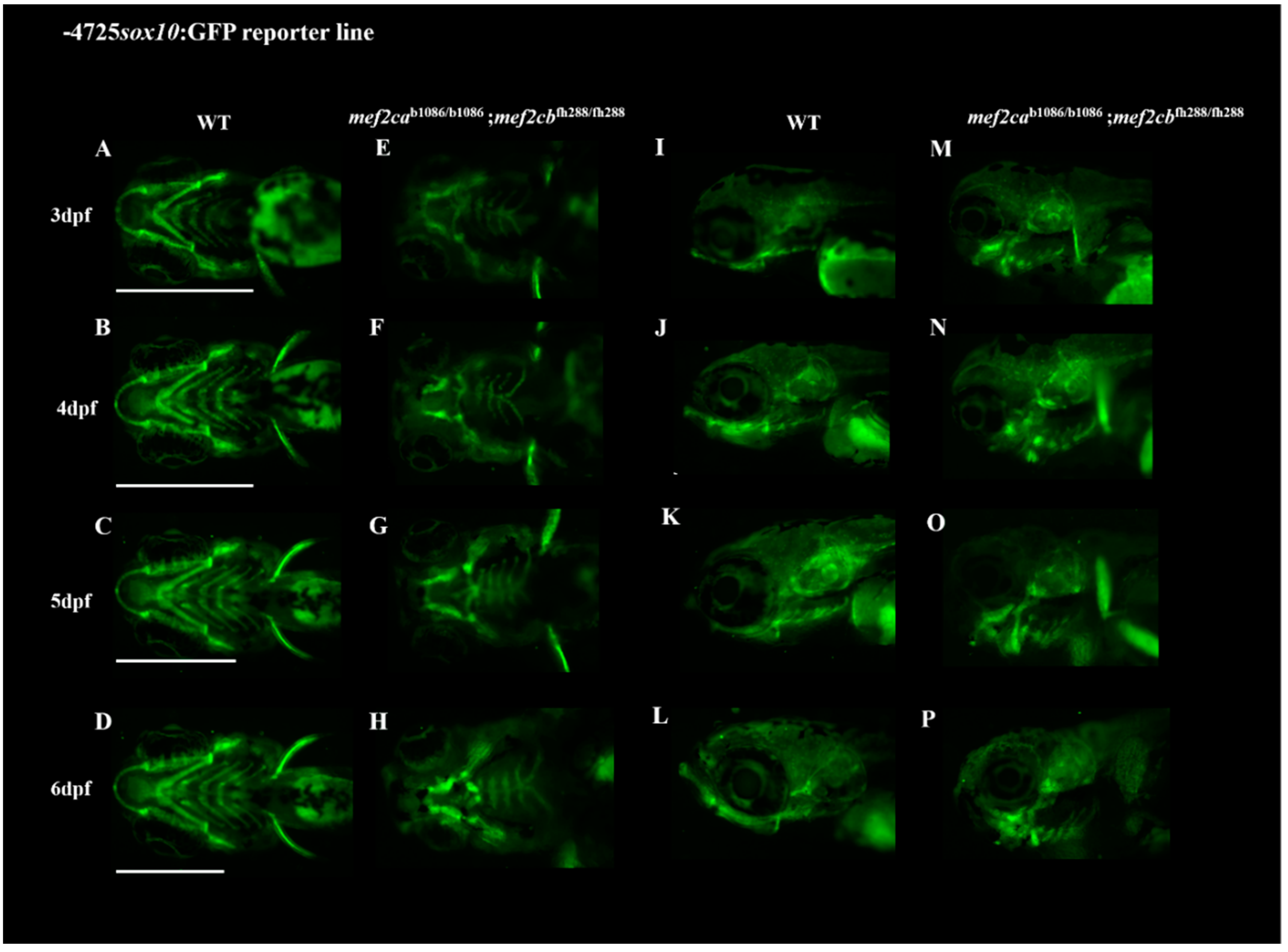
2.2. Tissue Labelling and Imaging
2.3. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qPCR)
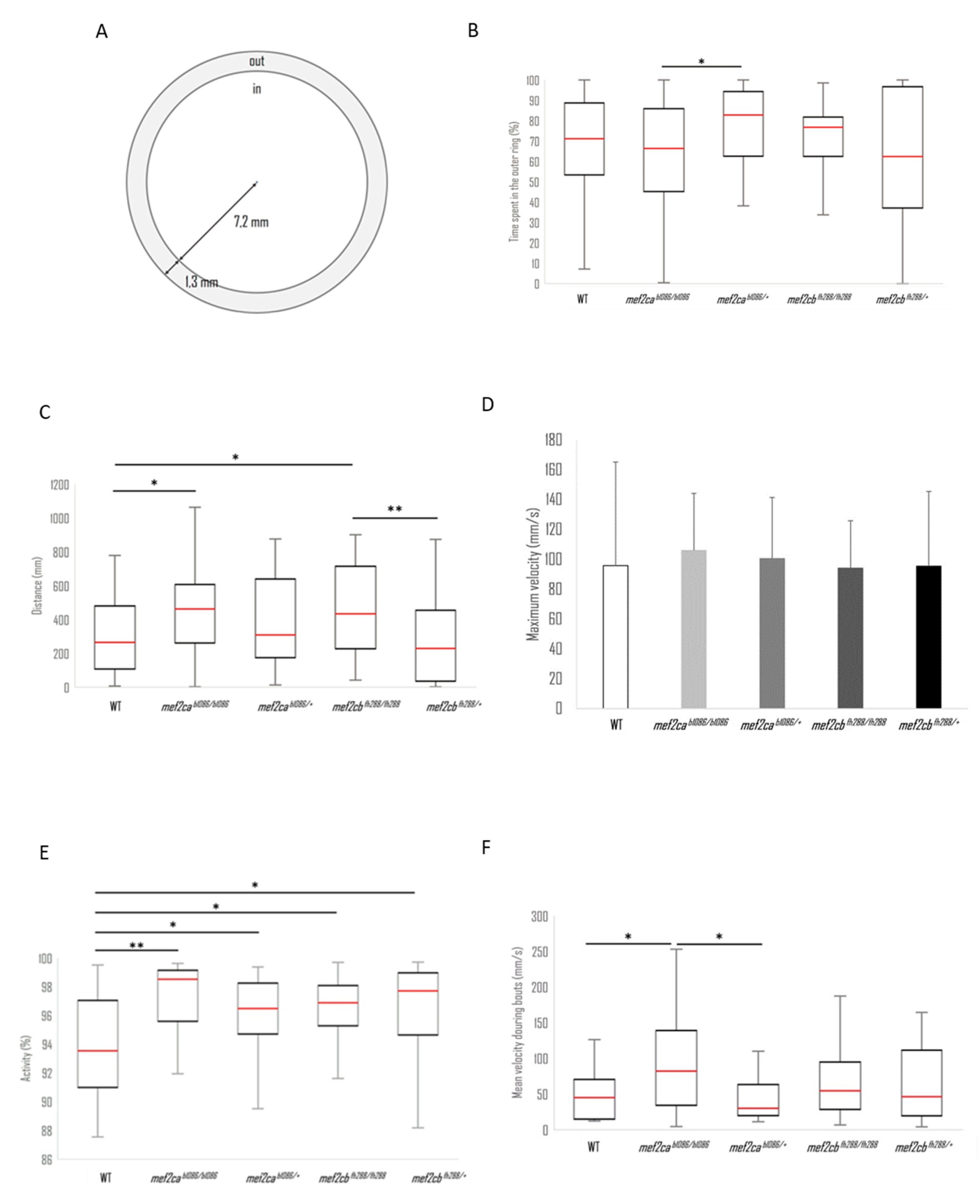
2.4. Analysis of Motor Behaviour
2.4.1. Early Spontaneous Motor Behaviour
2.4.2. Spontaneous Swimming Activity
2.5. Statistical Analysis
3. Results
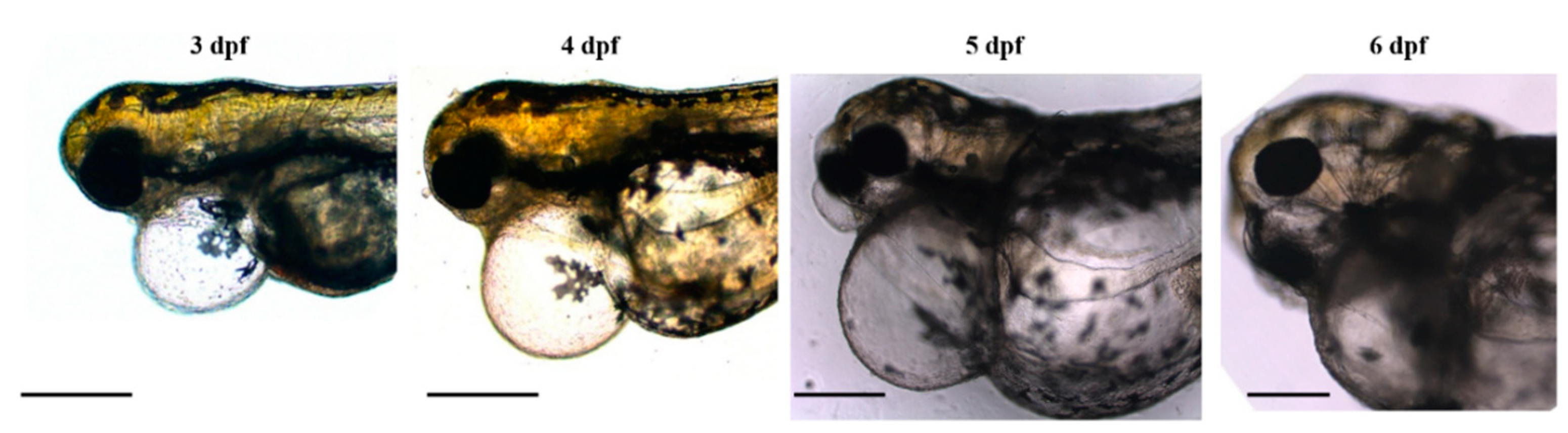
3.1. Disruption of Cardiac Oedema Causes the Death of Homozygous Double-Mutant Larvae
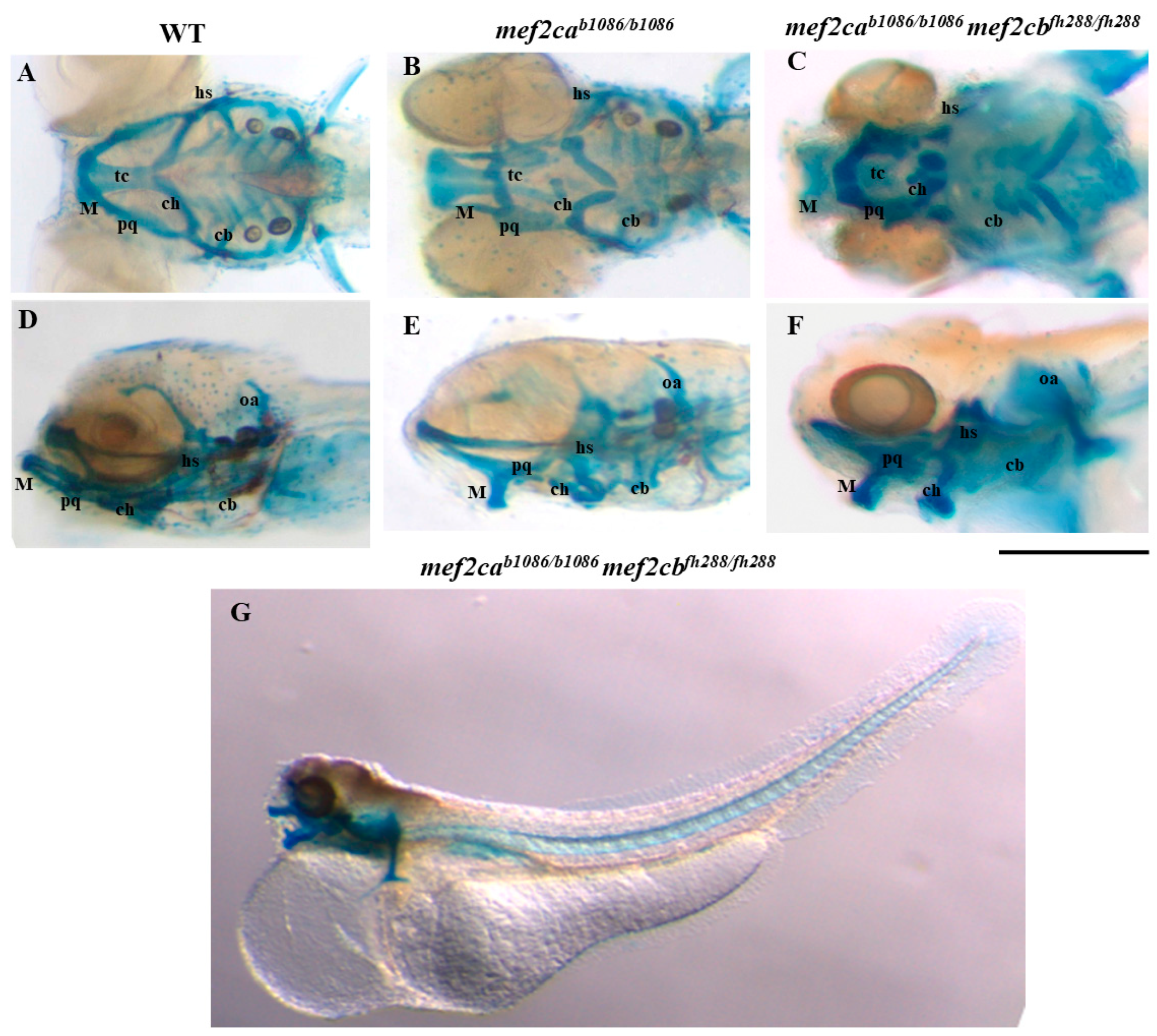
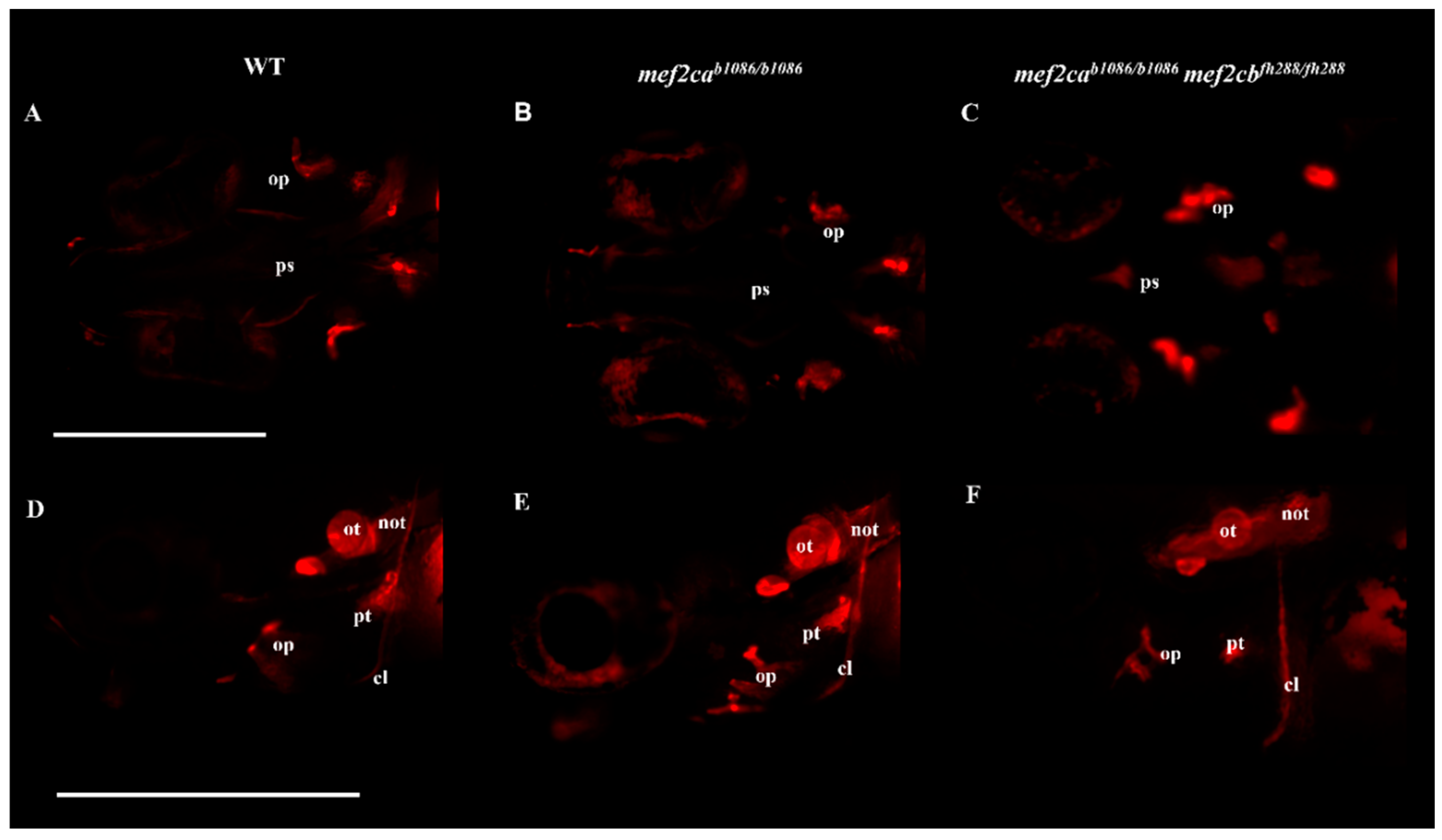
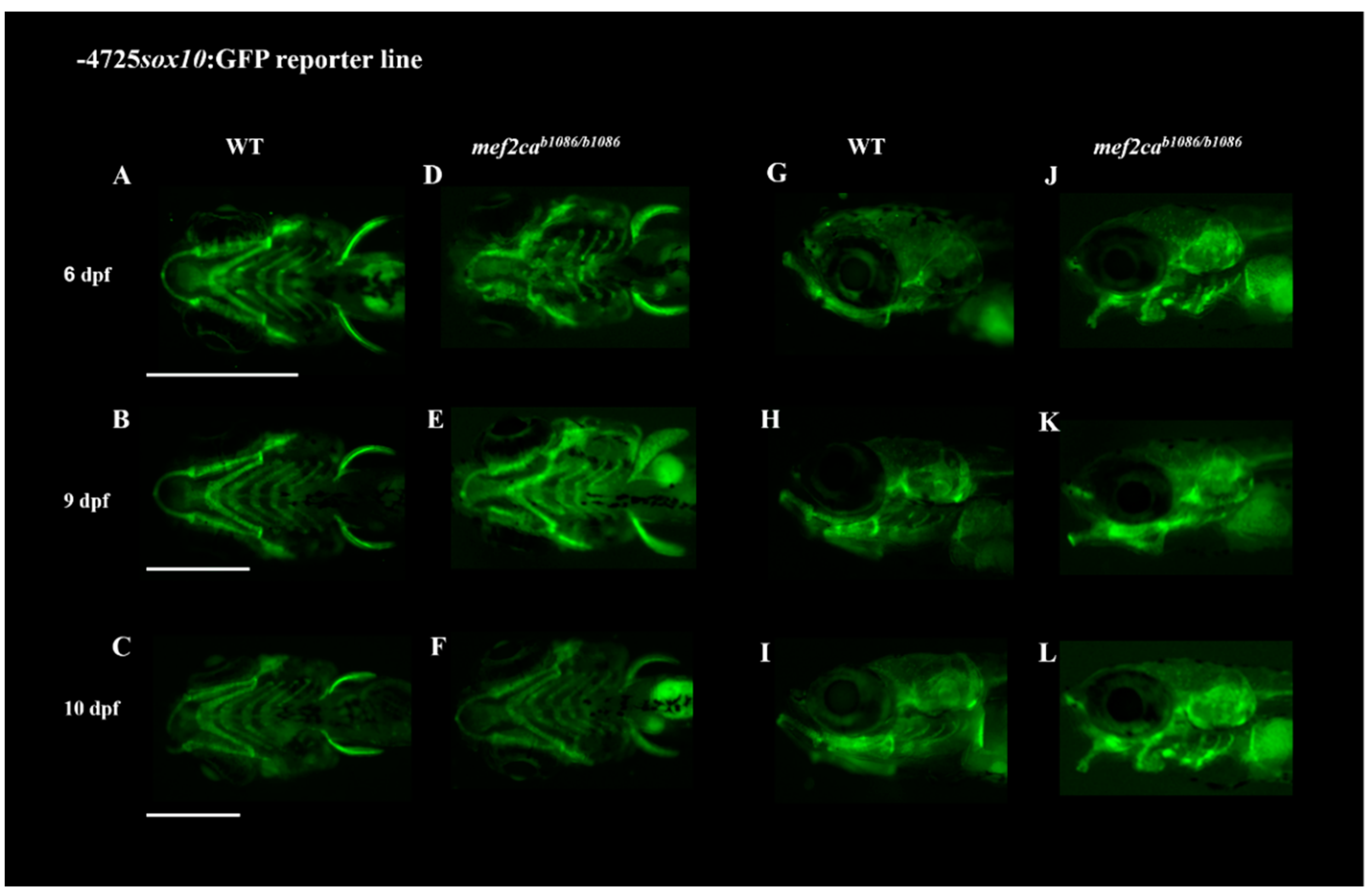
3.2. Homozygous Double Mutants Show a Severe Craniofacial Phenotype with Alterations in Cartilaginous and Bone Structures
3.3. Zebrafish mef2c Mutants Showed Alteration of mecp2 and cdkl5 Gene Expression
3.4. Early Motor Behaviour
3.5. Spontaneous Swimming Activity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Le Meur, N.; Holder-Espinasse, M.; Jaillard, S.; Goldenberg, A.; Joriot, S.; Amati-Bonneau, P.; Guichet, A.; Barth, M.; Charollais, A.; Journel, H.; et al. MEF2C haploinsufficiency caused by either microdeletion of the 5q14.3 region or mutation is responsible for severe mental retardation with stereotypic movements, epilepsy and/or cerebral malformations. J. Med. Genet. 2010, 47, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Verzi, M.P.; Agarwal, P.; Brown, C.; Mcculley, D.J.; John, J.; Black, B.L. The transcription factor MEF2C is required for craniofacial development. Dev. Cell 2007, 12, 645–652. [Google Scholar] [CrossRef]
- Hu, J.; Verzi, M.P.; Robinson, A.S.; Tang, P.L.-F.; Hua, L.L.; Xu, S.-M.; Kwok, P.-Y.; Black, B.L. Endothelin signaling activates Mef2c expression in the neural crest through a MEF2C-dependent positive-feedback transcriptional pathway. Development 2015, 142, 2775–2780. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.A.; Kim, Y.; Czubryt, M.P.; Phan, D.; McAnally, J.; Qi, X.; Shelton, J.M.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev. Cell 2007, 12, 377–389. [Google Scholar] [CrossRef]
- Choi, T.Y.; Choi, T.I.; Lee, Y.R.; Choe, S.K.; Kim, C.H. Zebrafish as an animal model for biomedical research. Exp. Mol. Med. 2021, 53, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Adrião, A.; Conceição, N.; Cancela, M.L. MEF2C orthologues from zebrafish: Evolution, expression and promoter regulation. Arch. Biochem. Biophys. 2016, 591, 43–56. [Google Scholar] [CrossRef]
- Miller, C.T.; Swartz, M.E.; Khuu, P.A.; Walker, M.B.; Eberhart, J.K.; Kimmel, C.B. mef2ca is required in cranial neural crest to effect Endothelin1 signaling in zebrafish. Dev. Biol. 2007, 308, 144–157. [Google Scholar] [CrossRef]
- Hinits, Y.; Pan, L.; Walker, C.; Dowd, J.; Moens, C.B.; Hughes, S.M. Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Dev. Biol. 2012, 369, 199–210. [Google Scholar] [CrossRef]
- DeLaurier, A.; Huycke, T.R.; Nichols, J.T.; Swartz, M.E.; Larsen, A.; Walker, C.; Dowd, J.; Pan, L.; Moens, C.B.; Kimmel, C.B. Role of mef2ca in developmental buffering of the zebrafish larval hyoid dermal skeleton. Dev. Biol. 2014, 385, 189–199. [Google Scholar] [CrossRef]
- Chadwick, L.H.; Wade, P.A. MeCP2 in Rett syndrome: Transcriptional repressor or chromatin architectural protein? Curr. Opin. Genet. Dev. 2007, 17, 121–125. [Google Scholar] [CrossRef]
- Amendola, E.; Zhan, Y.; Mattucci, C.; Castroflorio, E.; Calcagno, E.; Fuchs, C.; Lonetti, G.; Silingardi, D.; Vyssotski, A.L.; Farley, D.; et al. Mapping pathological phenotypes in a mouse model of CDKL5 disorder. PLoS ONE 2014, 9, e91613. [Google Scholar] [CrossRef] [PubMed]
- Zweier, M.; Gregor, A.; Zweier, C.; Engels, H.; Sticht, H.; Wohlleber, E.; Bijlsma, E.K.; Holder, S.E.; Zenker, M.; Rossier, E.; et al. Mutations in MEF2C from the 5q14.3q15 microdeletion syndrome region are a frequent cause of severe mental retardation and diminish MECP2 and CDKL5 expression. Hum. Mutat. 2010, 31, 722–733. [Google Scholar] [CrossRef] [PubMed]
- Coverdale, L.E.; Martyniuk, C.J.; Trudeau, V.L.; Martin, C.C. Differential expression of the methyl-cytosine binding protein 2 gene in embryonic and adult brain of zebrafish. Dev. Brain Res. 2004, 153, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Pietri, T.; Roman, A.-C.; Guyon, N.; Romano, S.A.; Washbourne, P.; Moens, C.B.; de Polavieja, G.G.; Sumbre, G. The first mecp2-null zebrafish model shows altered motor behaviors. Front. Neural Circuits 2013, 7, 118. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, M.; Cunha, N.; Conceição, N.; Cancela, M.L. Expression pattern of cdkl5 during zebrafish early development: Implications for use as model for atypical Rett syndrome. Mol. Biol. Rep. 2018, 45, 445–451. [Google Scholar] [CrossRef]
- Varela, T.; Varela, D.; Martins, G.; Conceição, N.; Cancela, M.L. Cdkl5 mutant zebrafish shows skeletal and neuronal alterations mimicking human CDKL5 deficiency disorder. Sci. Rep. 2022, 12, 9325. [Google Scholar] [CrossRef]
- Dutton, J.R.; Antonellis, A.; Carney, T.J.; Rodrigues, F.S.L.M.; Pavan, W.J.; Ward, A.; Kelsh, R.N. An evolutionarily conserved intronic region controls the spatiotemporal expression of the transcription factor Sox10. BMC Dev. Biol. 2008, 8, 105. [Google Scholar] [CrossRef]
- Walker, M.B.; Kimmel, C.B. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 2007, 82, 23–28. [Google Scholar] [CrossRef]
- Cubbage, C.C.; Mabee, P.M. Development of the cranium and paired fins in the zebrafish Danio rerio (Ostariophysi, Cyprinidae). J. Morphol. 1996, 229, 121–160. [Google Scholar] [CrossRef]
- DeLaurier, A.; Eames, B.F.; Blanco-sánchez, B.; Peng, G.; He, X.; Swartz, E.; Ullmann, B.; Westerfield, M.; Kimmel, C.B. Zebrafish sp7:EGFP: A transgenic for studying otic vesicle formation, skeletogenesis, and bone regeneration. Genesis 2011, 48, 505–511. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-Step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. Quantification strategies in real-time PCR. In The Real-Time PCR Encyclopedia A–Z of Quantitative PCR; Bustin, S.A., Ed.; International University Line: La Jolla, CA, USA, 2004; pp. 87–112. [Google Scholar]
- McDonald, J.H. Handbook of Biological Statistics, 3rd ed.; Sparky House Publishing: Baltimore, MD, USA, 2014. [Google Scholar]
- Chen, Q.; Zhu, Y.-C.; Yu, J.; Miao, S.; Zheng, J.; Xu, L.; Zhou, Y.; Li, D.; Zhang, C.; Tao, J.; et al. CDKL5, a protein associated with Rett syndrome, regulates neuronal morphogenesis via Rac1 signaling. J. Neurosci. 2010, 30, 12777–12786. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Schwarz, J.; Bucana, C.; Olson, E.N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 1997, 276, 1404–1407. [Google Scholar] [CrossRef] [PubMed]
- Thirumalai, V.; Cline, H.T. Endogenous dopamine suppresses initiation of swimming in prefeeding zebrafish larvae. J. Neurophysiol. 2008, 100, 1635–1648. [Google Scholar] [CrossRef] [PubMed]
- Simon, P.; Dupuis, R.; Costentin, J. Thigmotaxis as an index of anxiety in mice. Influence of dopaminergic transmissions. Behav. Brain Res. 1994, 61, 59–64. [Google Scholar] [CrossRef]
- Schnorr, S.J.; Steenbergen, P.J.; Richardson, M.K.; Champagne, D.L. Measuring thigmotaxis in larval zebrafish. Behav. Brain Res. 2012, 228, 367–374. [Google Scholar] [CrossRef]
- Paylor, R.; Nguyen, M.; Crawley, J.N.; Patrick, J.; Beaudet, A.; Orr-Urtreger, A. α7 nicotinic receptor subunits are not necessary for hippocampal-dependent learning or sensorimotor gating: A behavioral characterization of acra7-deficient mice. Learn. Mem. 1998, 5, 302–316. [Google Scholar] [CrossRef]




| Gene | Accession ID | Sequence 5′-3′ |
|---|---|---|
| methyl CpG binding protein 2 (mecp2) | AY298900.2 | Fw: AGAGACCTTTGAGAAACGACTG Rev: TCTTCTTGTGACTCTTCGGTG |
| Cyclin-dependent kinase-like 5 (cdkl5) | NM_001145768.1 | Fw: AGATGAACCGAAGCCTACTGA Rev: GGTGTATCCAAGACCGTAA |
| glyceraldehyde 3 phosphate dehydrogenase (gapdh) | NM_001115114.1 | Fw: GCCCACCAGAACATCATCCCTGC Rev: TGACAGACTCCTTGATGTTGGCGTAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adrião, A.; Mariano, S.; Mariano, J.; Gavaia, P.J.; Cancela, M.L.; Vitorino, M.; Conceição, N. mef2ca and mef2cb Double Mutant Zebrafish Show Altered Craniofacial Phenotype and Motor Behaviour. Biomolecules 2023, 13, 805. https://doi.org/10.3390/biom13050805
Adrião A, Mariano S, Mariano J, Gavaia PJ, Cancela ML, Vitorino M, Conceição N. mef2ca and mef2cb Double Mutant Zebrafish Show Altered Craniofacial Phenotype and Motor Behaviour. Biomolecules. 2023; 13(5):805. https://doi.org/10.3390/biom13050805
Chicago/Turabian StyleAdrião, Andreia, Sara Mariano, José Mariano, Paulo J. Gavaia, M. Leonor Cancela, Marta Vitorino, and Natércia Conceição. 2023. "mef2ca and mef2cb Double Mutant Zebrafish Show Altered Craniofacial Phenotype and Motor Behaviour" Biomolecules 13, no. 5: 805. https://doi.org/10.3390/biom13050805
APA StyleAdrião, A., Mariano, S., Mariano, J., Gavaia, P. J., Cancela, M. L., Vitorino, M., & Conceição, N. (2023). mef2ca and mef2cb Double Mutant Zebrafish Show Altered Craniofacial Phenotype and Motor Behaviour. Biomolecules, 13(5), 805. https://doi.org/10.3390/biom13050805







