Phospholipases in Gliomas: Current Knowledge and Future Perspectives from Bench to Bedside
Abstract
1. Introduction
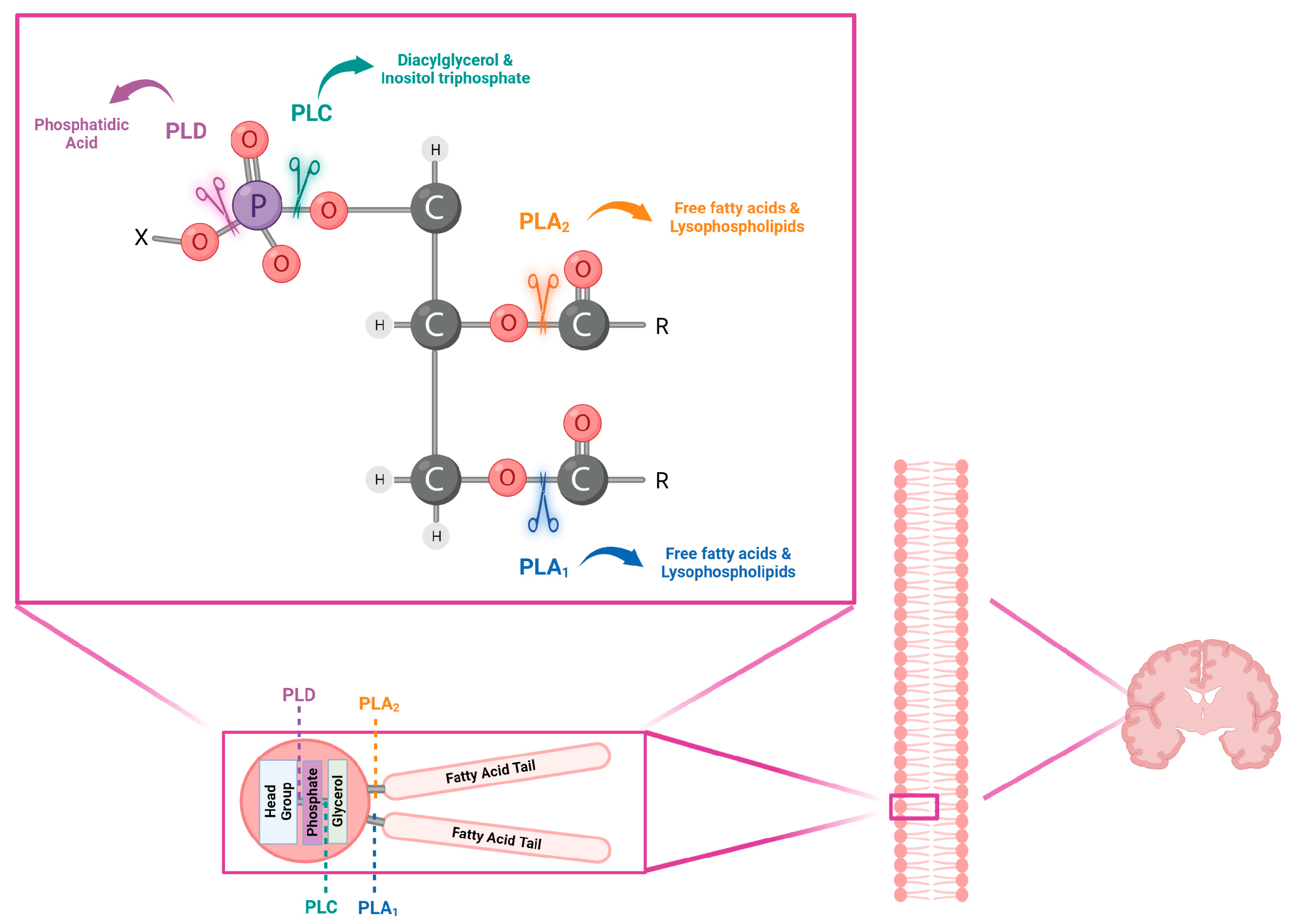
2. Phospholipases Characteristics
3. Phospholipase Signaling
4. Phospholipases in Brain Tumors
4.1. PLA2 Function and Involvement in Brain Tumors
4.2. PLCs’ Potential Role in the Aggressiveness of Low- and High-Grade Gliomas
4.3. PLD Involvement in Glioblastoma
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
| ATP | adenosine triphosphate |
| adPLA2 | adipose-specific phospholipase A2 |
| ATRX | ATRX chromatin remodeler |
| Ca2+ | calcium ions |
| CNS | Central Nervous System |
| CDKN2A−B | cyclin-dependent kinase inhibitors 2A and 2B |
| cPLA2 | cytosolic phospholipase A2 |
| DAG | diacylglycerol |
| EF-H | EF-hand |
| EGF | epidermal growth factor |
| EGFR | epidermal growth factor receptor |
| ERK | extracellular signal-regulated kinase |
| FGF | fibroblast growth factor |
| GSCs | glioma stem cells |
| GPCR | G-protein-coupled receptor |
| GTP | guanosine triphosphate |
| IP3 | inositol 1,4,5-trisphosphate |
| IGF | insulin-like growth factor |
| IDH | isocitrate dehydrogenase |
| LGG | low-grade glioma |
| LPA | lysophosphatidic acid |
| LPLA2 | lysosomal phospholipase A2 |
| MMPs | matrix metalloproteinases |
| MGMT | O-6-methylguanine-DNA methyltransferase |
| PTEN | phosphatase and tensin homolog |
| PA | phosphatidic acid |
| Ptdins(4,5)P2 | phosphatidylinositol 4,5-bisphosphate |
| PI-PLC | phosphatidylinositol-specific PLC |
| PI3K | phosphoinositide 3-kinases |
| PLA1 | phospholipase A1 |
| PLA2 | phospholipase A2 |
| PLB | phospholipase B |
| PLC | phospholipase C |
| PLD | phospholipase D |
| PLs | phospholipids |
| PX | phox consensus sequence |
| PDGF | platelet-derived growth factor |
| PH | pleckstrin homology |
| PTRF | polymerase I and transcript release factor |
| PKC | protein kinase C |
| Akt | RAC(Rho family)-alpha serine/threonine-protein kinase |
| ROS | reactive oxygen species |
| RTK | receptor tyrosine kinase |
| sPLA2 | secretory phospholipase A2 |
| SMase | sphingomyelinases |
| S1P | sphingosine-1-phosphate |
| SH2 | Src Homology 2 |
| sn-1 | stereospecific number-1 |
| sn-2 | stereospecific number-2 |
| sn-3 | stereospecific number-3 |
| TERT | telomerase reverse transcriptase |
| TMZ | temozolomide |
| TIM | triose phosphate isomerase |
| TP53 | tumor protein p53 |
| VEGF | vascular endothelial growth factor |
| WHO | World Health Organization |
References
- Joensuu, M.; Wallis, T.P.; Saber, S.H.; Meunier, F.A. Phospholipases in neuronal function: A role in learning and memory? J. Neurochem. 2020, 153, 300–333. [Google Scholar] [CrossRef]
- Lee, Y.J.; Shin, K.J.; Jang, H.J.; Noh, D.Y.; Ryu, S.H.; Suh, P.G. Phospholipase Signaling in Breast Cancer. Adv. Exp. Med. Biol. 2021, 1187, 23–52. [Google Scholar] [CrossRef]
- Park, J.B.; Lee, C.S.; Jang, J.H.; Ghim, J.; Kim, Y.J.; You, S.; Hwang, D.; Suh, P.G.; Ryu, S.H. Phospholipase signalling networks in cancer. Nat. Rev. Cancer 2012, 12, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lee, M.; Fairn, G.D. Phospholipid subcellular localization and dynamics. J. Biol. Chem. 2018, 293, 6230–6240. [Google Scholar] [CrossRef] [PubMed]
- Aloulou, A.; Rahier, R.; Arhab, Y.; Noiriel, A.; Abousalham, A. Phospholipases: An Overview. Methods Mol. Biol. 2018, 1835, 69–105. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Cerminati, S.; Paoletti, L.; Aguirre, A.; Peirú, S.; Menzella, H.G.; Castelli, M.E. Industrial uses of phospholipases: Current state and future applications. Appl. Microbiol. Biotechnol. 2019, 103, 2571–2582. [Google Scholar] [CrossRef] [PubMed]
- Owusu Obeng, E.; Rusciano, I.; Marvi, M.V.; Fazio, A.; Ratti, S.; Follo, M.Y.; Xian, J.; Manzoli, L.; Billi, A.M.; Mongiorgi, S.; et al. Phosphoinositide-Dependent Signaling in Cancer: A Focus on Phospholipase C Isozymes. Int. J. Mol. Sci. 2020, 21, 2581. [Google Scholar] [CrossRef]
- Ratti, S.; Mongiorgi, S.; Ramazzotti, G.; Follo, M.Y.; Mariani, G.A.; Suh, P.G.; McCubrey, J.A.; Cocco, L.; Manzoli, L. Nuclear Inositide Signaling Via Phospholipase, C. J. Cell. Biochem. 2017, 118, 1969–1978. [Google Scholar] [CrossRef]
- Gassama-Diagne, A.; Rogalle, P.; Fauvel, J.; Willson, M.; Klaébé, A.; Chap, H. Substrate specificity of phospholipase B from guinea pig intestine. A glycerol ester lipase with broad specificity. J. Biol. Chem. 1992, 267, 13418–13424. [Google Scholar] [CrossRef] [PubMed]
- Ali, U.; Lu, S.; Fadlalla, T.; Iqbal, S.; Yue, H.; Yang, B.; Hong, Y.; Wang, X.; Guo, L. The functions of phospholipases and their hydrolysis products in plant growth, development and stress responses. Prog. Lipid Res. 2022, 86, 101158. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Baba, T.; Sato, S.; Ohtsuki, R.; Takemori, A.; Watanabe, T.; Tagaya, M.; Tani, K. Roles of SAM and DDHD domains in mammalian intracellular phospholipase A1 KIAA0725p. Biochim. Biophys. Acta 2012, 1823, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Cocco, L.; Follo, M.Y.; Manzoli, L.; Suh, P.G. Phosphoinositide-specific phospholipase C in health and disease. J. Lipid Res. 2015, 56, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Béziau, D.M.; Toussaint, F.; Blanchette, A.; Dayeh, N.R.; Charbel, C.; Tardif, J.C.; Dupuis, J.; Ledoux, J. Expression of phosphoinositide-specific phospholipase C isoforms in native endothelial cells. PLoS ONE 2015, 10, e0123769. [Google Scholar] [CrossRef]
- Xian, J.; Owusu Obeng, E.; Ratti, S.; Rusciano, I.; Marvi, M.V.; Fazio, A.; De Stefano, A.; Mongiorgi, S.; Cappellini, A.; Ramazzotti, G.; et al. Nuclear Inositides and Inositide-Dependent Signaling Pathways in Myelodysplastic Syndromes. Cells 2020, 9, 697. [Google Scholar] [CrossRef] [PubMed]
- Ratti, S.; Follo, M.Y.; Ramazzotti, G.; Faenza, I.; Fiume, R.; Suh, P.G.; McCubrey, J.A.; Manzoli, L.; Cocco, L. Nuclear phospholipase C isoenzyme imbalance leads to pathologies in brain, hematologic, neuromuscular, and fertility disorders. J. Lipid Res. 2019, 60, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Fazio, A.; Owusu Obeng, E.; Rusciano, I.; Marvi, M.V.; Zoli, M.; Mongiorgi, S.; Ramazzotti, G.; Follo, M.Y.; McCubrey, J.A.; Cocco, L.; et al. Subcellular Localization Relevance and Cancer-Associated Mechanisms of Diacylglycerol Kinases. Int. J. Mol. Sci. 2020, 21, 5297. [Google Scholar] [CrossRef] [PubMed]
- Mongiorgi, S.; Follo, M.Y.; Yang, Y.R.; Ratti, S.; Manzoli, L.; McCubrey, J.A.; Billi, A.M.; Suh, P.G.; Cocco, L. Selective Activation of Nuclear PI-PLCbeta1 During Normal and Therapy-Related Differentiation. Curr. Pharm. Des. 2016, 22, 2345–2348. [Google Scholar] [CrossRef]
- Manzoli, L.; Billi, A.M.; Gilmour, R.S.; Martelli, A.M.; Matteucci, A.; Rubbini, S.; Weber, G.; Cocco, L. Phosphoinositide signaling in nuclei of Friend cells: Tiazofurin down-regulates phospholipase C beta 1. Cancer Res. 1995, 55, 2978–2980. [Google Scholar]
- García del Caño, G.; Montaña, M.; Aretxabala, X.; González-Burguera, I.; López de Jesús, M.; Barrondo, S.; Sallés, J. Nuclear phospholipase C-β1 and diacylglycerol LIPASE-α in brain cortical neurons. Adv. Biol. Regul. 2014, 54, 12–23. [Google Scholar] [CrossRef]
- Nelson, R.K.; Frohman, M.A. Physiological and pathophysiological roles for phospholipase D. J. Lipid Res. 2015, 56, 2229–2237. [Google Scholar] [CrossRef]
- Gresset, A.; Sondek, J.; Harden, T.K. The phospholipase C isozymes and their regulation. Subcell. Biochem. 2012, 58, 61–94. [Google Scholar] [CrossRef]
- Poli, A.; Ratti, S.; Finelli, C.; Mongiorgi, S.; Clissa, C.; Lonetti, A.; Cappellini, A.; Catozzi, A.; Barraco, M.; Suh, P.G.; et al. Nuclear translocation of PKC-α is associated with cell cycle arrest and erythroid differentiation in myelodysplastic syndromes (MDSs). FASEB J. 2018, 32, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Han, J.M.; Park, J.B.; Lee, S.D.; Oh, Y.S.; Chung, C.; Lee, T.G.; Kim, J.H.; Park, S.K.; Yoo, J.S.; et al. Phosphorylation and activation of phospholipase D1 by protein kinase C in vivo: Determination of multiple phosphorylation sites. Biochemistry 1999, 38, 10344–10351. [Google Scholar] [CrossRef] [PubMed]
- Rusciano, I.; Marvi, M.V.; Owusu Obeng, E.; Mongiorgi, S.; Ramazzotti, G.; Follo, M.Y.; Zoli, M.; Morandi, L.; Asioli, S.; Fabbri, V.P.; et al. Location-dependent role of phospholipase C signaling in the brain: Physiology and pathology. Adv. Biol. Regul. 2021, 79, 100771. [Google Scholar] [CrossRef]
- Balboa, M.A.; Varela-Nieto, I.; Killermann Lucas, K.; Dennis, E.A. Expression and function of phospholipase A(2) in brain. FEBS Lett. 2002, 531, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.; Martín, R.; García-Cubillas, M.D.; Maeso-Hernández, P.; Nieto, M.L. Secreted PLA2 induces proliferation in astrocytoma through the EGF receptor: Another inflammation-cancer link. Neuro-oncology 2010, 12, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Ferro, F.; Servais, S.; Besson, P.; Roger, S.; Dumas, J.F.; Brisson, L. Autophagy and mitophagy in cancer metabolic remodelling. Semin. Cell. Dev. Biol. 2020, 98, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Wang, L.; Jin, W.; Liu, X.; Wang, Q.; Wu, Y.; Tan, Y.; Wang, Y.; Cui, X.; Zhao, J.; et al. PTRF/Cavin-1 enhances chemo-resistance and promotes temozolomide efflux through extracellular vesicles in glioblastoma. Theranostics 2022, 12, 4330–4347. [Google Scholar] [CrossRef]
- Yi, K.; Zhan, Q.; Wang, Q.; Tan, Y.; Fang, C.; Wang, Y.; Zhou, J.; Yang, C.; Li, Y.; Kang, C. PTRF/cavin-1 remodels phospholipid metabolism to promote tumor proliferation and suppress immune responses in glioblastoma by stabilizing cPLA2. Neuro-oncology 2021, 23, 387–399. [Google Scholar] [CrossRef]
- Zhan, Q.; Yi, K.; Cui, X.; Li, X.; Yang, S.; Wang, Q.; Fang, C.; Tan, Y.; Li, L.; Xu, C.; et al. Blood exosomes-based targeted delivery of cPLA2 siRNA and metformin to modulate glioblastoma energy metabolism for tailoring personalized therapy. Neuro-oncology 2022, 24, 1871–1883. [Google Scholar] [CrossRef]
- Tsuji, S.; Ohno, Y.; Nakamura, S.; Yamada, T.; Noda, Y.; Saio, M.; Iwama, T.; Shimazawa, M.; Hara, H. Temozolomide has anti-tumor effects through the phosphorylation of cPLA. Brain Res. 2019, 1723, 146396. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Baranowska-Bosiacka, I.; Gutowska, I.; Chlubek, D. The effect of reactive oxygen species on the synthesis of prostanoids from arachidonic acid. J. Physiol. Pharmacol. 2013, 64, 409–421. [Google Scholar] [PubMed]
- Vento, R.; D’Alessandro, N.; Giuliano, M.; Lauricella, M.; Carabillò, M.; Tesoriere, G. Induction of apoptosis by arachidonic acid in human retinoblastoma Y79 cells: Involvement of oxidative stress. Exp. Eye Res. 2000, 70, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Ratti, S.; Marvi, M.V.; Mongiorgi, S.; Obeng, E.O.; Rusciano, I.; Ramazzotti, G.; Morandi, L.; Asioli, S.; Zoli, M.; Mazzatenta, D.; et al. Impact of phospholipase C β1 in glioblastoma: A study on the main mechanisms of tumor aggressiveness. Cell. Mol. Life Sci. 2022, 79, 195. [Google Scholar] [CrossRef]
- Marvi, M.V.; Mongiorgi, S.; Ramazzotti, G.; Follo, M.Y.; Billi, A.M.; Zoli, M.; Mazzatenta, D.; Morandi, L.; Asioli, S.; Papa, V.; et al. Role of PLCγ1 in the modulation of cell migration and cell invasion in glioblastoma. Adv. Biol. Regul. 2021, 83, 100838. [Google Scholar] [CrossRef]
- Lu, G.; Chang, J.T.; Liu, Z.; Chen, Y.; Li, M.; Zhu, J.J. Phospholipase C Beta 1: A Candidate Signature Gene for Proneural Subtype High-Grade Glioma. Mol. Neurobiol. 2016, 53, 6511–6525. [Google Scholar] [CrossRef]
- Engebraaten, O.; Bjerkvig, R.; Pedersen, P.H.; Laerum, O.D. Effects of EGF, bFGF, NGF and PDGF(bb) on cell proliferative, migratory and invasive capacities of human brain-tumour biopsies in vitro. Int. J. Cancer 1993, 53, 209–214. [Google Scholar] [CrossRef]
- Khoshyomn, S.; Penar, P.L.; Rossi, J.; Wells, A.; Abramson, D.L.; Bhushan, A. Inhibition of phospholipase C-gamma1 activation blocks glioma cell motility and invasion of fetal rat brain aggregates. Neurosurgery 1999, 44, 568–577, discussion 577–568. [Google Scholar] [CrossRef]
- Li, T.; Yang, Z.; Li, H.; Zhu, J.; Wang, Y.; Tang, Q.; Shi, Z. Phospholipase Cγ1 (PLCG1) overexpression is associated with tumor growth and poor survival in IDH wild-type lower-grade gliomas in adult patients. Lab. Investig. 2022, 102, 143–153. [Google Scholar] [CrossRef]
- Brat, D.J.; Verhaak, R.G.; Aldape, K.D.; Yung, W.K.; Salama, S.R.; Cooper, L.A.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; Morozova, O.; et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef]
- Okada, M.; Nakagawa-Saito, Y.; Mitobe, Y.; Sugai, A.; Togashi, K.; Suzuki, S.; Kitanaka, C. Inhibition of the Phospholipase Cε-c-Jun N-Terminal Kinase Axis Suppresses Glioma Stem Cell Properties. Int. J. Mol. Sci. 2022, 23, 8785. [Google Scholar] [CrossRef]
- Zepecki, J.P.; Snyder, K.M.; Moreno, M.M.; Fajardo, E.; Fiser, A.; Ness, J.; Sarkar, A.; Toms, S.A.; Tapinos, N. Regulation of human glioma cell migration, tumor growth, and stemness gene expression using a Lck targeted inhibitor. Oncogene 2019, 38, 1734–1750. [Google Scholar] [CrossRef]
- Ramazzotti, G.; Ratti, S.; Fiume, R.; Follo, M.Y.; Billi, A.M.; Rusciano, I.; Owusu Obeng, E.; Manzoli, L.; Cocco, L.; Faenza, I. Phosphoinositide 3 Kinase Signaling in Human Stem Cells from Reprogramming to Differentiation: A Tale in Cytoplasmic and Nuclear Compartments. Int. J. Mol. Sci. 2019, 20, 2026. [Google Scholar] [CrossRef]
- Hawkins, C.C.; Ali, T.; Ramanadham, S.; Hjelmeland, A.B. Sphingolipid Metabolism in Glioblastoma and Metastatic Brain Tumors: A Review of Sphingomyelinases and Sphingosine-1-Phosphate. Biomolecules 2020, 10, 1357. [Google Scholar] [CrossRef] [PubMed]
- Bruntz, R.C.; Taylor, H.E.; Lindsley, C.W.; Brown, H.A. Phospholipase D2 mediates survival signaling through direct regulation of Akt in glioblastoma cells. J. Biol. Chem. 2014, 289, 600–616. [Google Scholar] [CrossRef]
- McDowell, K.A.; Riggins, G.J.; Gallia, G.L. Targeting the AKT pathway in glioblastoma. Curr. Pharm. Des. 2011, 17, 2411–2420. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.W.; Hwang, W.C.; Noh, Y.N.; Park, K.S.; Min, D.S. Phospholipase D1 inhibition sensitizes glioblastoma to temozolomide and suppresses its tumorigenicity. J. Pathol. 2020, 252, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Shoji, T.; Hayashi, M.; Sumi, C.; Kusunoki, M.; Uba, T.; Matsuo, Y.; Hirota, K. Sevoflurane Does Not Promote the Colony-Forming Ability of Human Mesenchymal Glioblastoma Stem Cells In Vitro. Medicina 2022, 58, 1614. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Lv, X.; Lu, C.; Ye, X.; Chen, X.; Fu, J.; Luo, C.; Zhao, Y. Prognostic factors of patients with Gliomas—An analysis on 335 patients with Glioblastoma and other forms of Gliomas. BMC Cancer 2020, 20, 35. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Cheng, S.Y. Targeting phospholipid metabolism for glioblastoma therapy. Neuro-oncology 2021, 23, 343–344. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Bhujwalla, Z.M.; Glunde, K. Targeting Phospholipid Metabolism in Cancer. Front. Oncol. 2016, 6, 266. [Google Scholar] [CrossRef] [PubMed]
| Phospholipase | Brain Tumor Type | Expression | Clinical Potential | References |
|---|---|---|---|---|
| sPLA2 | Astrocytoma | Increased | Therapeutic target | [27] |
| cPLA2 | Glioblastoma | Increased | Therapeutic target | [30,31] |
| PLCβ1 | Glioblastoma | Decreased | Prognostic biomarker | [36,38] |
| PLCγ1 | Glioblastoma and LGG (IDH-wt) | Increased | Therapeutic target | [37,40,41] |
| PLCε | Glioblastoma | Increased | Therapeutic target | [43] |
| PLD2 | Glioblastoma | Increased | Therapeutic target | [47] |
| PLD1 | Glioblastoma | Increased | Therapeutic target | [49] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marvi, M.V.; Neri, I.; Evangelisti, C.; Ramazzotti, G.; Asioli, S.; Zoli, M.; Mazzatenta, D.; Neri, N.; Morandi, L.; Tonon, C.; et al. Phospholipases in Gliomas: Current Knowledge and Future Perspectives from Bench to Bedside. Biomolecules 2023, 13, 798. https://doi.org/10.3390/biom13050798
Marvi MV, Neri I, Evangelisti C, Ramazzotti G, Asioli S, Zoli M, Mazzatenta D, Neri N, Morandi L, Tonon C, et al. Phospholipases in Gliomas: Current Knowledge and Future Perspectives from Bench to Bedside. Biomolecules. 2023; 13(5):798. https://doi.org/10.3390/biom13050798
Chicago/Turabian StyleMarvi, Maria Vittoria, Irene Neri, Camilla Evangelisti, Giulia Ramazzotti, Sofia Asioli, Matteo Zoli, Diego Mazzatenta, Niccolò Neri, Luca Morandi, Caterina Tonon, and et al. 2023. "Phospholipases in Gliomas: Current Knowledge and Future Perspectives from Bench to Bedside" Biomolecules 13, no. 5: 798. https://doi.org/10.3390/biom13050798
APA StyleMarvi, M. V., Neri, I., Evangelisti, C., Ramazzotti, G., Asioli, S., Zoli, M., Mazzatenta, D., Neri, N., Morandi, L., Tonon, C., Lodi, R., Franceschi, E., McCubrey, J. A., Suh, P.-G., Manzoli, L., & Ratti, S. (2023). Phospholipases in Gliomas: Current Knowledge and Future Perspectives from Bench to Bedside. Biomolecules, 13(5), 798. https://doi.org/10.3390/biom13050798