The Crystal Structure of the Hsp90-LA1011 Complex and the Mechanism by Which LA1011 May Improve the Prognosis of Alzheimer’s Disease
Abstract
1. Introduction
2. Methods and Materials
2.1. Protein Purification
2.2. Crystallization and Refinement
2.3. Isothermal Titration Calorimetry
3. Results and Conclusions
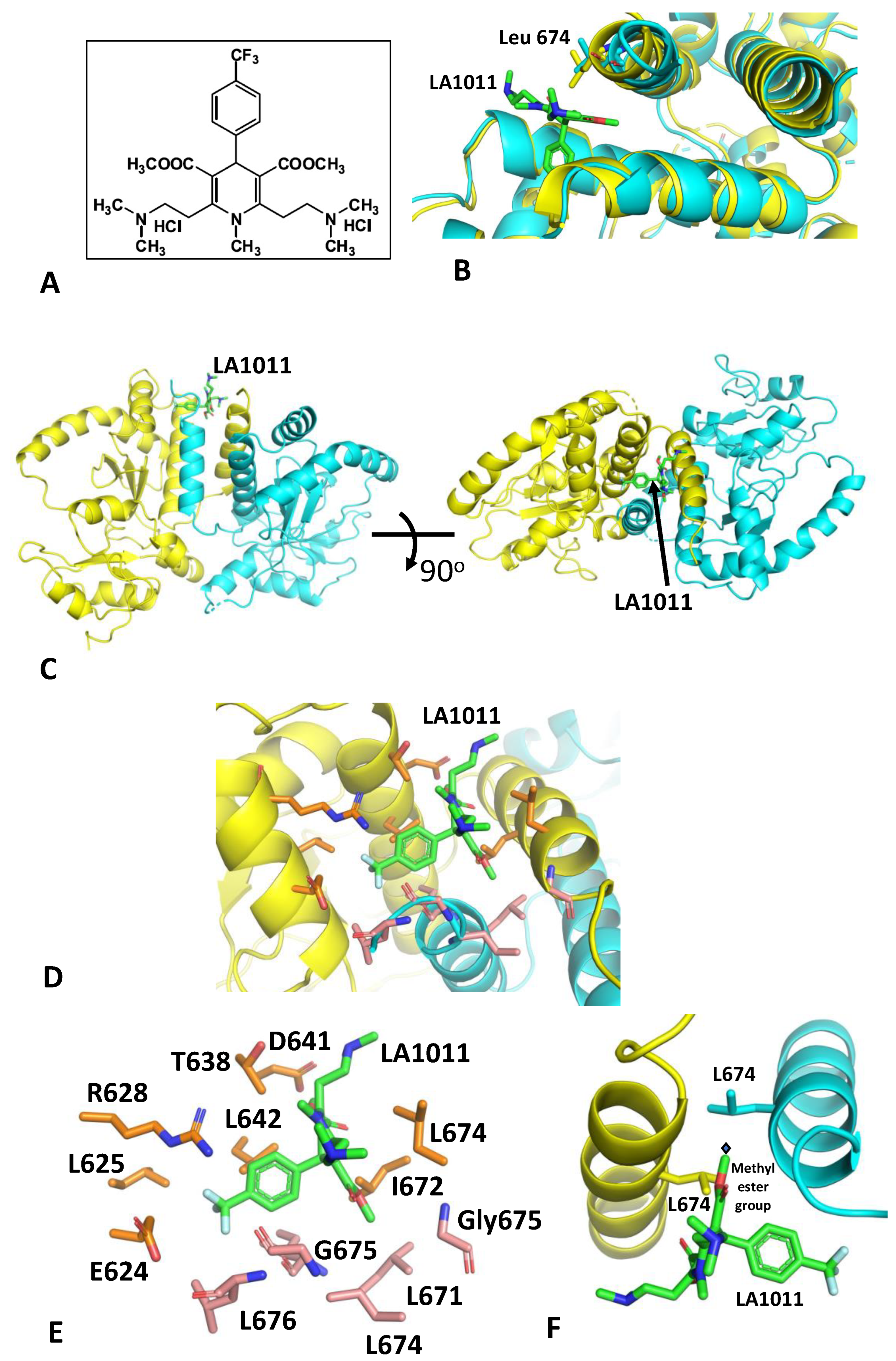
3.1. The Crystal Structure of Hsp90-LA1011
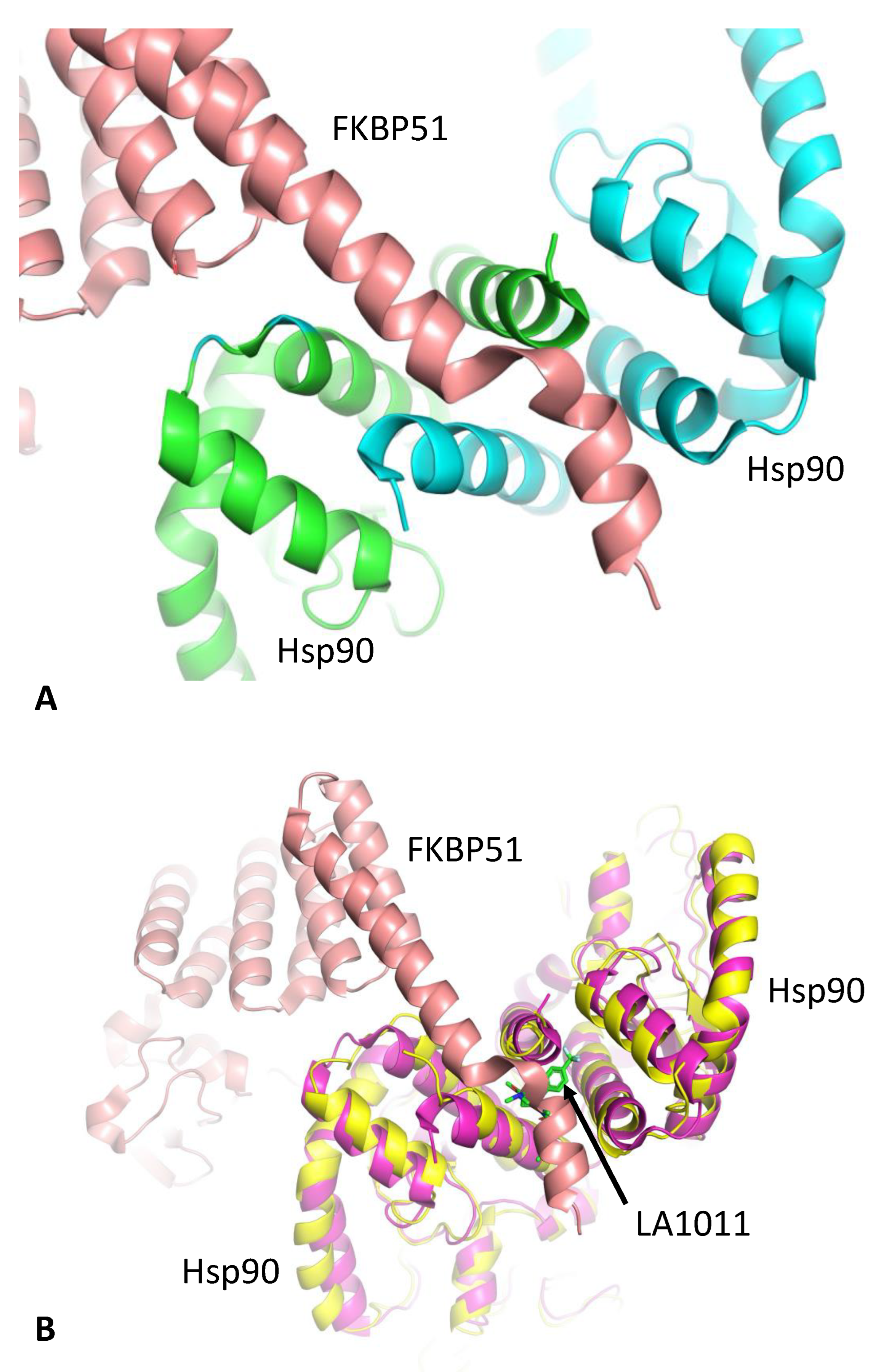
3.2. LA1011 and FKBP51 Competitively Bind to Hsp90
3.3. Effects of LA1011 on the Binding of Other TPR-Domain-Containing Co-Chaperones of Hsp90
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prodromou, C.; Bjorklund, D.M. Advances towards Understanding the Mechanism of Action of the Hsp90 Complex. Biomolecules 2022, 12, 600. [Google Scholar] [CrossRef]
- Whitesell, L.; Lindquist, S.L. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 2005, 5, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Pearl, L.H.; Prodromou, C.; Workman, P. The Hsp90 molecular chaperone: An open and shut case for treatment. Biochem. J. 2008, 410, 439–453. [Google Scholar] [CrossRef] [PubMed]
- Rawat, P.; Sehar, U.; Bisht, J.; Selman, A.; Culberson, J.; Reddy, P.H. Phosphorylated Tau in Alzheimer’s Disease and Other Tauopathies. Int. J. Mol. Sci. 2022, 23, 12841. [Google Scholar] [CrossRef]
- Choi, S.H.; Kim, Y.H.; D’avanzo, C.; Aronson, J.; Tanzi, R.E.; Kim, O.Y. Recapitulating amyloid β and tau pathology in human neural cell culture models: Clinical implications. US Neurol. 2015, 11, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.-R.; Tan, M.-S.; Xie, A.-M.; Yu, J.-T.; Tan, L. Heat Shock Protein 90 in Alzheimer’s Disease. BioMed Res. Int. 2014, 2014, 796869. [Google Scholar] [CrossRef] [PubMed]
- Koopman, M.B.; Rüdiger, S.G.D. Alzheimer Cells on Their Way to Derailment Show Selective Changes in Protein Quality Control Network. Front. Mol. Biosci. 2020, 7, 214. [Google Scholar] [CrossRef]
- Karagöz, G.E.; Duarte, A.M.; Akoury, E.; Ippel, H.; Biernat, J.; Luengo, T.M.; Radli, M.; Didenko, T.; Nordhues, B.A.; Veprintsev, D.B.; et al. Hsp90-Tau Complex Reveals Molecular Basis for Specificity in Chaperone Action. Cell 2014, 156, 963–974. [Google Scholar] [CrossRef]
- Weickert, S.; Wawrzyniuk, M.; John, L.H.; Rüdiger, S.G.D.; Drescher, M. The mechanism of Hsp90-induced oligomerizaton of Tau. Sci. Adv. 2020, 6, eaax6999. [Google Scholar] [CrossRef]
- Blair, L.J.; Sabbagh, J.J.; Dickey, C.A. Targeting Hsp90 and its co-chaperones to treat Alzheimer’s disease. Expert Opin. Ther. Targets 2014, 18, 1219–1232. [Google Scholar] [CrossRef]
- Jinwal, U.K.; Koren, J., 3rd; Dickey, C.A. Reconstructing the Hsp90/Tau Machine. Curr. Enzym. Inhib. 2013, 9, 41–45. [Google Scholar] [CrossRef]
- Jinwal, U.K.; Trotter, J.H.; Abisambra, J.F.; Koren, J., 3rd; Lawson, L.Y.; Vestal, G.D.; O’Leary, J.C., 3rd; Johnson, A.G.; Jin, Y.; Jones, J.R.; et al. The Hsp90 Kinase Co-chaperone Cdc37 Regulates Tau Stability and Phosphorylation Dynamics. J. Biol. Chem. 2011, 286, 16976–16983. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Selkoe, D.J. The Amyloid Hypothesis of Alzheimer’s Disease: Progress and Problems on the Road to Therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef]
- Selkoe, D.J. Alzheimer’s Disease Is a Synaptic Failure. Science 2002, 298, 789–791. [Google Scholar] [CrossRef] [PubMed]
- Oddo, S.; Caccamo, A.; Shepherd, J.D.; Murphy, M.P.; Golde, T.E.; Kayed, R.; Metherate, R.; Mattson, M.P.; Akbari, Y.; LaFerla, F.M. Triple-Transgenic Model of Alzheimer’s Disease with Plaques and Tangles: Intracellular Abeta and Synaptic Dysfunction. Neuron 2003, 39, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Tanzi, R.E.; Bertram, L. Twenty Years of the Alzheimer’s Disease Amyloid Hypothesis: A Genetic Perspective. Cell 2005, 120, 545–555. [Google Scholar] [CrossRef]
- Annaert, W.; De Strooper, B. Alzheimer’s Disease Neurons Fail the Acid Test. Cell 2010, 141, 1112–1114. [Google Scholar] [CrossRef]
- Karran, E.; Mercken, M.; De Strooper, B. The amyloid cascade hypothesis for Alzheimer’s disease: An appraisal for the development of therapeutics. Nat. Rev. Drug Discov. 2011, 10, 698–712. [Google Scholar] [CrossRef]
- Benilova, I.; Karran, E.; De Strooper, B. The toxic Aβ oligomer and Alzheimer’s disease: An emperor in need of clothes. Nat. Neurosci. 2012, 15, 349–357. [Google Scholar] [CrossRef]
- Munari, F.; Mollica, L.; Valente, C.; Parolini, F.; Kachoie, E.A.; Arrigoni, G.; D’Onofrio, M.; Capaldi, S.; Assfalg, M. Structural Basis for Chaperone-Independent Ubiquitination of Tau Protein by Its E3 Ligase CHIP. Angew. Chem. Int. Ed. Engl. 2022, 61, e202112374. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, Z.-W.; Mao, C.-Y.; Shi, C.-H.; Xu, Y.-M. CHIP as a therapeutic target for neurological diseases. Cell Death Dis. 2020, 11, 727. [Google Scholar] [CrossRef] [PubMed]
- Shelton, L.B.; Koren, J., 3rd; Blair, L.J. Imbalances in the Hsp90 Chaperone Machinery: Implications for Tauopathies. Front. Neurosci. 2017, 11, 724. [Google Scholar] [CrossRef]
- Baker, J.D.; Shelton, L.B.; Zheng, D.; Favretto, F.; Nordhues, B.A.; Darling, A.; Sullivan, L.E.; Sun, Z.; Solanki, P.K.; Martin, M.D.; et al. Human cyclophilin 40 unravels neurotoxic amyloids. PLoS Biol. 2017, 15, e2001336. [Google Scholar] [CrossRef] [PubMed]
- Brehme, M.; Voisine, C.; Rolland, T.; Wachi, S.; Soper, J.H.; Zhu, Y.; Orton, K.; Villella, A.; Garza, D.; Vidal, M.; et al. A Chaperome Subnetwork Safeguards Proteostasis in Aging and Neurodegenerative Disease. Cell Rep. 2014, 9, 1135–1150. [Google Scholar] [CrossRef]
- Blair, L.J.; Nordhues, B.A.; Hill, S.E.; Scaglione, K.M.; O’leary, J.C., 3rd; Fontaine, S.N.; Breydo, L.; Zhang, B.; Li, P.; Wang, L.; et al. Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. J. Clin. Investig. 2013, 123, 4158–4169. [Google Scholar] [CrossRef] [PubMed]
- Jinwal, U.K.; Koren, J., 3rd; Borysov, S.I.; Schmid, A.B.; Abisambra, J.F.; Blair, L.J.; Johnson, A.G.; Jones, J.R.; Shults, C.L.; O’Leary, J.C., 3rd; et al. The Hsp90 Cochaperone, FKBP51, Increases Tau Stability and Polymerizes Microtubules. J. Neurosci. 2010, 30, 591–599. [Google Scholar] [CrossRef]
- Sabbagh, J.J.; O’Leary, J.C., 3rd; Blair, L.J.; Klengel, T.; Nordhues, B.A.; Fontaine, S.N.; Binder, E.B.; Dickey, C.A. Age-Associated Epigenetic Upregulation of the FKBP5 Gene Selectively Impairs Stress Resiliency. PLoS ONE 2014, 9, e107241. [Google Scholar] [CrossRef]
- Kuznetsova, I.L.; Ponomareva, N.V.; Alemastseva, E.A.; Manakhov, A.D.; Andreeva, T.V.; Gusev, F.E.; Rogaev, E.I. The Interactive Effect of Genetic and Epigenetic Variations in FKBP5 and ApoE Genes on Anxiety and Brain EEG Parameters. Genes 2022, 13, 164. [Google Scholar] [CrossRef]
- Justice, N.J. The relationship between stress and Alzheimer’s disease. Neurobiol. Stress 2018, 8, 127–133. [Google Scholar] [CrossRef]
- Lee, K.; Thwin, A.C.; Nadel, C.M.; Tse, E.; Gates, S.N.; Gestwicki, J.E.; Southworth, D.R. The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state. Mol. Cell 2021, 81, 3496–3508.e3495. [Google Scholar] [CrossRef]
- Kamah, A.; Cantrelle, F.X.; Huvent, I.; Giustiniani, J.; Guillemeau, K.; Byrne, C.; Jacquot, Y.; Landrieu, I.; Baulieu, E.; Smet, C.; et al. Isomerization and Oligomerization of Truncated and Mutated Tau Forms by FKBP52 are Independent Processes. J. Mol. Biol. 2016, 428, 1080–1090. [Google Scholar] [CrossRef] [PubMed]
- Giustiniani, J.; Guillemeau, K.; Dounane, O.; Sardin, E.; Huvent, I.; Schmitt, A.; Hamdane, M.; Buée, L.; Landrieu, I.; Lippens, G.; et al. The FK506-binding protein FKBP52 in vitro induces aggregation of truncated Tau forms with prion-like behavior. FASEB J. 2015, 29, 3171–3181. [Google Scholar] [CrossRef] [PubMed]
- Meduri, G.; Guillemeau, K.; Dounane, O.; Sazdovitch, V.; Duyckaerts, C.; Chambraud, B.; Baulieu, E.E.; Giustiniani, J. Caspase-cleaved Tau-D421 is colocalized with the immunophilin FKBP52 in the autophagy-endolysosomal system of Alzheimer’s disease neurons. Neurobiol. Aging 2016, 46, 124–137. [Google Scholar] [CrossRef]
- Conde, R.; Xavier, J.; McLoughlin, C.; Chinkers, M.; Ovsenek, N. Protein phosphatase 5 Is a negative modulator of heat shock factor 1. J. Biol. Chem. 2005, 280, 28989–28996. [Google Scholar] [CrossRef]
- Gong, C.-X.; Liu, F.; Wu, G.; Rossie, S.; Wegiel, J.; Li, L.; Grundke-Iqbal, I.; Iqbal, K. Dephosphorylation of microtubule-associated protein tau by protein phosphatase 5. J. Neurochem. 2004, 88, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Grundke-Iqbal, I.; Iqbal, K.; Gong, C.-X. Contributions of protein phosphatases PP1, PP2A, PP2B and PP5 to the regulation of tau phosphorylation. Eur. J. Neurosci. 2005, 22, 1942–1950. [Google Scholar] [CrossRef] [PubMed]
- Hamos, J.E.; Oblas, B.; Pulaski-Salo, D.; Welch, W.J.; Bole, D.G.; Drachman, D.A. Expression of heat shock proteins in Alzheimer’s disease. Neurology 1991, 41, 345–350. [Google Scholar] [CrossRef]
- Alavez, S.; Vantipalli, M.C.; Zucker, D.J.S.; Klang, I.M.; Lithgow, G.J. Amyloid-binding compounds maintain protein homeostasis during ageing and extend lifespan. Nature 2011, 472, 226–229. [Google Scholar] [CrossRef]
- Dickey, C.A.; Kamal, A.; Lundgren, K.; Klosak, N.; Bailey, R.M.; Dunmore, J.; Ash, P.; Shoraka, S.; Zlatkovic, J.; Eckman, C.B.; et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Investig. 2007, 117, 648–658. [Google Scholar] [CrossRef]
- Luo, W.; Dou, F.; Rodina, A.; Chip, S.; Kim, J.; Zhao, Q.; Moulick, K.; Aguirre, J.; Wu, N.; Greengard, P.; et al. Roles of heat-shock protein 90 in maintaining and facilitating the neurodegenerative phenotype in tauopathies. Proc. Natl. Acad. Sci. USA 2007, 104, 9511–9516. [Google Scholar] [CrossRef]
- Kasza, A.; Hunya, A.; Frank, Z.; Fülöp, F.; Török, Z.; Balogh, G.; Sántha, M.; Bálind, A.; Bernáth, S.; Blundell, K.L.; et al. Dihydropyridine Derivatives Modulate Heat Shock Responses and have a Neuroprotective Effect in a Transgenic Mouse Model of Alzheimer’s Disease. J. Alzheimer’s Dis. 2016, 53, 557–571. [Google Scholar] [CrossRef] [PubMed]
- Roe, M.S.; Wahab, B.; Török, Z.; Horváth, I.; Vigh, L.; Prodromou, C. Dihydropyridines Allosterically Modulate Hsp90 Providing a Novel Mechanism for Heat Shock Protein Co-induction and Neuroprotection. Front. Mol. Biosci. 2018, 5, 51. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.M.; Hernández-Ramírez, L.C.; Trivellin, G.; Zhou, L.; Roe, S.M.; Korbonits, M.; Prodromou, C. Structure of the TPR Domain of AIP: Lack of Client Protein Interaction with the C-Terminal α-7 Helix of the TPR Domain of AIP Is Sufficient for Pituitary Adenoma Predisposition. PLoS ONE 2012, 7, e53339. [Google Scholar] [CrossRef] [PubMed]
- Panaretou, B.; Siligardi, G.; Meyer, P.; Maloney, A.; Sullivan, J.K.; Singh, S.; Millson, S.H.; Clarke, P.A.; Naaby-Hansen, S.; Stein, R.; et al. Activation of the ATPase activity of Hsp90 by the stress-regulated cochaperone aha1. Mol. Cell 2002, 10, 1307–1318. [Google Scholar] [CrossRef]
- Vonrhein, C.; Flensburg, C.; Keller, P.; Sharff, A.; Smart, O.; Paciorek, W.; Womack, T.; Bricogne, G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 293–302. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Struct. Biol. Crystallogr. 2004, D60, 2126–2132. [Google Scholar] [CrossRef]
- Oberoi, J.; Guiu, X.A.; Outwin, E.A.; Schellenberger, P.; Roumeliotis, T.I.; Choudhary, J.S.; Pearl, L.H. HSP90-CDC37-PP5 forms a structural platform for kinase dephosphorylation. Nat. Commun. 2022, 13, 7343. [Google Scholar] [CrossRef]
- Jaime-Garza, M.; Nowotny, C.A.; Coutandin, D.; Wang, F.; Tabios, M.; Agard, D.A. Hsp90 provides a platform for kinase dephosphorylation by PP5. Nat. Commun. 2023, 14, 2197. [Google Scholar] [CrossRef]
- Karran, E.; De Strooper, B. The amyloid hypothesis in Alzheimer disease: New insights from new therapeutics. Nat. Rev. Drug Discov. 2022, 21, 306–318. [Google Scholar] [CrossRef] [PubMed]


| Data Set (Highest Shell in Parentheses) | AutoPROC | STARANISO |
|---|---|---|
| a (Å) | 97.71 | |
| b (Å) | 97.71 | |
| c (Å) | 275.57 | |
| α (°) | 90.0 | |
| β (°) | 90.0 | |
| γ (°) | 90.0 | |
| Space group | P 41 21 21 | |
| Wavelength (Å) | 0.96864 | |
| Resolution limit (Å) | 92.1–3.18 (3.23–3.18) | 92.1–2.94 (3.19–2.94) |
| Number of unique obs. | 22,841 (1103) | 23,054 (1153) |
| Completeness (%) | 97.4 (97.0) | 91.9 (58.1) |
| Multiplicity | 9.3 (5.9) | 9.2 (4.0) |
| Rmerge (I) % | 0.218 (1.867) | 0.217 (1.215) |
| Rpim (I) % | 0.073 (0.803) | 0.073 (0.666) |
| CC1/2 | 0.995 (0.408) | 0.995 (0.361) |
| I/σI | 9.0 (1.1) | 9.0 (1.4) |
| Refinement | ||
| Resolution range (Å) | 79.7–2.94 | |
| Rcryst | 0.2470 | |
| Rfree | 0.3062 | |
| Number of protein atoms | 6816 | |
| Number of ligand atoms | 0 | |
| Number of solvent atoms | 1 | |
| Mean B | 70.11 | |
| Rmsd bond lengths (Å) | 0.005 | |
| Rmsd bond angles (°) | 1.051 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roe, S.M.; Török, Z.; McGown, A.; Horváth, I.; Spencer, J.; Pázmány, T.; Vigh, L.; Prodromou, C. The Crystal Structure of the Hsp90-LA1011 Complex and the Mechanism by Which LA1011 May Improve the Prognosis of Alzheimer’s Disease. Biomolecules 2023, 13, 1051. https://doi.org/10.3390/biom13071051
Roe SM, Török Z, McGown A, Horváth I, Spencer J, Pázmány T, Vigh L, Prodromou C. The Crystal Structure of the Hsp90-LA1011 Complex and the Mechanism by Which LA1011 May Improve the Prognosis of Alzheimer’s Disease. Biomolecules. 2023; 13(7):1051. https://doi.org/10.3390/biom13071051
Chicago/Turabian StyleRoe, S. Mark, Zsolt Török, Andrew McGown, Ibolya Horváth, John Spencer, Tamás Pázmány, László Vigh, and Chrisostomos Prodromou. 2023. "The Crystal Structure of the Hsp90-LA1011 Complex and the Mechanism by Which LA1011 May Improve the Prognosis of Alzheimer’s Disease" Biomolecules 13, no. 7: 1051. https://doi.org/10.3390/biom13071051
APA StyleRoe, S. M., Török, Z., McGown, A., Horváth, I., Spencer, J., Pázmány, T., Vigh, L., & Prodromou, C. (2023). The Crystal Structure of the Hsp90-LA1011 Complex and the Mechanism by Which LA1011 May Improve the Prognosis of Alzheimer’s Disease. Biomolecules, 13(7), 1051. https://doi.org/10.3390/biom13071051








