Abstract
Lysosomes are membrane-bound organelles with an acidic lumen and are traditionally characterized as a recycling center in cells. Lysosomal ion channels are integral membrane proteins that form pores in lysosomal membranes and allow the influx and efflux of essential ions. Transmembrane protein 175 (TMEM175) is a unique lysosomal potassium channel that shares little sequence similarity with other potassium channels. It is found in bacteria, archaea, and animals. The prokaryotic TMEM175 consists of one six-transmembrane domain that adopts a tetrameric architecture, while the mammalian TMEM175 is comprised of two six-transmembrane domains that function as a dimer in lysosomal membranes. Previous studies have demonstrated that the lysosomal K+ conductance mediated by TMEM175 is critical for setting membrane potential, maintaining pH stability, and regulating lysosome–autophagosome fusion. AKT and B-cell lymphoma 2 regulate TMEM175’s channel activity through direct binding. Two recent studies reported that the human TMEM175 is also a proton-selective channel under normal lysosomal pH (4.5–5.5) as the K+ permeation dramatically decreased at low pH while the H+ current through TMEM175 greatly increased. Genome-wide association studies and functional studies in mouse models have established that TMEM175 is implicated in the pathogenesis of Parkinson’s disease, which sparks more research interests in this lysosomal channel.
1. Introduction
Lysosomes (and equivalent vacuoles in plant and yeast cells) are membrane-enclosed cytoplasmic organelles that contribute to the breakdown and recycling of biological macromolecules in nearly all eukaryotic cells. The cargo vesicles originating from endocytic, phagocytic, and autophagic pathways are sent to lysosomes for degradation, and the resulting catabolic products are exported to the cytoplasm for subsequent reutilization [1,2,3]. Growing evidence shows that lysosomes are more than a terminal recycling center in cells. They serve as a signaling hub that integrates signals from multiple metabolic pathways and cellular structures, which further associates this acidic organelle with a plethora of cellular functions [4,5]. More than 60 hydrolases are found in their acidic lumen, which is a hallmark of lysosomes. The luminal low pH environment is limited by a single-lipid bilayer membrane embedded with a wide variety of important proteins including transporters, pumps, and ion channels, which control the influx/efflux of metabolites and ions [6]. For example, the vacuolar H+-ATPase (V-ATPase) is an ATP-dependent proton pump that transports protons across the lysosomal membrane to establish and maintain the acidic pH of the lysosomal lumen [7].
Ion channels are integral pore-forming membrane proteins that facilitate the passive movement of ions across the membrane. They play a critical role in nearly all fundamental physiological processes, including muscle contraction, synaptic transmission, and action potential propagation. Therefore, it is not surprising that mutations in lysosomal channels give rise to a wide range of diseases, such as renal disorders, heart diseases, and neurodegenerative diseases [8,9]. Several types of ion channels have been identified on lysosomal membranes [10,11]. The still-expanding list includes the non-selective cation channels, transient receptor potential mucolipins (TRPMLs) [12,13,14], volume-regulated Cl−/anion channel, leucine-rich repeat-containing family 8 (LRRC8) [15], endolysosome-localized chloride channel, H+/Cl− exchanger 7 (CLC-7) [16], Na+/Ca2+-permeable P2 × 4 purinoceptor [17], Na+-selective two-pore channels (TPCs) [18,19,20], K+-selective channels such as the large conductance Ca2+-activated K+ channel (big potassium, BK) [21,22], and transmembrane protein 175 (TMEM175) [23].
Potassium ions are the most abundant cations inside the cytosol and potassium channels are widely distributed in numerous cell types. Based on their gating mechanisms and structural characteristics, the K+ channels are classified into several categories including Ca2+-activated K+ channels (KCa), voltage-gated K+ channels (Kv), inwardly rectifying K+ channels (Kir), tandem pore domain K+ channels (K2P), and Na+-activated K+ channels [24,25,26]. TWIK2, BK, and TMEM175 are the three lysosomal K+ channels that have been discovered thus far. The K2P family member TWIK2 (tandem of pore domains in a weakly inward rectifying K+ channel 2) generates weak background K+ currents in lysosomes [27]. BK channels exhibit a high K+ selectivity and a large single-channel conductance, which confer a significant physiological mechanism to modulate lysosomal membrane potential and intracellular Ca2+ homeostasis [21,22]. TMEM175 has been identified as a K+-selective channel on endosomes and lysosomes, which remarkably contributes to the total K+ conductance in lysosomes [23,28]. Crystal structures of prokaryotic TMEM175 from Chamaesiphon minutus [29] and Marivirga tractuosa [30] and cryo-EM structures of human TMEM175 [31,32] have suggested that TMEM175 shows a different structure and K+ selectivity mechanism from canonical K+ channels. As a major functional K+ channel of lysosomes, TMEM175 plays a vital role in setting lysosomal membrane potential and maintaining lysosomal pH stability [23].
TMEM175 is a growth-factor-activated and AKT (protein kinase B)-gated lysosomal ion channel. It forms a complex with AKT and conformational changes in AKT are sufficient to activate TMEM175 [28]. A recent study reported that the apoptosis regulator Bcl-2 (B-cell lymphoma 2) binds to TMEM175 and suppresses its ion channel activity, which reveals the involvement of TMEM175 in apoptotic pathways [33]. Further data point to TMEM175 being more than just a K+ channel. It exhibits proton selectivity and conductivity, generating a constitutive proton current and acting as a lysosomal H+-leak channel [32,34].
TMEM175 has been recognized as a genetic risk factor for Parkinson’s disease (PD) by genome-wide association studies [35,36]. PD is the second most prevalent neurodegenerative disorder in the elderly, and it is clinically characterized by motor disabilities of rigidity, tremor, and bradykinesia as well as many non-motor symptoms including cognitive decline, sleep disorder, depression, olfactory loss, and autonomic dysfunction. The primary neuropathology of PD features the loss of dopaminergic neurons in the substantia nigra pars compacta and α-synuclein accumulation [37,38,39]. TMEM175 deficiency causes accumulation of α-synuclein in neurons and loss of dopaminergic neurons, which are prominent pathological characteristics of PD [28,34,40].
2. Structures of TMEM175
TMEM175 channels are found in archaea, bacteria, and animals, but are not present in fungi and plants. They differ from canonical K+ channels in several aspects. First, they share little amino acid sequence homology with other known potassium channels, rendering them evolutionarily distinct. Second, most canonical K+ channels feature a conserved P-loop selectivity filter composed of TVGYG-like signature sequences [41,42], but this conserved motif is missing in TMEM175 channels, which may indicate that they show a great difference in ion permeation properties. Third, unlike the canonical K+ channels that assemble the C-terminal helix as a pore-lining helix, TMEM175 channels assemble their subunits in an inverted way, with the N-terminal helix serving as a pore-lining helix [43,44]. Lastly, TMEM175 channels and canonical K+ channels show distinct pharmaceutical properties.
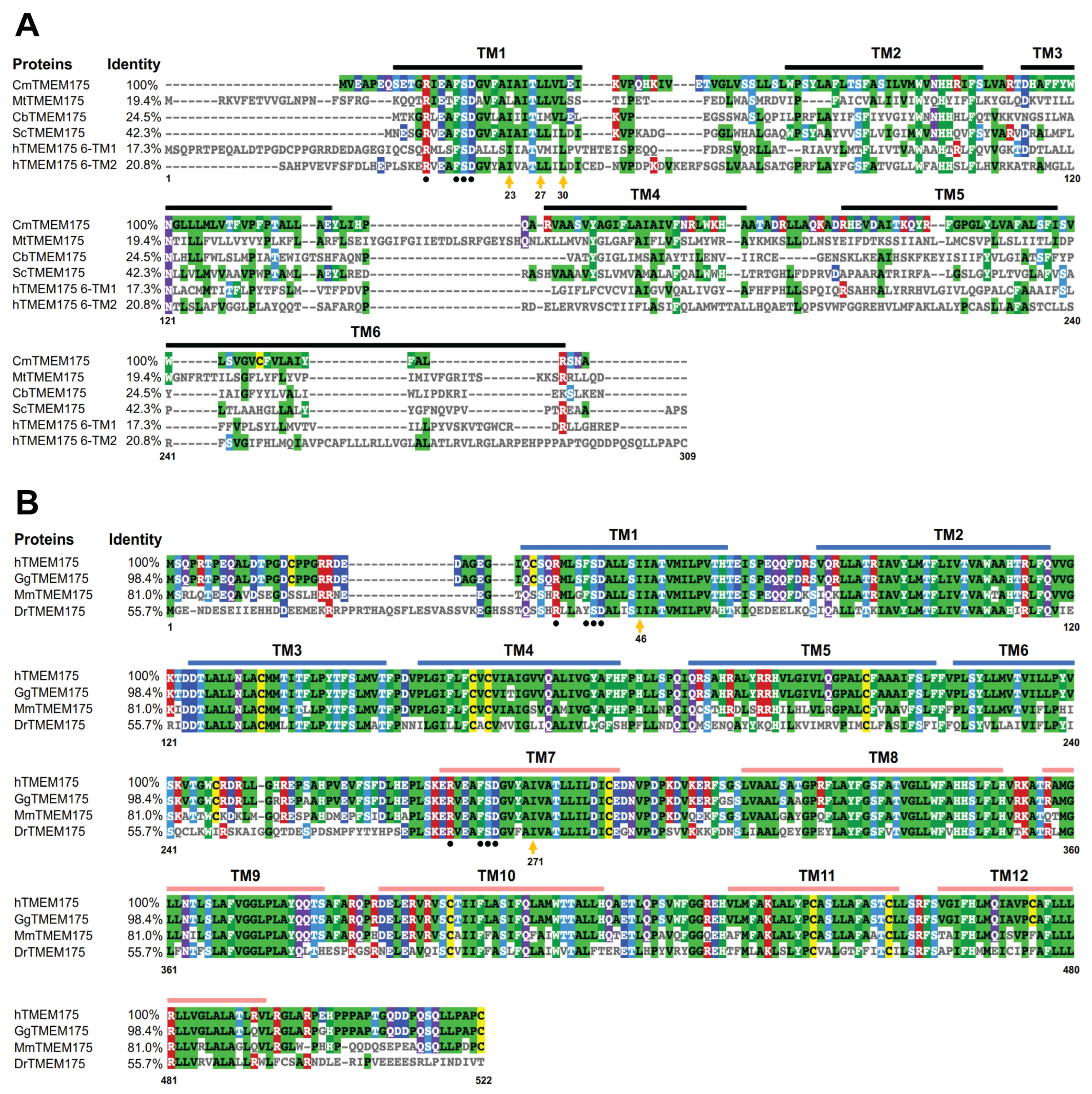
Prokaryotic TMEM175 proteins display a low sequence identity (Figure 1A). The sequence identity between CmTMEM175 and MtTMEM175 is lower than 20%. The most conserved region in prokaryotic TMEM175 proteins is the transmembrane 1 (TM1) helix that lines the channel pore. In contrast, mammalian TMEM175 channels show a much higher sequence identity. The human TMEM175 and the gorilla TMEM175 almost have an identical protein sequence. Additionally, the sequence identity between human TMEM175 and mouse TMEM175 is as high as 81% (Figure 1B). However, the level of conservation between prokaryotic and eukaryotic TMEM175 homologs is very low. The sequence identity between CmTMEM175 and hMEM175 repeat I or repeat II is 17.3% and 20.8%, respectively (Figure 1A).
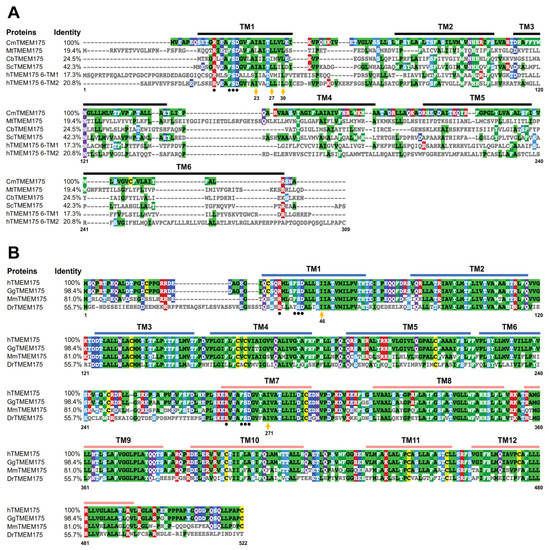
Figure 1.
Sequence alignment analysis of prokaryotic and eukaryotic TMEM175 proteins. (A) Sequence alignment analysis of prokaryotic TMEM175 proteins and the human TMEM175 repeat I and repeat II. (B) Sequence alignment analysis of eukaryotic TMEM175 proteins. The letters before TMEM175 indicate the species where it comes from, and UniProtKB reference numbers for these TMEM175 proteins are shown. Cm, Chamaesiphon minutus, K9UJK2; Mt, Marivirga tractuosa, E4TN31; Cb, Chryseobacterium sp., A0A086F3E3; Sc, Streptomyces collinus, S5VBU1; h: Homo sapiens, Q9BSA9; Gg, Gorilla gorilla, G3R453; Mm, Mus musculus, Q9CXY1; Dr, Danio rerio, A5PN43. The Clustal Omega sequence alignment program was used to align amino acid sequences and the sequence alignment result was shown and colored using MView.
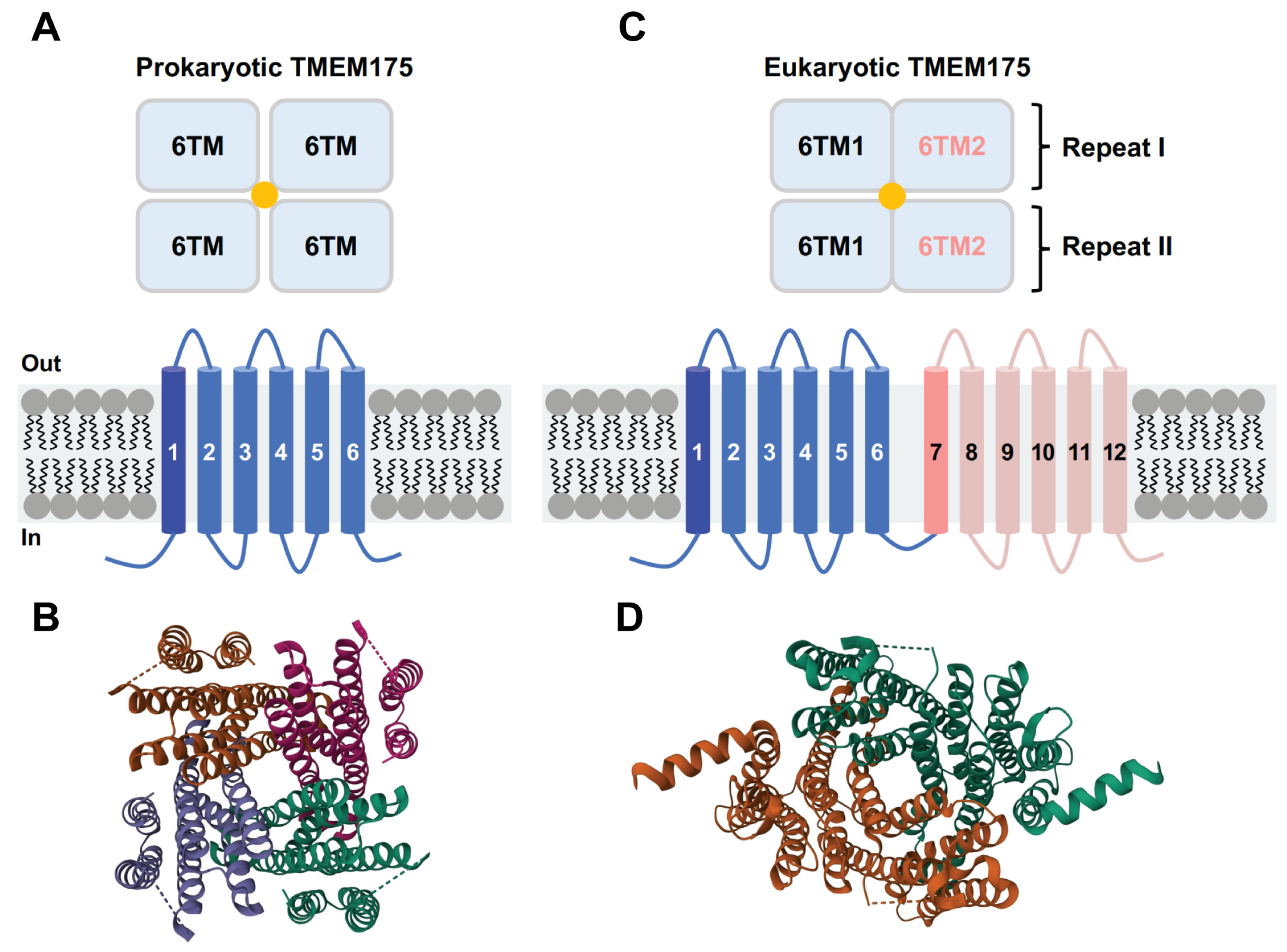
The structures of three different TMEM175 proteins have been resolved so far: two prokaryotic TMEM175 homologs from Chamaesiphon minutus (CmTMEM175) [29] and Marivirga tractuosa (MtTMEM175) [30], and the human homolog (hTMEM175) [31,32]. Even though prokaryotic and eukaryotic TMEM175 proteins do not share many similarities in their amino acid sequences, these three TMEM175 variants display a similar structural topology. The prokaryotic TMEM175 proteins exhibit a homotetrameric architecture, with each protomer containing one repeat of six transmembrane (6-TM) helices (Figure 2A,B). The first TM helix of each of the four protomers forms a central ion permeation channel, which is in line with the finding that the TM1 helix exhibits the highest level of conservation [29,30]. The human TMEM175, on the other hand, is a homodimer and each subunit contains two repeats of 6-TM helices (repeat I and repeat II) (Figure 2C,D) [31,32]. The structures of repeat I and repeat II highly resemble the monomeric structures of prokaryotic TMEM175 channels. The first transmembrane helix of repeat I (TM1) and repeat II (TM7) together form the ion-permeation pore. The ion-conduction pathway is situated in the middle of the channel along the pseudo-four-fold axis of the human TMEM175, which adopts a pseudo-four-fold symmetric architecture [31,32].
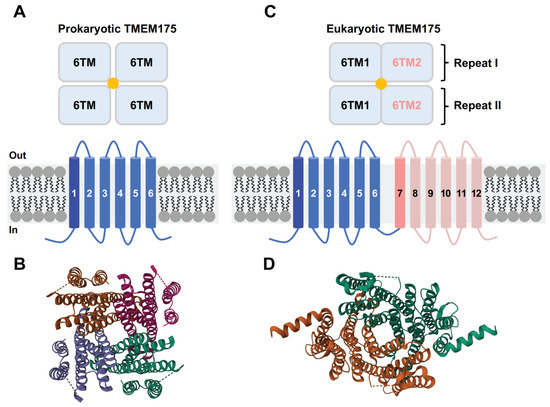
Figure 2.
Topological arrangement of prokaryotic and eukaryotic TMEM175 channels. (A) Schematic diagram of the prokaryotic TMEM175 channel structures. (B) Structural model of CmTMEM175 (PDB code: 5VRE). (C) Schematic diagram of the eukaryotic TMEM175 channel structures. (D) Structural model of hTMEM175 (PDB code: 6WC9, open state TMEM175 in KCl).
As TMEM175 channels lack the canonical P-loop selectivity filter, it raises the question of how they achieve K+ permeation and selectivity. In CmTMEM175, four TM1 helices form a bundle crossing that contains three layers of highly conserved hydrophobic residues. It was proposed that the conserved Ile23, Leu27, and Leu30 residues from each pore-lining TM1 helix create an ion permeation pathway, with the Ile23 residue playing a key role in defining K+ permeability and selectivity [29]. In line with this proposal, two independent studies have demonstrated that the conserved Ile46 from TM1 and the conserved Ile271 from TM7 constitute a narrow hydrophobic isoleucine constriction that acts as a physical gate and ion selectivity filter in hTMEM175 [31,32,45].
However, the structural and functional investigation of MtTMEM175 offers a divergent hypothesis for how TMEM175 channels selectively permeate potassium ions, despite having somewhat similar overall structures. Instead of hydrophobic residues, a highly conserved layer of threonine (Thr38) was discovered to be responsible for the K+ selectivity of MtTMEM175. The bulky hydrophobic residue Leu35 at the homologous position of Ile23 in CmTMEM175 or Ile46/Ile271 in hTMEM175 serves as a physical gate rather than a selectivity filter for ion permeation [30]. Furthermore, a layer of threonine residues (Thr49 and Thr274) and a serine residue (Ser45) also contribute to the K+ selectivity of hTMEM175. Selective permeation of ions through ion channels largely depends on the interplay between channel–ion interactions and water–ion interactions. The behavior of water within an ion channel pore is also very important to its conductive status. A “hydrophobic gating” mechanism may exist in TMEM175 channels, through which the hydrophobic pore spontaneously dehydrates and thereby functionally closes the channel [46,47].
The human TMEM175 shows decreased K+ permeation and increased H+ conductance at an acidic pH. It has been shown via molecular dynamics simulation and structure-guided mutagenesis that protons and potassium ions may share a permeation pathway. Besides the conserved amino acid residues that determine the K+ selectivity of TMEM175 channels, the N-terminal region of TM1 has a conserved RxxxFSD motif (Figure 1A,B). These residues are involved in inter- and intra-subunit interaction networks, which play a crucial role in maintaining the quaternary structure of TMEM175 channels [29,31]. Mutations in this signature motif abolished the channel activity [23].
3. The Ion Selectivity of TMEM175
3.1. The Human TMEM175 Is Characterized as a Lysosomal K+-Selective Channel
TMEM175 was first identified as a multi-spanning transmembrane protein of unknown functions (Domain-of-Unknown-Function DUF1211) through comparative proteomic analysis of integral and associated lysosomal membrane proteins [48]. The subcellular localization of TMEM175 was confirmed via transient expression of YFP-fusion protein in HeLa cells and compared with the lysosomal/late endosomal marker LAMP1 [49]. A whole-organelle patch-clamp-based candidate gene screening was utilized to identify the lysosomal K+ channel that is responsible for the K+ conductance in endosomes and lysosomes. Lysosomal currents were recorded from candidate genes-transfected HEK293T (human embryonic kidney 293T) cells. Of the 16 screened candidates, only TMEM175 showed a large increase in potassium current. TMEM175 contributes to the major lysosomal K+ conductance. The knock-down of mouse TMEM175 (mTMEM175) in RAW264.7 cells (a murine macrophage cell line commonly used in lysosomal studies) and human TMEM175 (hTMEM175) in HEK293T cells with short hairpin interfering RNAs significantly decrease the native K+ conductance. Furthermore, the knock-out of mTMEM175 in RAW264.7 cells with the CRISPR/Cas9 technique abolished the K+ conductance, which suggests that TMEM175 confers the lysosomal K+ permeability [23]. Ion selectivity is a fundamental property of ion channels and crucial to their physiological functions. hTMEM175 is permeable to Rb+, Cs+, and K+. It shows a better permeability for Cs+ than K+, which is consistent with the ion permeability of the lysosomal membrane [50]. It is minimally permeable to NMDG+ (N-methyl-D-glucamine, a large organic cation), Na+, and Ca2+ (Table 1).
TMEM175 homologs are also found in bacteria and archaea. The cellular localization of prokaryotic TMEM175 channels is the plasm membrane as prokaryotes do not have intracellular organelles. The ion selectivity of several prokaryotic TMEM175s has been analyzed through overexpressing bacterial TMEM175 proteins in HEK293T cells and using whole-cell patch clamp recording in electrophysiological characterization. Chryseobacterium sp. TMEM175 (CbTMEM175) and Streptomyces collinus TMEM175 (ScTMEM175) are more permeable to Na+ and less permeable to Cs+ than hTMEM175 [23]. Similar to CbTMEM175 (PK/PNa ≈ 2.4 ± 0.5, Table 1) and ScTMEM175 (PK/PNa ≈ 4.4 ± 1.4, Table 1), the Marivirga tractuosa TMEM175 (MtTMEM175) is a weakly K+ selective channel (PK/PNa ≈ 4.3, Table 1) [30]. Using 86Rb assay, another prokaryotic TMEM175 channel, Chamaesiphon minutus TMEM175 (CmTMEM175), was found to be selective for K+, Rb+, and Cs+. This channel is not permeable to NMDG+ or Na+, which is consistent with the ion selectivity of hTMEM175 (Table 1) [29]. The physiological functions of prokaryotic TMEM175 channels remain unknown but they are supposed to be responsible for the influx/efflux of specific ions depending on their ion selectivity. Overall, hTMEM175 shows higher selectivity for K+ and lower Na+ permeability than prokaryotic TMEM175s.

Table 1.
The ion selectivity of TMEM175.
Table 1.
The ion selectivity of TMEM175.
| TMEM175s | Ion Selectivity | Permeable Ions | References |
|---|---|---|---|
| hTMEM175 | PK/PNa = 36.0 ± 4.4 PK/PCa = 141.6 ± 27.7 PK/PCs = 0.51 ± 0.03 | K+, Cs+, Rb+ | [23] |
| hTMEM175 | PK/PNa ≈ 9.0 | K+, Cs+ | [31] |
| CbTMEM175 | PK/PNa = 2.4 ± 0.5 PK/PCs = 2.1 ± 0.7 | K+, Cs+, Na+ | [23] |
| ScTMEM175 | PK/PNa = 4.4 ± 1.4 PK/PCs = 2.3 ± 0.3 | K+, Cs+, Na+ | [23] |
| CmTMEM175 | PK > PNa | K+, Cs+, Rb+ | [29] |
| MtTMEM175 | PK/PNa ≈ 4.3 | K+, Cs+, Rb+, Na+, Li+ | [30] |
| hTMEM175 | PH/PK ≈ 3.3 × 104 | H+, Cs+, K+ | [32] |
| hTMEM175 | PH/PK ≈ 4.8 × 104 | H+, K+ | [34] |
Abbreviations: hTMEM175: human TMEM175; CbTMEM175, Chryseobacterium sp. TMEM175; ScTMEM175, Streptomyces collinus TMEM175; CmTMEM175, Chamaesiphon minutus TMEM175; MtTMEM175, Marivirga tractuosa TMEM175. PK, PNa, PCa, PCs, and PH are the permeabilities to K+, Na+, Ca2+, Cs+, and H+, respectively.
3.2. The Human TMEM175 Emerges as a Lysosomal H+-Leak Channel
The catabolic function of lysosomes is accomplished by more than 60 digestive enzymes including proteases, peptidases, lipases, nucleases, glycosidases, and sulfatases that break down biological macromolecules into small building-block molecules. These hydrolytic enzymes require an acidic pH of ~4.7 to carry out their catalytic activities. The acidity of the lysosomal lumen is dynamically and meticulously regulated in order to achieve pH homeostasis. Disturbance of lysosomal pH can cause lysosomal dysfunctions which are associated with multiple diseases such as lysosomal storage disorders, Parkinson’s disease, and Alzheimer’s disease [51,52,53]. Many ion channels, pumps, and transporters may contribute to the pH homeostasis of lysosomes in different ways.
It is well established that the vacuolar H+-ATPase, also known as the V-ATPase, is an ATP-dependent proton pump that mediates the active transport of protons from the cytoplasm to the lysosomal lumen and propels lysosome acidification [54,55,56]. A transmembrane electrical potential difference forms between the lysosome and cytosol when a significant number of protons have been pumped into the lysosomal lumen, which precludes additional proton pumping and consequently restricts the acidification of lysosomes. To circumvent this self-limiting behavior, proton influx must be accompanied by the movement of a counterion to dissipate the transmembrane voltage. Either the influx of cytosolic anions into the lysosomal lumen, the efflux of luminal cations into the cytosol, or the concurrent activity of both pathways can divert the transmembrane voltage generated by V-ATPases and facilitate lysosomal acidification [54,57]. The Cl−/H+ antiporter ClC-7 transports chloride ions into the lysosomal lumen and moves protons out of lysosomes with a fixed stoichiometry of 2Cl−:1H+, which has an important role in lysosomal pH regulation [16,57,58]. Na+ and K+ channels may also participate in counterion pathways and give rise to the effective acidification of lysosomes. For instance, an endolysosomal ATP-sensitive Na+ channel formed by two-pore channels (TPCs) can regulate lysosomal pH stability [19,59].
Early studies on the acidification of endosomes and lysosomes have revealed a proton leak current in these cellular compartments [60]. Additionally, a proton leak current was also detected in organelles of the regulated secretory pathway [61], which indicates that an unidentified proton-leak channel is present on endolysosomal membranes. Proton leak is proposed to be an important determinant of lysosomal pH, but the molecular identity of this lysosomal proton leak channel remains a mystery [34,57].
Two recent studies have demonstrated that hTMEM175 is a proton-selective and proton-activated channel that mediates the proton leak current of lysosomes [32,34]. A “proton-leak” like current was observed in hTMEM175-overexpressing HEK293T cells, but not in hTMEM175-KO (knock-out) cells, which suggests that hTMEM175 is responsible for the proton conductance. hTMEM175 is highly selective and permeable to protons when the luminal side is exposed to a very acidic pH. In contrast, it displays decreased K+ permeation at low pH [32]. It is estimated that the permeability of hTMEM175 for H+ relative to K+ is at a ratio of about 0.5 × 105-fold (Table 1).
3.3. Pharmacological Properties of TMEM175
Reflecting their divergent pore structure and selectivity profile, TMEM175 channels show highly distinct pharmacological properties compared to canonical K+ channels. First, unlike many K+ channels on the plasma membrane, which are typically blocked by Cs+, TMEM175 channels are more permeable to Cs+ than to K+. Second, the ion conductance of TMEM175 channels is unaffected by various frequently used K+ channel blockers, including Ba2+, tetraethylammonium, and quinine. Third, in contrast to canonical K+ channels, Zn2+ modestly inhibits bacterial TMEM175 channels and blocks the human TMEM175 channel with an IC50 of ~38 μM [23,29].
4-aminopyridine (4-AP), a commonly used K+ channel blocker, is the only known small-molecule inhibitor that inhibits the human TMEM175 activity, whereas the bacterial TMEM175 channels are insensitive to 4-AP [23]. 4-AP inhibits the hTMEM175-mediated K+ conductance with an estimated IC50 of 35 μM [23] and the proton flux through hTMEM175 with an IC50 of 55 ± 13 μM [62]. 4-AP binds in the ion permeation channel close to the isoleucine constriction, precluding the flow of ions and water, as shown by the structure of hTMEM175 bound to 4-AP [62]. The proton current of hTMEM175 was also significantly inhibited by Zn2+ [32]. Therefore, both 4-AP and Zn2+ were found to inhibit the K+ and H+ conductance of hTMEM175, supporting the idea that K+ and H+ share a common ion permeation pathway.
4. Physiological Functions of TMEM175
4.1. TMEM175 Controls Lysosomal Membrane Potential
An electrical potential gradient (ΔΨ = Vcytosol − Vlumen) exists across the lysosomal membrane due to a stark difference in ion strength between cytosol and lysosomal lumen. Lysosomal ion channels play an essential role in controlling lysosomal membrane potential as they mediate the influx/efflux of various ions across the lysosomal membrane, which alters the ionic homeostasis of the lysosomal lumen [3,11,63]. TMEM175 controls lysosomal ΔΨ through its K+ conductance. Lysosomes from the TMEM175 knock-out RAW264.7 cells displayed a depolarized ΔΨ relative to the lysosomes from wild-type cells using a whole-organelle patch-clamp. When measured in the lysosomes from wild-type cells, the lysosomal membrane potential was directly correlated with K+ concentration, which indicates the ΔΨ’s sensitivity to K+. The lysosomes from the TMEM175 knock-out cells were devoid of K+ sensitivity [23]. Even though the membrane potential ΔΨ is widely acknowledged as a critical biophysical property of lysosomes, its cellular roles are not well understood. Presumably, the lysosomal membrane potential ΔΨ may regulate luminal pH and catabolite export [11,23].
4.2. TMEM175 Contributes to Lysosomal pH Stability
Lysosomes have an acidic lumen, and the luminal pH value is maintained within a narrow range (4.5–5.5). The delicate regulation of lysosomal pH homeostasis requires the functional orchestration of many ion channels and transporters [54,64]. TMEM175 was found to be critical for lysosomal pH stability. Under fed conditions, the lysosomal pH of TMEM175 knock-out RAW264.7 murine macrophages was comparable to that of wild-type cells. After starvation in Earle’s Balanced Salt Solution (EBSS) for two hours, lysosomes from the TMEM175 knock-out cells showed a significant increase in pH value, whereas wild-type cells maintained a normal lysosomal pH [23]. Similar observations were reported in the SH-SY5Y neuroblastoma cell line. The wild-type SH-SY5Y cells maintained a lysosomal pH in both fed and starvation conditions, while knocking out TMEM175 results in the alkalinization of lysosomes under nutrient-depleted conditions [40,65]. Additionally, lysosomal acidification is disrupted by TMEM175 loss in neurons subjected to oxygen–glucose deprivation/reoxygenation, leading to a marked rise in lysosomal pH. Lysosome alkalinization can be reversed via overexpressing TMEM175 [66]. These pieces of evidence unambiguously point to TMEM175’s involvement in the control of lysosomal pH stability.
It is hypothesized that TMEM175 modulates lysosomal pH stability through counterion pathways as it mediates the exit of potassium ions from the lysosomal lumen [23]. Recent studies have shown that TMEM175 functions as a proton leak channel on the lysosomal membrane that modulates the lysosomal pH homeostasis [34]. The steady-state lysosomal pH is determined by the equilibrium between the proton influx driven by V-ATPases and the proton outflow mediated by TMEM175. As more protons exit lysosomes, TMEM175 overexpression induces an alkaline shift in lysosomal pH, whereas TMEM175 deficiency results in hyper-acidification of lysosomes [34]. The role of TMEM175 in H+-leak appears to be inconsistent with the finding of hypo-acidification of lysosomes in TMEM175 knock-out cells during starvation and requires further clarification [23,28].
4.3. TMEM175 Regulates Autophagosome-Lysosome Fusion
Macroautophagy (autophagy) is a conserved cellular pathway that eukaryotic cells utilize to degrade and recycle misfolded proteins, protein aggregates, and damaged organelles. Autophagic cargoes are sequestered in double-membrane vesicles which are called autophagosomes. The subsequent fusion of autophagosomes and lysosomes results in the formation of autophagolysosomes, and their contents are digested by lysosomal acidic hydrolases [67,68]. Several potassium channels are proposed to play an important part in the regulation of autophagy, including KCNH7 (potassium voltage-gated channel subfamily H member 7) [69] and a mitochondrial K+ ATP channel [70].
It was reported that TMEM175 regulates autophagosome–lysosome fusion [23]. To monitor the number of autophagosomes before and after fusion with lysosomes, the autophagosome marker LC3 (microtube-associated protein light chain 3) was tagged with RFP (red fluorescent protein) and GFP (green fluorescent protein). In this assay, autophagosomes before fusion with lysosomes are marked by RFP and GFP signals, whereas autophagosomes after fusion with lysosomes are only indicated by the RFP signals because of the quenching of GFP signals in the lysosomal acidic milieu [71]. The RFP–GFP–LC3 construct was transiently transfected into RAW264.7 macrophages and the starvation condition was used to induce autophagy [23]. The total number of autophagosomes (indicated by RFP puncta) before and after autophagosome–lysosome fusion is similar in wild-type and TMEM175 knock-out cells, indicating that the formation of autophagosomes is not affected by the deficiency of TMEM175. However, TMEM175 knock-out cells showed a significantly decreased GFP/RFP ratio compared with wild-type cells, which suggests that a larger portion of autophagosomes were fused with lysosomes. Similar findings in mouse hippocampal neurons [28] and SH-SY5Y cells support the hypothesis that TMEM175 deficiency speeds up the fusion of autophagosomes with lysosomes. Furthermore, depletion of TMEM175 leads to stagnant clearance of autophagosomes because of impaired lysosomal degradation [40].
4.4. TMEM175 Modulates Lysosomal and Mitochondrial Function
As a lysosomal potassium/proton channel, TMEM175 regulates lysosome physiology in many facets, such as membrane potential and pH homeostasis. Perturbations in lysosomal pH markedly change the catalytic activity of lysosomal hydrolases. The enzymatic activity of cathepsin B, cathepsin D, and glucocerebrosidase significantly decreased in TMEM175 knock-out cells relative to wild-type cells [34,40]. Consistent with this observation, a loss-of-function mutant of TMEM175, p.M393T, was associated with decreased glucocerebrosidase activity [72]. Therefore, TMEM175 deficiency may result in impaired lysosomal degradation. Both cathepsin B and cathepsin D are essential in the lysosomal degradation of α-synuclein [73,74] and decreased glucocerebrosidase is related to the abnormal accumulation of α-synuclein in Parkinson’s disease [75]. Using a preformed α-synuclein fibril (PFF) model, several studies have demonstrated that TMEM175 deficiency causes α-synuclein accumulation in neurons [28,34,40,65]. The TMEM175 knock-out neurons showed much greater aggregation of phosphorylated α-synuclein compared with wild-type neurons, whereas the intensity of phosphorylated α-synuclein inclusions decreased in TMEM175-overexpressed cells, suggesting that TMEM175 has a potential to enhance lysosomal degradation of α-synuclein aggregates.
Besides, TMEM175 may also regulate mitochondrial function. Compared with wild-type rat hippocampal neurons, TMEM175-deficient neurons showed compromised mitochondrial respiration capacity, decreased ATP levels, and reduced migratory ability of active mitochondria [40]. Another study found that TMEM175 overexpression significantly improved mitochondrial activity and ameliorated mitochondrial dysfunction in the course of ischemic injury. Upregulation of the TMEM175 level dramatically reduced the production of reactive oxygen species (ROS), an indicator of mitochondrial dysfunction [66]. A different study, however, presented conflicting results. Overexpression of TMEM175 increased ROS levels and resulted in mitochondrial dysfunction [33]. TMEM175 is primarily localized to lysosomes and endosomes and does not show mitochondrial localization [40]. Accordingly, it is proposed that TMEM175 regulates mitochondrial function through lysosome-mediated mitophagy, the selective degradation of mitochondria via autophagy [76], which plays a key role in the clearance of damaged mitochondria [33,40].
5. Regulation of the TMEM175 Channel Activity
Numerous factors, such as ions and membrane potential across the lysosomal membrane, endogenous lipids, luminal acidic pH, and ion channel binding partners, can regulate the activity of lysosomal ion channels [3,11]. The proton conductance of TMEM175 is activated by the luminal acidic pH, which drives the efflux of H+ across lysosomal membranes and thereby avoids the over-acidification of lysosomes. An endogenous lipid (arachidonic acid) and two other synthetic chemicals (DCPIB and ML 67–33) were also shown to activate the TMEM175 channel, causing an alkaline shift in the set-point pH of lysosomes [34].
TMEM175 is a growth-factor-activated K+ channel and is gated by AKT [28]. AKT binds to the TM2-TM3 linker region of repeat II, which is close to the cytosolic end of the pore-forming TM1 helix and forms a complex with TMEM175 (Figure 3A). AKT regulates TMEM175 channel activity in a kinase-independent manner. Instead, conformational changes in TMEM175-bound AKT are sufficient to activate TMEM175. AKT has a pleckstrin homology (PH) domain at the N-terminus, a kinase domain (KD) in the middle, and a hydrophobic motif at the C-terminus. It is proposed that in the inactive (“PH-in”) conformation, the AKT PH domain negatively regulates AKT kinase activity through intramolecularly interacting with the KD. Upon growth factor activation, the phosphatidylinositol (3,4,5)-trisphosphate [PtdIns(3,4,5)P3] level increases on the plasma membrane. PtdIns(3,4,5)P3 binding via the AKT PH domain causes conformational changes in AKT, leading to its active (“PH-out”) conformation [77]. A bath application of PtdIns(3,4,5)P3 was sufficient to activate TMEM175 current. In contrast, allosteric inhibitors such as MK-2206 and AKT inhibitor VIII that inhibit this conformational change blocked TMEM175 activity [28].
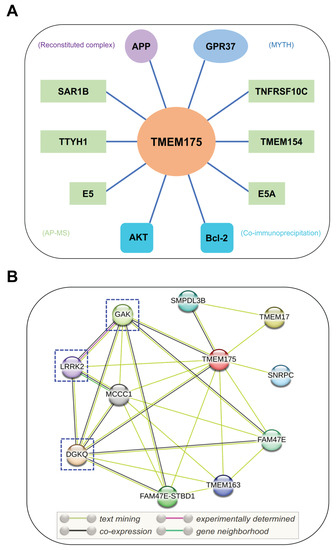
Figure 3.
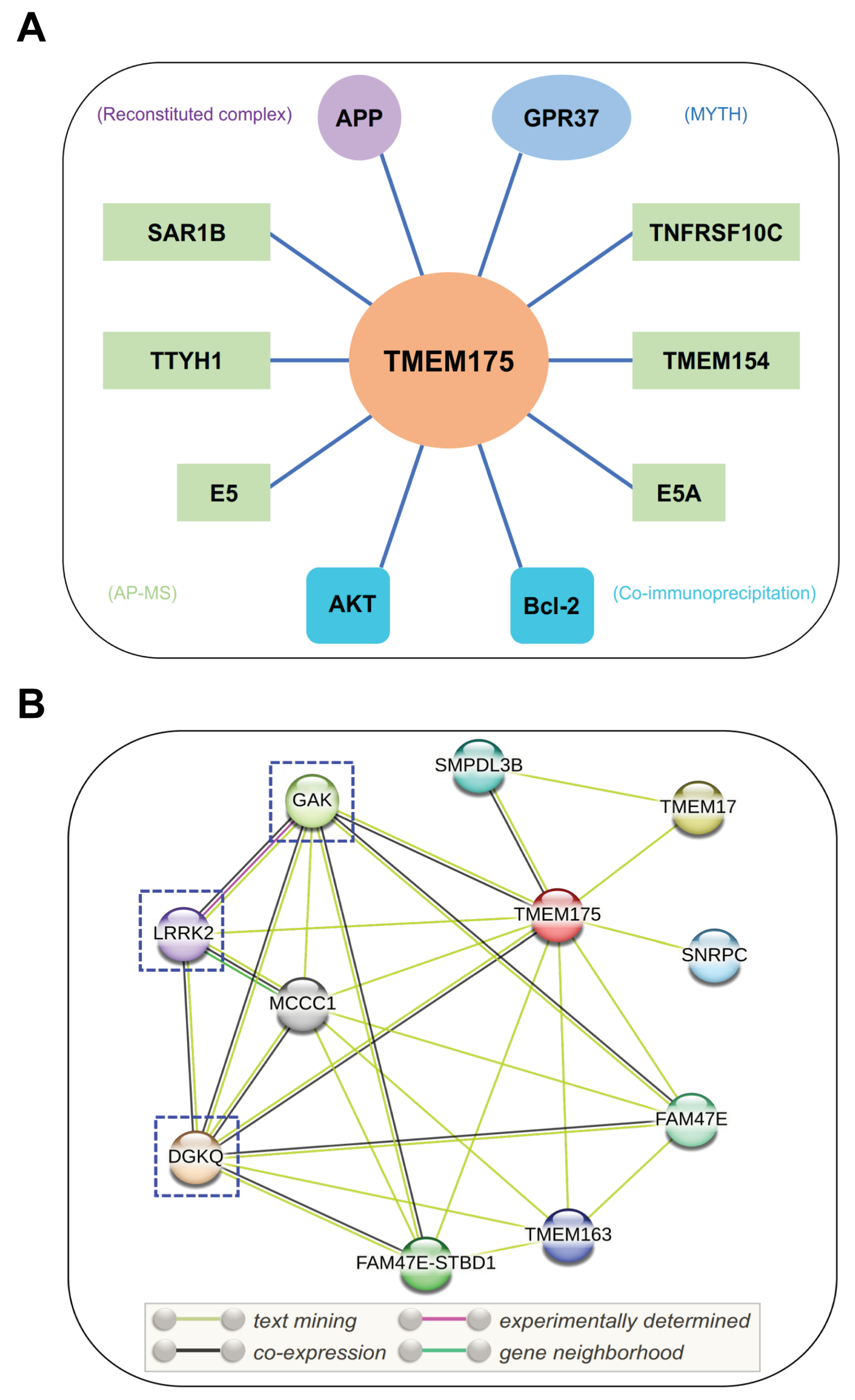
Interacting partners of TMEM175. (A) Potential binding partners of TMEM175. The experimental methods used to identify these TMEM175-protein interactions are shown in brackets. GPR37, G-protein-coupled receptor 37; APP, amyloid-beta precursor protein; E5 and E5A, viral proteins of human papillomavirus; SAR1B, secretion-associated Ras-related GTPase 1B; TMEM154, transmembrane protein 154; TNFRSF10C, tumor necrosis factor receptor superfamily member 10c; TTYH1, Tweety Homolog 1. MYTH, modified membrane yeast two-hybrid; AP-MS, affinity purification mass spectrometry. (B) STRING analysis of TMEM175-protein interaction networks. DGKQ, diacylglycerol kinase theta; FAM47E, family with sequence similarity 47 member E; GAK, cyclin G-associated kinase; LRRK2, leucine-rich repeat kinase 2; MCCC1, methylcrotonoyl-CoA carboxylase subunit alpha; SMPDL3B, acid sphingomyelinase-like phosphodiesterase 3b; SNRPC, U1 small nuclear ribonucleoprotein C; FAM47E-STBD1, FAM47E-STBD1 readthrough; TMEM17, transmembrane protein 17; and TMEM163, transmembrane protein 163. The PD-related binding partners of TMEM175 are highlighted by dashed boxes.
According to a recent study, Bcl-2, an antiapoptotic protein, interacts with TMEM175 and inhibits TMEM175 activity (Figure 3A) [33]. Bcl-2-specific inhibitors increase TMEM175-mediated K+ currents in a caspase-independent way. Overexpression of Bcl-2 in HEK293T cells significantly decreased whole-cell TMEM175 currents. Two amino acid residues of TMEM175, R377 and V145, are important to the association between TMEM175 and Bcl-2. Mutations of these two residues abolished their association and relieved the inhibitory effect of Bcl-2 on TMEM175 [33]. Bcl-2 is a critical regulator of mitochondrial apoptosis and is mainly localized on mitochondria. The interactions between mitochondrial Bcl-2 and lysosomal TMEM175 provide additional evidence of communication between mitochondria and lysosomes [78,79]. Additionally, beyond its functional roles in the endosomal–lysosomal pathway, TMEM175 may be implicated in the regulation of mitochondrial functions.
Besides AKT and Bcl-2, TMEM175 has other binding partners that may regulate its function (Figure 3A). According to the BioPlex 3.0 Interactome [80], a human interactome derived from HEK293T and HCT116 cells, TMEM175 was found to interact with SAR1B, TMEM154, TNFRSF10C, and TTYH1. Other potential binding partners include APP [81], GPR37 [82], and two viral proteins (E5 and E5A) [83]. A Calmodulin Target Database scan revealed that TMEM175 contains a single calmodulin-binding domain [84], which indicates that TMEM175 may interact with calmodulin. Moreover, the STRING database (https://string-db.org/ (accessed on 23 December 2022)) was used to analyze the protein–protein interaction networks of TMEM175 (Figure 3B) [85]. TMEM175 has connections with DGKQ, FAM47E, GAK, LRRK2, MCCC1, SMPDL3B, SNRPC, TMEM17, and TMEM163. As previously noted, the TMEM175/GAK/DGKQ locus has been identified as a risk locus in PD. Notably, TMEM175 may interact with LRRK2 (leucine-rich repeat kinase 2), an extensively studied PD causative protein [86]. However, their interactions need to be experimentally confirmed. Machine learning algorithms were utilized to identify gene–gene interactions in the Parkinson’s Progression Markers Initiative dataset. A variant rs3822019 in gene TMEM175 showed a statistically significant interaction with a variant rs17022452 in gene GAPDHP25 (glyceraldehyde-3 phosphate dehydrogenase pseudogene 25) [87], but it is unclear if they associate with each other at a protein level.
The human TMEM175 is described as a lysosomal potassium/proton channel but HEK293T cells overexpressing GFP-TMEM175 showed detectable GFP signals on their plasma membranes and functional TMEM175 currents can be measured on the plasma membrane [33]. Additionally, the heterologous expression of mouse TMEM175 in Xenopus laevis oocytes results in very small currents through the plasma membrane of oocytes. Dynasore compounds, which are potent dynamin inhibitors and disrupt clathrin-dependent endocytosis, have been reported to control the cellular localization of TMEM175 proteins [88]. TMEM175 currents in the plasma membrane were greatly increased in the cells treated with dynasore, with an estimated EC50 of at least 30 μM [88]. Dyngo-4a, a hydroxylated derivative of dynasore, induces more pronounced TMEM175 currents with an estimated EC50 of 2.3 μM [88]. Following the removal of dynasore chemicals, TMEM175 currents rapidly decreased, indicating the utilization of an effective internalization mechanism to translocate TMEM175 proteins from the plasma membrane to intracellular organelles.
6. The Pathological Relevance of TMEM175
6.1. TMEM175 Is Identified as a Risk Gene in Parkinson’s Disease
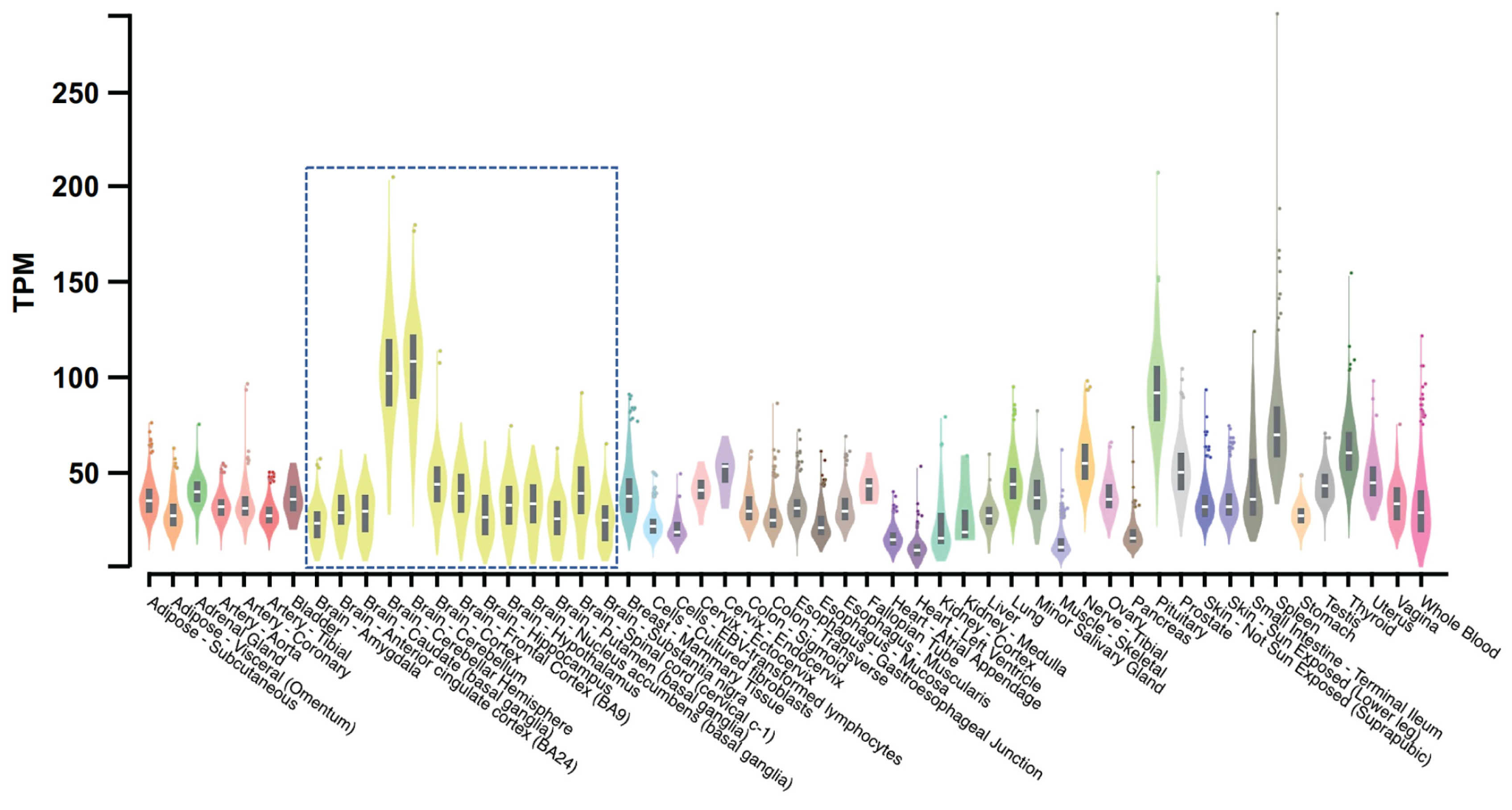
Expression of TMEM175 has been detected in several human tissues with its top three highest expression levels estimated in the cerebellar hemisphere, cerebellum, and pituitary gland [89] (Figure 4). Brain proteome-wide and transcriptome-wide association studies found that TMEM175 shows enrichment in glutamatergic neurons [90]. The high expression level in brains and enrichment in neurons may indicate that TMEM175 is crucial for the neural system’s functions.
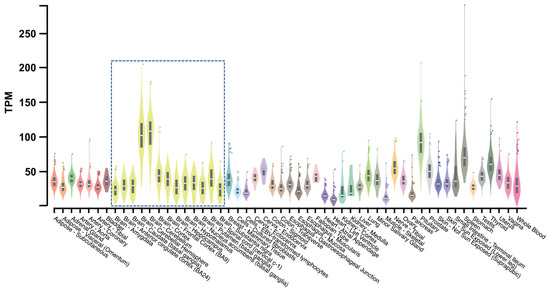
Figure 4.
Bulk tissue gene expression for TMEM175. This figure is processed from the GTEx portal (https://www.gtexportal.org (accessed on 1 December 2022)) [89]. Data source: GTEx Analysis Release V8 (dbGaP Accession phs000424.v8.p2). The gene expression level is shown in TPM (transcripts per million kilobases), calculated from a gene model with isoforms collapsed to a single gene. The gene expression level in different tissues is indicated by different colors with yellow violin plots (boxed) showing expression levels in brain tissues.
Genome-wide association studies (GWASs) investigate the genome of a large group of people and search for single nucleotide polymorphisms (SNPs) that are associated with a particular disease. A GWAS of Parkinson’s disease has identified the TMEM175/GAK/DGKQ region at chromosome 4p16.3 as a significant risk locus of this disease [35,36,91,92]. GAK (cyclin G-associated kinase) is a serine/threonine kinase involved in clathrin-mediated membrane trafficking and maintenance of proper centrosome maturation [93,94]. Additionally, GAK was identified as a binding partner of LRRK2 [95]. GAK and a few other interactors of LRRK2 form a complex with LRRK2 promoting removal of trans-Golgi-derived vesicles through an autophagy-dependent mechanism. The DGKQ gene encodes diacylglycerol kinase theta, which converts diacylglycerol into phosphatidic acid and regulates the respective levels of these two lipids [96]. Using short hairpin RNAs (shRNA) to knock down each of the genes in this locus, TMEM175 was found to be the only gene that is consistently associated with changes in levels of phosphorylated α-synuclein.
TMEM175 may have a neuroprotective effect, as homozygous TMEM175 knock-out mice at 18–22 months of age showed a significant loss of dopaminergic neurons in the substantia nigra pars compacta, which is a pathological feature of PD [28]. In this study, mouse brains from naturally aging mice without any drug treatment were used for neuron counting analysis. However, a recent study presented contradictory results, showing that more dopaminergic neurons were found in TMEM175 knock-out mice compared with wild-type mice when challenged with the neurotoxin MPTP, which suggests that the knock-out of TMEM175 has a neuroprotective effect [33]. Conflicting results reported in these studies may be attributed to the use of different mouse models. The naturally aging mouse model mimics the Parkinsonism caused by genetic factors, whereas the MPTP-induced mouse model mimics the Parkinsonism caused by environmental factors. Further studies are needed to investigate the pathological roles of TMEM175 in different types of PD.
6.2. Two Important SNPs of TMEM175 in Parkinson’s Disease
Further investigation of the chromosome 4: 951,947 GWAS signal in this PD-related region reveals a missense SNP in TMEM175, rs34311866 (p.M393T), which is the most significant coding variant in this locus. Conditional analysis of this main SNP results in the identification of a secondary genome-wide significant SNP, rs34884217 (p.Q65P) (Table 2) [65,72,97].
The p.M393T and p.Q65P variants are independent as they are not in linkage disequilibrium [72]. TMEM175 (M393T) is a loss-of-function mutant. Compared with the wild-type TMEM175, the M393T mutation decreases lysosomal K+ currents by about 40% in HEK293T cells [28]. In contrast, TMEM175 (Q65P) is a gain-of-function mutant under stress. One hour of starvation is sufficient to abolish the K+ current in wild-type TMEM175- and TMEM175 (M393T)-expressing HEK293T cells, whereas it has little effect on the K+ current of TMEM175 (Q65P)-expressing HEK293T cells. Additionally, these two mutations also have different effects on the proton conductance of TMEM175. The Q65P mutation promotes the proton conductance of TMEM175 while the M393T mutation decreases it [32]. As the proton conductivity of TMEM175 is critical to maintaining lysosomal pH stability and lysosomal hydrolases require a narrow pH for their optimal activity, it may explain why the M393T mutant shows decreased glucocerebrosidase activity relative to wild-type TMEM175 [65,72], whereas the Q65P mutation does not affect the glucocerebrosidase activity [72].
According to a meta-analysis of variants in three PD cohorts, the M393T mutation in TMEM175 is associated with an increased risk for PD, while the Q65P mutation is associated with a decreased risk for PD [72]. It has been reported that the TMEM175 M393T mutation affects the progression of PD. Through studying a cohort of 341 longitudinally followed PD patients, the M393T mutation was found to be associated with the rate of cognitive decline [28] and the rate of motor decline in PD, with p.M393T carriers declining more rapidly than patients without this mutation [28,98]. Moreover, multiple GWASs and meta-analysis of age at onset of PD have identified the TMEM175 rs34311866 (p.M393T) as a genome-wide significant variant that is associated with earlier age at onset of PD [72,92,99,100]. In a cohort of 1526 Danish PD patients, the TMEM175 rs34311866 variant showed a decrease in onset age by 1.2 years (95% confidence interval, 2.0–0.4) per PD risk allele [101].
Structural analysis of TMEM175 M393T and Q65P mutants revealed that these two mutations may have an impact on the structure of TMEM175. The C-terminal transmembrane domain of TMEM175 is proposed to be significantly destabilized by the M393T mutation, whereas the Q65P mutation only induces a minor global destabilization [72]. More structural and functional studies are needed to fully decipher how these two mutations influence the functions of TMEM175.
6.3. SNPs of TMEM175 in Other Diseases
The TMEM175 p.M393T mutation, in addition to being associated with a higher risk for PD, is also an independent risk locus for rapid eye movement sleep behavior disorder, a prodromal syndrome of alpha-synucleinopathies (Table 2) [72,102]. Other SNPs of TMEM175 were shown to be associated with early-onset and late-onset PD [103,104]. Genome-wide association studies and mutation analysis have identified important SNPs of TMEM175 in many other diseases such as type 2 diabetes [105], breast cancer [106], systemic sclerosis [107], Lewy body dementia [108], and short QT syndrome [109] (Table 2). However, the functional role of TMEM175 in these diseases remains unclear. Remarkably, TMEM175 is a critical genetic risk locus for Parkinson’s disease, Lewy body dementia, and rapid eye movement sleep behavior disorder, showcasing its significance in maintaining the proper functions of neurons.

Table 2.
SNPs of TMEM175 in diseases.
Table 2.
SNPs of TMEM175 in diseases.
| SNP/Mutation | Genomic Position (bp) | Reference Allele | Alternate Allele | Disease | References |
|---|---|---|---|---|---|
| rs34311866 | Chr.4:951947 | T | C | PD | [35] |
| rs34311866 | Chr.4:951947 | T | C | RBD | [72,102] |
| rs34884217 | Chr.4:950422 | A | C | PD | [35] |
| rs2290402 | Chr.4:931518 | T | C | Type 2 diabetes | [105] |
| rs2290405 | Chr.4:953186 | G | A, C | Breast cancer | [106] |
| rs2290405 | Chr.4:953186 | G | A, C | Systemic sclerosis | [107] |
| rs6599388 | Chr.4:945299 | C | T | Lewy body dementia | [108] |
| c.A526G | Chr.4:949608 | A | G | Early-onset PD | [103] |
| c.C547T | Chr.4:947062 | C | T | Late-onset PD | [104] |
| p.Arg377His | - | - | - | Short QT Syndrome | [109] |
Abbreviations: PD, Parkinson’s disease; RBD, rapid eye movement sleep behavior disorder.
7. Conclusions and Perspectives
TMEM175 is a pore-forming protein that was initially characterized as a K+-selective channel in endosomal–lysosomal membranes. Its channel activity is essential for the regulation of lysosomal membrane potential, lysosomal pH stability, and lysosome–autophagosome fusion. Dysfunction or loss of TMEM175 leads to impaired lysosomal degradation, resulting in compromised hydrolase activity, accelerated lysosome–autophagosome fusion, and accumulated pathogenic α-synuclein. Recent studies have shown that TMEM175 also mediates proton conductance and functions as a lysosomal H+-leak channel, modulating the acidity of lysosomes.
Human and bacterial TMEM175 channels show distinct cellular localization. When expressed in HEK293 cells, the bacterial MtTMEM175 enters the secretory pathway and is finally sorted into the plasma membrane [30]. Overexpression of human TMEM175 in HEK293T cells [33] or mouse TMEM175 in Xenopus laevis oocytes [88] generates detectable currents on the plasma membrane. Moreover, dynasore, an endocytosis inhibitor, is able to translocate lysosomal TMEM175 channels to the plasma membrane [88]. It is therefore tempting to hypothesize that TMEM175 is also found on the plasma membrane of native cells and goes through an internalization process before arriving at its target organelles, endosomes and lysosomes. Further research should be undertaken to investigate why TMEM175 is translocated to the plasma membrane, how TMEM175 gets sorted into endosomes and lysosomes, and whether it performs any biological functions there.
The pathological roles of TMEM175 in PD are primarily underpinned by its modulation of the endo-lysosomal pathway because it is a lysosomal potassium and proton channel that regulates the physiology of lysosomes. TMEM175 is also shown to be involved in the apoptotic pathway. The apoptosis regulator Bcl-2 inhibits the TMEM175 channel activity through direct binding. TMEM175 is a proapoptotic ion channel that plays an important role in the death of neurons [33]. Protein–protein interaction analysis revealed that TMEM175 has other potential binding partners. Therefore, it is important to find out whether they are TMEM175 modulators as this could shed light on TMEM175’s roles in other cellular pathways.
Potassium channels play a significant role in PD pathophysiology and represent an appealing therapeutic target for PD [110,111]. The TMEM175 gene has been identified as a risk locus of PD. TMEM175 or its knock-out exhibit neuroprotective effect in different PD mouse models. Finding specific activators or blockers may be a prospective strategy for PD treatment. It’s important to note that TMEM175 has been linked to a number of different neurological conditions, including Lewy body dementia and rapid eye movement sleep behavior disorder. Additionally, many SNPs of TMEM175 are shown to be associated with other diseases, such as breast cancer and systemic sclerosis. Further work is required to confirm the involvement of TMEM175 variants in these diseases and define their pathological roles.
Author Contributions
T.T., writing—original draft preparation, review and editing; B.J., writing—review; Z.L., writing—review, Open Access fee acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
The article processing charge (APC) for this manuscript was supported by the National Natural Science Foundation of China, grant number 32200782, and the National Agriculture Key Science & Technology Project, grant number NK20221201.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in Figure 1 are openly available in GTEx Portal (https://www.gtexportal.org/home/ (accessed on 1 December 2022)). For the STRING protein–protein interaction network analysis, the data are available in the STRING database (https://string-db.org/ (accessed on 23 December 2022)).
Acknowledgments
The authors thank Dejian Ren for reading and making comments on the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lawrence, R.E.; Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 2019, 21, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Settembre, C.; Fraldi, A.; Medina, D.L.; Ballabio, A. Signals from the lysosome: A control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 2013, 14, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Ren, D. Lysosomal Physiology. Annu. Rev. Physiol. 2015, 77, 57–80. [Google Scholar] [CrossRef] [PubMed]
- Ballabio, A.; Bonifacino, J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 101–118. [Google Scholar] [CrossRef] [PubMed]
- Lamming, D.W.; Bar-Peled, L. Lysosome: The metabolic signaling hub. Traffic 2019, 20, 27–38. [Google Scholar] [CrossRef]
- Bagshaw, R.D.; Mahuran, D.J.; Callahan, J.W. A Proteomic Analysis of Lysosomal Integral Membrane Proteins Reveals the Diverse Composition of the Organelle. Mol. Cell. Proteom. 2005, 4, 133–143. [Google Scholar] [CrossRef]
- Nishi, T.; Forgac, M. The vacuolar (H+)-ATPases—nature’s most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 2002, 3, 94–103. [Google Scholar] [CrossRef]
- Hübner, C.A.; Jentsch, T.J. Ion channel diseases. Hum. Mol. Genet. 2002, 11, 2435–2445. [Google Scholar] [CrossRef]
- Imbrici, P.; Liantonio, A.; Camerino, G.M.; De Bellis, M.; Camerino, C.; Mele, A.; Giustino, A.; Pierno, S.; De Luca, A.; Tricarico, D.; et al. Therapeutic Approaches to Genetic Ion Channelopathies and Perspectives in Drug Discovery. Front. Pharmacol. 2016, 7, 121. [Google Scholar] [CrossRef]
- Riederer, E.; Cang, C.; Ren, D. Lysosomal Ion Channels: What Are They Good For and Are They Druggable Targets? Annu. Rev. Pharmacol. Toxicol. 2023, 63, 19–41. [Google Scholar] [CrossRef]
- Li, P.; Gu, M.; Xu, H. Lysosomal Ion Channels as Decoders of Cellular Signals. Trends Biochem. Sci. 2019, 44, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.-P.; Cheng, X.; Mills, E.; Delling, M.; Wang, F.; Kurz, T.; Xu, H. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature 2008, 455, 992–996. [Google Scholar] [CrossRef] [PubMed]
- Lev, S.; Zeevi, D.A.; Frumkin, A.; Offen-Glasner, V.; Bach, G.; Minke, B. Constitutive Activity of the Human TRPML2 Channel Induces Cell Degeneration. J. Biol. Chem. 2010, 285, 2771–2782. [Google Scholar] [CrossRef]
- Hirschi, M.; Herzik Jr, M.A.; Wie, J.; Suo, Y.; Borschel, W.F.; Ren, D.; Lander, G.C.; Lee, S.-Y. Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3. Nature 2017, 550, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Deneka, D.; Sawicka, M.; Lam, A.K.M.; Paulino, C.; Dutzler, R. Structure of a volume-regulated anion channel of the LRRC8 family. Nature 2018, 558, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Graves, A.R.; Curran, P.K.; Smith, C.L.; Mindell, J.A. The Cl−/H+ antiporter ClC-7 is the primary chloride permeation pathway in lysosomes. Nature 2008, 453, 788–792. [Google Scholar] [CrossRef]
- Huang, P.; Zou, Y.; Zhong, X.Z.; Cao, Q.; Zhao, K.; Zhu, M.X.; Murrell-Lagnado, R.; Dong, X.-P. P2X4 Forms Functional ATP-activated Cation Channels on Lysosomal Membranes Regulated by Luminal pH. J. Biol. Chem. 2014, 289, 17658–17667. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Dong, X.-P.; Samie, M.; Li, X.; Cheng, X.; Goschka, A.; Shen, D.; Zhou, Y.; Harlow, J.; et al. TPC Proteins Are Phosphoinositide-Activated Sodium-Selective Ion Channels in Endosomes and Lysosomes. Cell 2012, 151, 372–383. [Google Scholar] [CrossRef]
- Cang, C.; Zhou, Y.; Navarro, B.; Seo, Y.-j.; Aranda, K.; Shi, L.; Battaglia-Hsu, S.; Nissim, I.; Clapham, D.E.; Ren, D. mTOR Regulates Lysosomal ATP-Sensitive Two-Pore Na+ Channels to Adapt to Metabolic State. Cell 2013, 152, 778–790. [Google Scholar] [CrossRef]
- She, J.; Guo, J.; Chen, Q.; Zeng, W.; Jiang, Y.; Bai, X.-C. Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel. Nature 2018, 556, 130–134. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, X.; Gao, Q.; Lawas, M.; Yu, L.; Cheng, X.; Gu, M.; Sahoo, N.; Li, X.; Li, P.; et al. A voltage-dependent K+ channel in the lysosome is required for refilling lysosomal Ca2+ stores. J. Cell Biol. 2017, 216, 1715–1730. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Zhong, X.Z.; Zou, Y.; Zhang, Z.; Toro, L.; Dong, X.-P. BK Channels Alleviate Lysosomal Storage Diseases by Providing Positive Feedback Regulation of Lysosomal Ca2+ Release. Dev. Cell 2015, 33, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Cang, C.; Aranda, K.; Seo, Y.-J.; Gasnier, B.; Ren, D. TMEM175 Is an Organelle K+ Channel Regulating Lysosomal Function. Cell 2015, 162, 1101–1112. [Google Scholar] [CrossRef]
- Feng, X.; Zhao, Z.; Li, Q.; Tan, Z. Lysosomal Potassium Channels: Potential Roles in Lysosomal Function and Neurodegenerative Diseases. CNS Neurol. Disord.-Drug Targets 2018, 17, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xu, M.; Wang, P.; Syeda, A.K.R.; Huang, P.; Dong, X.-P. Lysosomal potassium channels. Cell Calcium 2022, 102, 102536. [Google Scholar] [CrossRef] [PubMed]
- Dryer, S.E. Na+-activated K+ channels: A new family of large-conductance ion channels. Trends Neurosci. 1994, 17, 155–160. [Google Scholar] [CrossRef]
- Bobak, N.; Feliciangeli, S.; Chen, C.-C.; Ben Soussia, I.; Bittner, S.; Pagnotta, S.; Ruck, T.; Biel, M.; Wahl-Schott, C.; Grimm, C.; et al. Recombinant tandem of pore-domains in a Weakly Inward rectifying K+ channel 2 (TWIK2) forms active lysosomal channels. Sci. Rep. 2017, 7, 649. [Google Scholar] [CrossRef]
- Wie, J.; Liu, Z.; Song, H.; Tropea, T.F.; Yang, L.; Wang, H.; Liang, Y.; Cang, C.; Aranda, K.; Lohmann, J.; et al. A growth-factor-activated lysosomal K+ channel regulates Parkinson’s pathology. Nature 2021, 591, 431–437. [Google Scholar] [CrossRef]
- Lee, C.; Guo, J.; Zeng, W.; Kim, S.; She, J.; Cang, C.; Ren, D.; Jiang, Y. The lysosomal potassium channel TMEM175 adopts a novel tetrameric architecture. Nature 2017, 547, 472–475. [Google Scholar] [CrossRef]
- Brunner, J.D.; Jakob, R.P.; Schulze, T.; Neldner, Y.; Moroni, A.; Thiel, G.; Maier, T.; Schenck, S. Structural basis for ion selectivity in TMEM175 K+ channels. eLife 2020, 9, e53683. [Google Scholar] [CrossRef]
- Oh, S.; Paknejad, N.; Hite, R.K. Gating and selectivity mechanisms for the lysosomal K+ channel TMEM175. eLife 2020, 9, e53430. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Shen, C.; Wang, L.; Rawson, S.; Xie, W.J.; Nist-Lund, C.; Wu, J.; Shen, Z.; Xia, S.; Holt, J.R.; et al. pH regulates potassium conductance and drives a constitutive proton current in human TMEM175. Sci. Adv. 2022, 8, eabm1568. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Lin, B.; Zeng, W.; Fan, C.; Wu, H.; Ge, Y.; Li, Q.; Li, C.; Wei, Y.; Xin, J.; et al. Lysosomal K+ channel TMEM175 promotes apoptosis and aggravates symptoms of Parkinson’s disease. EMBO Rep. 2022, 23, e53234. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Li, P.; Wang, C.; Feng, X.; Geng, Q.; Chen, W.; Marthi, M.; Zhang, W.; Gao, C.; Reid, W.; et al. Parkinson’s disease-risk protein TMEM175 is a proton-activated proton channel in lysosomes. Cell 2022, 185, 2292–2308.e20. [Google Scholar] [CrossRef]
- Nalls, M.A.; Pankratz, N.; Lill, C.M.; Do, C.B.; Hernandez, D.G.; Saad, M.; DeStefano, A.L.; Kara, E.; Bras, J.; Sharma, M.; et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014, 46, 989–993. [Google Scholar] [CrossRef]
- Chang, D.; Nalls, M.A.; Hallgrímsdóttir, I.B.; Hunkapiller, J.; Van Der Brug, M.; Cai, F.; International Parkinson’s Disease Genomics Consortium; 23 and Me Research Team; Kerchner, G.A.; Ayalon, G.; et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 2017, 49, 1511–1516. [Google Scholar] [CrossRef]
- Funayama, M.; Nishioka, K.; Li, Y.; Hattori, N. Molecular genetics of Parkinson’s disease: Contributions and global trends. J. Hum. Genet. 2023, 68, 125–130. [Google Scholar] [CrossRef]
- Rudakou, U.; Yu, E.; Krohn, L.; Ruskey, J.A.; Asayesh, F.; Dauvilliers, Y.; Spiegelman, D.; Greenbaum, L.; Fahn, S.; Waters, C.H.; et al. Targeted sequencing of Parkinson’s disease loci genes highlights SYT11, FGF20 and other associations. Brain 2020, 144, 462–472. [Google Scholar] [CrossRef]
- Udayar, V.; Chen, Y.; Sidransky, E.; Jagasia, R. Lysosomal dysfunction in neurodegeneration: Emerging concepts and methods. Trends Neurosci. 2022, 45, 184–199. [Google Scholar] [CrossRef]
- Jinn, S.; Drolet, R.E.; Cramer, P.E.; Wong, A.H.-K.; Toolan, D.M.; Gretzula, C.A.; Voleti, B.; Vassileva, G.; Disa, J.; Tadin-Strapps, M.; et al. TMEM175 deficiency impairs lysosomal and mitochondrial function and increases α-synuclein aggregation. Proc. Natl. Acad. Sci. USA 2017, 114, 2389–2394. [Google Scholar] [CrossRef]
- Doyle, D.A.; Cabral, J.M.; Pfuetzner, R.A.; Kuo, A.; Gulbis, J.M.; Cohen, S.L.; Chait, B.T.; MacKinnon, R. The Structure of the Potassium Channel: Molecular Basis of K+ Conduction and Selectivity. Science 1998, 280, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Lee, A.; Chen, J.; Ruta, V.; Cadene, M.; Chait, B.T.; MacKinnon, R. X-ray structure of a voltage-dependent K+ channel. Nature 2003, 423, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Q.; Purhonen, P.; Hebert, H. Structure of potassium channels. Cell. Mol. Life Sci. 2015, 72, 3677–3693. [Google Scholar] [CrossRef] [PubMed]
- Sancho, M.; Kyle, B.D. The Large-Conductance, Calcium-Activated Potassium Channel: A Big Key Regulator of Cell Physiology. Front. Physiol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Marinelli, F.; Zhou, W.; Lee, J.; Choi, H.J.; Kim, M.; Faraldo-Gómez, J.D.; Hite, R.K. Differential ion dehydration energetics explains selectivity in the non-canonical lysosomal K+ channel TMEM175. eLife 2022, 11, e75122. [Google Scholar] [CrossRef]
- Rao, S.; Lynch, C.I.; Klesse, G.; Oakley, G.E.; Stansfeld, P.J.; Tucker, S.J.; Sansom, M.S.P. Water and hydrophobic gates in ion channels and nanopores. Faraday Discuss. 2018, 209, 231–247. [Google Scholar] [CrossRef]
- Lynch, C.I.; Klesse, G.; Rao, S.; Tucker, S.J.; Sansom, M.S.P. Water Nanoconfined in a Hydrophobic Pore: Molecular Dynamics Simulations of Transmembrane Protein 175 and the Influence of Water Models. ACS Nano 2021, 15, 19098–19108. [Google Scholar] [CrossRef]
- Schröder, B.; Wrocklage, C.; Pan, C.; Jäger, R.; Kösters, B.; Schäfer, H.; Elsässer, H.-P.; Mann, M.; Hasilik, A. Integral and Associated Lysosomal Membrane Proteins. Traffic 2007, 8, 1676–1686. [Google Scholar] [CrossRef]
- Chapel, A.; Kieffer-Jaquinod, S.; Sagné, C.; Verdon, Q.; Ivaldi, C.; Mellal, M.; Thirion, J.; Jadot, M.; Bruley, C.; Garin, J.; et al. An Extended Proteome Map of the Lysosomal Membrane Reveals Novel Potential Transporters. Mol. Cell. Proteom. 2013, 12, 1572–1588. [Google Scholar] [CrossRef]
- Casey, R.P.; Hollemans, M.; Tager, J.M. The permeability of the lysosomal membrane to small ions. Biochim. Biophys. Acta (BBA)-Biomembr. 1978, 508, 15–26. [Google Scholar] [CrossRef]
- Senkevich, K.; Gan-Or, Z. Autophagy lysosomal pathway dysfunction in Parkinson’s disease; Evidence from human genetics. Park. Relat. Disord. 2020, 73, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Platt, F.M.; d’Azzo, A.; Davidson, B.L.; Neufeld, E.F.; Tifft, C.J. Lysosomal storage diseases. Nat. Rev. Dis. Prim. 2018, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Gan-Or, Z.; Dion, P.A.; Rouleau, G.A. Genetic perspective on the role of the autophagy-lysosome pathway in Parkinson disease. Autophagy 2015, 11, 1443–1457. [Google Scholar] [CrossRef] [PubMed]
- Mindell, J.A. Lysosomal Acidification Mechanisms. Annu. Rev. Physiol. 2012, 74, 69–86. [Google Scholar] [CrossRef]
- Jefferies, K.C.; Cipriano, D.J.; Forgac, M. Function, structure and regulation of the vacuolar (H+)-ATPases. Arch. Biochem. Biophys. 2008, 476, 33–42. [Google Scholar] [CrossRef]
- Forgac, M. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007, 8, 917–929. [Google Scholar] [CrossRef]
- Ishida, Y.; Nayak, S.; Mindell, J.A.; Grabe, M. A model of lysosomal pH regulation. J. Gen. Physiol. 2013, 141, 705–720. [Google Scholar] [CrossRef]
- Leray, X.; Hilton, J.K.; Nwangwu, K.; Becerril, A.; Mikusevic, V.; Fitzgerald, G.; Amin, A.; Weston, M.R.; Mindell, J.A. Tonic inhibition of the chloride/proton antiporter ClC-7 by PI(3,5)P2 is crucial for lysosomal pH maintenance. eLife 2022, 11, e74136. [Google Scholar] [CrossRef]
- Steinberg, B.E.; Huynh, K.K.; Brodovitch, A.; Jabs, S.; Stauber, T.; Jentsch, T.J.; Grinstein, S. A cation counterflux supports lysosomal acidification. J. Cell Biol. 2010, 189, 1171–1186. [Google Scholar] [CrossRef]
- Van Dyke, R.W. Acidification of rat liver lysosomes: Quantitation and comparison with endosomes. Am. J. Physiol.-Cell Physiol. 1993, 265, C901–C917. [Google Scholar] [CrossRef]
- Wu, M.M.; Grabe, M.; Adams, S.; Tsien, R.Y.; Moore, H.-P.H.; Machen, T.E. Mechanisms of pH Regulation in the Regulated Secretory Pathway. J. Biol. Chem. 2001, 276, 33027–33035. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Stix, R.; Zhou, W.; Faraldo-Gómez, J.D.; Hite, R.K. Mechanism of 4-aminopyridine inhibition of the lysosomal channel TMEM175. Proc. Natl. Acad. Sci. USA 2022, 119, e2208882119. [Google Scholar] [CrossRef] [PubMed]
- Sterea, A.M.; Almasi, S.; El Hiani, Y. The hidden potential of lysosomal ion channels: A new era of oncogenes. Cell Calcium 2018, 72, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Chen, J.; Liu, S.; Xu, H. The Acid Gate in the Lysosome. Autophagy 2022, 19, 1368–1370. [Google Scholar] [CrossRef]
- Jinn, S.; Blauwendraat, C.; Toolan, D.; Gretzula, C.A.; Drolet, R.E.; Smith, S.; Nalls, M.A.; Marcus, J.; Singleton, A.B.; Stone, D.J. Functionalization of the TMEM175 p.M393T variant as a risk factor for Parkinson disease. Hum. Mol. Genet. 2019, 28, 3244–3254. [Google Scholar] [CrossRef]
- Zhang, M.; Lu, H.; Xie, X.; Shen, H.; Li, X.; Zhang, Y.; Wu, J.; Ni, J.; Li, H.; Chen, G. TMEM175 mediates Lysosomal function and participates in neuronal injury induced by cerebral ischemia-reperfusion. Mol. Brain 2020, 13, 113. [Google Scholar] [CrossRef]
- Rubinsztein, D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 2006, 443, 780–786. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Emr, S.D. Autophagy as a Regulated Pathway of Cellular Degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef]
- Perez-Neut, M.; Haar, L.; Rao, V.; Santha, S.; Lansu, K.; Rana, B.; Jones, W.K.; Gentile, S. Activation of hERG3 channel stimulates autophagy and promotes cellular senescence in melanoma. Oncotarget 2016, 7, 21991–22004. [Google Scholar] [CrossRef]
- Yu, K.-Y.; Wang, Y.-P.; Wang, L.-H.; Jian, Y.; Zhao, X.-D.; Chen, J.-W.; Murao, K.; Zhu, W.; Dong, L.; Wang, G.-Q.; et al. Mitochondrial KATP channel involvement in angiotensin II-induced autophagy in vascular smooth muscle cells. Basic Res. Cardiol. 2014, 109, 416. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in Mammalian Autophagy Research. Cell 2010, 140, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Krohn, L.; Öztürk, T.N.; Vanderperre, B.; Bencheikh, B.O.A.; Ruskey, J.A.; Laurent, S.B.; Spiegelman, D.; Postuma, R.B.; Arnulf, I.; Hu, M.T.M.; et al. Genetic, Structural, and Functional Evidence Link TMEM175 to Synucleinopathies. Ann. Neurol. 2020, 87, 139–153. [Google Scholar] [CrossRef] [PubMed]
- McGlinchey, R.P.; Lee, J.C. Cysteine cathepsins are essential in lysosomal degradation of α-synuclein. Proc. Natl. Acad. Sci. USA 2015, 112, 9322–9327. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Hamamichi, S.; Caldwell, K.A.; Caldwell, G.A.; Yacoubian, T.A.; Wilson, S.; Xie, Z.-L.; Speake, L.D.; Parks, R.; Crabtree, D.; et al. Lysosomal enzyme cathepsin D protects against α-synuclein aggregation and toxicity. Mol. Brain 2008, 1, 17. [Google Scholar] [CrossRef]
- Murphy, K.E.; Gysbers, A.M.; Abbott, S.K.; Tayebi, N.; Kim, W.S.; Sidransky, E.; Cooper, A.; Garner, B.; Halliday, G.M. Reduced glucocerebrosidase is associated with increased α-synuclein in sporadic Parkinson’s disease. Brain 2014, 137, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Kroemer, G.; Kepp, O. Mitophagy: An Emerging Role in Aging and Age-Associated Diseases. Front. Cell Dev. Biol. 2020, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- Ebner, M.; Lučić, I.; Leonard, T.A.; Yudushkin, I. PI(3,4,5)P3 Engagement Restricts Akt Activity to Cellular Membranes. Mol. Cell 2017, 65, 416–431.e6. [Google Scholar] [CrossRef]
- Audano, M.; Schneider, A.; Mitro, N. Mitochondria, lysosomes, and dysfunction: Their meaning in neurodegeneration. J. Neurochem. 2018, 147, 291–309. [Google Scholar] [CrossRef]
- Plotegher, N.; Duchen, M.R. Crosstalk between Lysosomes and Mitochondria in Parkinson’s Disease. Front. Cell Dev. Biol. 2017, 5, 110. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Bruckner, R.J.; Navarrete-Perea, J.; Cannon, J.R.; Baltier, K.; Gebreab, F.; Gygi, M.P.; Thornock, A.; Zarraga, G.; Tam, S.; et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell 2021, 184, 3022–3040.e28. [Google Scholar] [CrossRef]
- Oláh, J.; Vincze, O.; Virók, D.; Simon, D.; Bozsó, Z.; Tőkési, N.; Horváth, I.; Hlavanda, E.; Kovács, J.; Magyar, A.; et al. Interactions of Pathological Hallmark Proteins: TUBULIN POLYMERIZATION PROMOTING PROTEIN/p25, β-AMYLOID, AND α-SYNUCLEIN. J. Biol. Chem. 2011, 286, 34088–34100. [Google Scholar] [CrossRef] [PubMed]
- Sokolina, K.; Kittanakom, S.; Snider, J.; Kotlyar, M.; Maurice, P.; Gandía, J.; Benleulmi-Chaachoua, A.; Tadagaki, K.; Oishi, A.; Wong, V.; et al. Systematic protein–protein interaction mapping for clinically relevant human GPCRs. Mol. Syst. Biol. 2017, 13, 918. [Google Scholar] [CrossRef] [PubMed]
- Rozenblatt-Rosen, O.; Deo, R.C.; Padi, M.; Adelmant, G.; Calderwood, M.A.; Rolland, T.; Grace, M.; Dricot, A.; Askenazi, M.; Tavares, M.; et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 2012, 487, 491–495. [Google Scholar] [CrossRef] [PubMed]
- O’Day, D.H. Calmodulin Binding Domains in Critical Risk Proteins Involved in Neurodegeneration. Curr. Issues Mol. Biol. 2022, 44, 5802–5814. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2020, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Parisiadou, L.; Gu, X.-L.; Wang, L.; Shim, H.; Sun, L.; Xie, C.; Long, C.-X.; Yang, W.-J.; Ding, J.; et al. Leucine-Rich Repeat Kinase 2 Regulates the Progression of Neuropathology Induced by Parkinson’s-Disease-Related Mutant α-synuclein. Neuron 2009, 64, 807–827. [Google Scholar] [CrossRef]
- Cope, J.L.; Baukmann, H.A.; Klinger, J.E.; Ravarani, C.N.J.; Böttinger, E.P.; Konigorski, S.; Schmidt, M.F. Interaction-Based Feature Selection Algorithm Outperforms Polygenic Risk Score in Predicting Parkinson’s Disease Status. Front. Genet. 2021, 12. [Google Scholar] [CrossRef]
- Pergel, E.; Veres, I.; Csigi, G.I.; Czirják, G. Translocation of TMEM175 Lysosomal Potassium Channel to the Plasma Membrane by Dynasore Compounds. Int. J. Mol. Sci. 2021, 22, 10515. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Zhou, S.; Tian, Y.; Song, X.; Xiong, J.; Cheng, G. Brain Proteome-Wide and Transcriptome-Wide Association Studies, Bayesian Colocalization, and Mendelian Randomization Analyses Reveal Causal Genes of Parkinson’s Disease. J. Gerontol. Ser. A 2022, 78, 563–568. [Google Scholar] [CrossRef]
- Nalls, M.A.; Blauwendraat, C.; Vallerga, C.L.; Heilbron, K.; Bandres-Ciga, S.; Chang, D.; Tan, M.; Kia, D.A.; Noyce, A.J.; Xue, A.; et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019, 18, 1091–1102. [Google Scholar] [CrossRef] [PubMed]
- Blauwendraat, C.; Heilbron, K.; Vallerga, C.L.; Bandres-Ciga, S.; von Coelln, R.; Pihlstrøm, L.; Simón-Sánchez, J.; Schulte, C.; Sharma, M.; Krohn, L.; et al. Parkinson’s disease age at onset genome-wide association study: Defining heritability, genetic loci, and α-synuclein mechanisms. Mov. Disord. 2019, 34, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Nagamori, I.; Yabuta, N.; Nojima, H. GAK, a regulator of clathrin-mediated membrane traffic, also controls centrosome integrity and chromosome congression. J. Cell Sci. 2009, 122, 3145–3152. [Google Scholar] [CrossRef] [PubMed]
- Nagle, M.W.; Latourelle, J.C.; Labadorf, A.; Dumitriu, A.; Hadzi, T.C.; Beach, T.G.; Myers, R.H. The 4p16.3 Parkinson Disease Risk Locus Is Associated with GAK Expression and Genes Involved with the Synaptic Vesicle Membrane. PLoS ONE 2016, 11, e0160925. [Google Scholar] [CrossRef]
- Beilina, A.; Rudenko, I.N.; Kaganovich, A.; Civiero, L.; Chau, H.; Kalia, S.K.; Kalia, L.V.; Lobbestael, E.; Chia, R.; Ndukwe, K.; et al. Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc. Natl. Acad. Sci. USA 2014, 111, 2626–2631. [Google Scholar] [CrossRef]
- Houssa, B.; Schaap, D.; van der Wal, J.; Goto, K.; Kondo, H.; Yamakawa, A.; Shibata, M.; Takenawa, T.; van Blitterswijk, W.J. Cloning of a Novel Human Diacylglycerol Kinase (DGKθ) Containing Three Cysteine-rich Domains, a Proline-rich Region, and a Pleckstrin Homology Domain with an Overlapping Ras-associating Domain. J. Biol. Chem. 1997, 272, 10422–10428. [Google Scholar] [CrossRef]
- Langmyhr, M.; Henriksen, S.P.; Cappelletti, C.; van de Berg, W.D.J.; Pihlstrøm, L.; Toft, M. Allele-specific expression of Parkinson’s disease susceptibility genes in human brain. Sci. Rep. 2021, 11, 504. [Google Scholar] [CrossRef]
- Cao, L.-X.; Jiang, Y.; Piao, Y.-S.; Huang, Y. Rapid motor progression of Parkinson’s disease associates with clinical and genetic variants. Front. Biosci. 2021, 26, 1503–1512. [Google Scholar] [CrossRef]
- Iwaki, H.; Blauwendraat, C.; Leonard, H.L.; Liu, G.; Maple-Grødem, J.; Corvol, J.-C.; Pihlstrøm, L.; van Nimwegen, M.; Hutten, S.J.; Nguyen, K.-D.H.; et al. Genetic risk of Parkinson disease and progression. Neurol. Genet. 2019, 5, e348. [Google Scholar] [CrossRef]
- Grover, S.; Kumar Sreelatha, A.A.; Pihlstrom, L.; Domenighetti, C.; Schulte, C.; Sugier, P.-E.; Radivojkov-Blagojevic, M.; Lichtner, P.; Mohamed, O.; Portugal, B.; et al. Genome-wide Association and Meta-analysis of Age at Onset in Parkinson Disease. Neurology 2022, 99, e698–e710. [Google Scholar] [CrossRef]
- Lill, C.M.; Hansen, J.; Olsen, J.H.; Binder, H.; Ritz, B.; Bertram, L. Impact of Parkinson’s disease risk loci on age at onset. Mov. Disord. 2015, 30, 847–850. [Google Scholar] [CrossRef] [PubMed]
- Krohn, L.; Heilbron, K.; Blauwendraat, C.; Reynolds, R.H.; Yu, E.; Senkevich, K.; Rudakou, U.; Estiar, M.A.; Gustavsson, E.K.; Brolin, K.; et al. Genome-wide association study of REM sleep behavior disorder identifies polygenic risk and brain expression effects. Nat. Commun. 2022, 13, 7496. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ou, R.; Chen, Y.; Gu, X.; Wei, Q.; Cao, B.; Zhang, L.; Hou, Y.; Liu, K.; Chen, X.; et al. Mutation analysis of TMEM family members for early-onset Parkinson’s disease in Chinese population. Neurobiol. Aging 2021, 101, e291.e1–e299.e6. [Google Scholar] [CrossRef]
- Gialluisi, A.; Reccia, M.G.; Modugno, N.; Nutile, T.; Lombardi, A.; Di Giovannantonio, L.G.; Pietracupa, S.; Ruggiero, D.; Scala, S.; Gambardella, S.; et al. Identification of sixteen novel candidate genes for late onset Parkinson’s disease. Mol. Neurodegener. 2021, 16, 35. [Google Scholar] [CrossRef] [PubMed]
- Meitinger, T.; Shriner, D.; Chen, B.H.; Lindgren, C.; Chen, W.-M.; Guo, X.; Liu, J.; Bielinski, S.J.; Yanek, L.R.; Nalls, M.A.; et al. Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes. PLOS Genet. 2014, 10, e1004517. [Google Scholar] [CrossRef]
- Tan, J.; Fu, L.; Chen, H.; Guan, J.; Chen, Y.; Fang, J. Association study of genetic variation in the autophagy lysosome pathway genes and risk of eight kinds of cancers. Int. J. Cancer 2018, 143, 80–87. [Google Scholar] [CrossRef]
- Gorlova, O.Y.; Li, Y.; Gorlov, I.; Ying, J.; Chen, W.V.; Assassi, S.; Reveille, J.D.; Arnett, F.C.; Zhou, X.; Bossini-Castillo, L.; et al. Gene-level association analysis of systemic sclerosis: A comparison of African-Americans and White populations. PLoS ONE 2018, 13, e0189498. [Google Scholar] [CrossRef]
- Chia, R.; Sabir, M.S.; Bandres-Ciga, S.; Saez-Atienzar, S.; Reynolds, R.H.; Gustavsson, E.; Walton, R.L.; Ahmed, S.; Viollet, C.; Ding, J.; et al. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat. Genet. 2021, 53, 294–303. [Google Scholar] [CrossRef]
- Jáimez, J.J.; Bermúdez, F.; Moreno-Manuel, A.; Monteros, L.G.E.d.l.; Vera-Pedrosa, M.L.; Álvarez, M.; Tercedor, L. Short qt syndrome and sudden cardiac death due to a mutation in the tmem175 endolysosomal potassium channel. J. Am. Coll. Cardiol. 2020, 75, 469. [Google Scholar] [CrossRef]
- Chen, X.; Xue, B.; Wang, J.; Liu, H.; Shi, L.; Xie, J. Potassium Channels: A Potential Therapeutic Target for Parkinson’s Disease. Neurosci. Bull. 2018, 34, 341–348. [Google Scholar] [CrossRef]
- Chen, X.; Feng, Y.; Quinn, R.J.; Pountney, D.L.; Richardson, D.R.; Mellick, G.D.; Ma, L. Potassium Channels in Parkinson’s Disease: Potential Roles in its Pathogenesis and Innovative Molecular Targets for Treatment. Pharmacol. Rev. 2023. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).