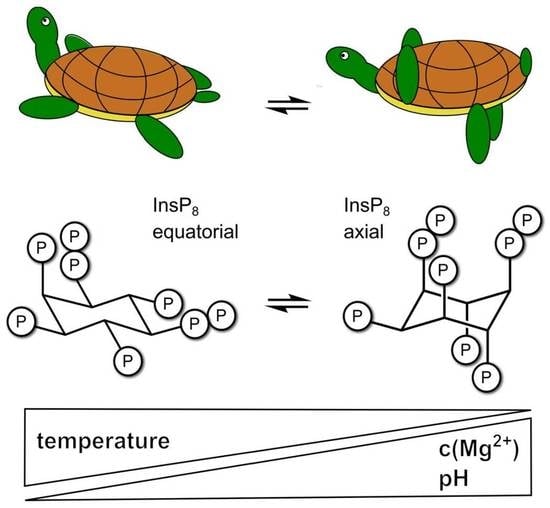
One Scaffold, Two Conformations: The Ring-Flip of the Messenger InsP8 Occurs under Cytosolic Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthesis and BIRD-HMQC NMR Analysis of InsPs
2.2. Assignment of HMQC-NMR Spectra of Axial Conformations
2.3. Van’t Hoff Thermodynamic Analysis
2.4. NMR Titrations
2.5. Isothermal Titration Calorimetry
2.6. DFT Calculations
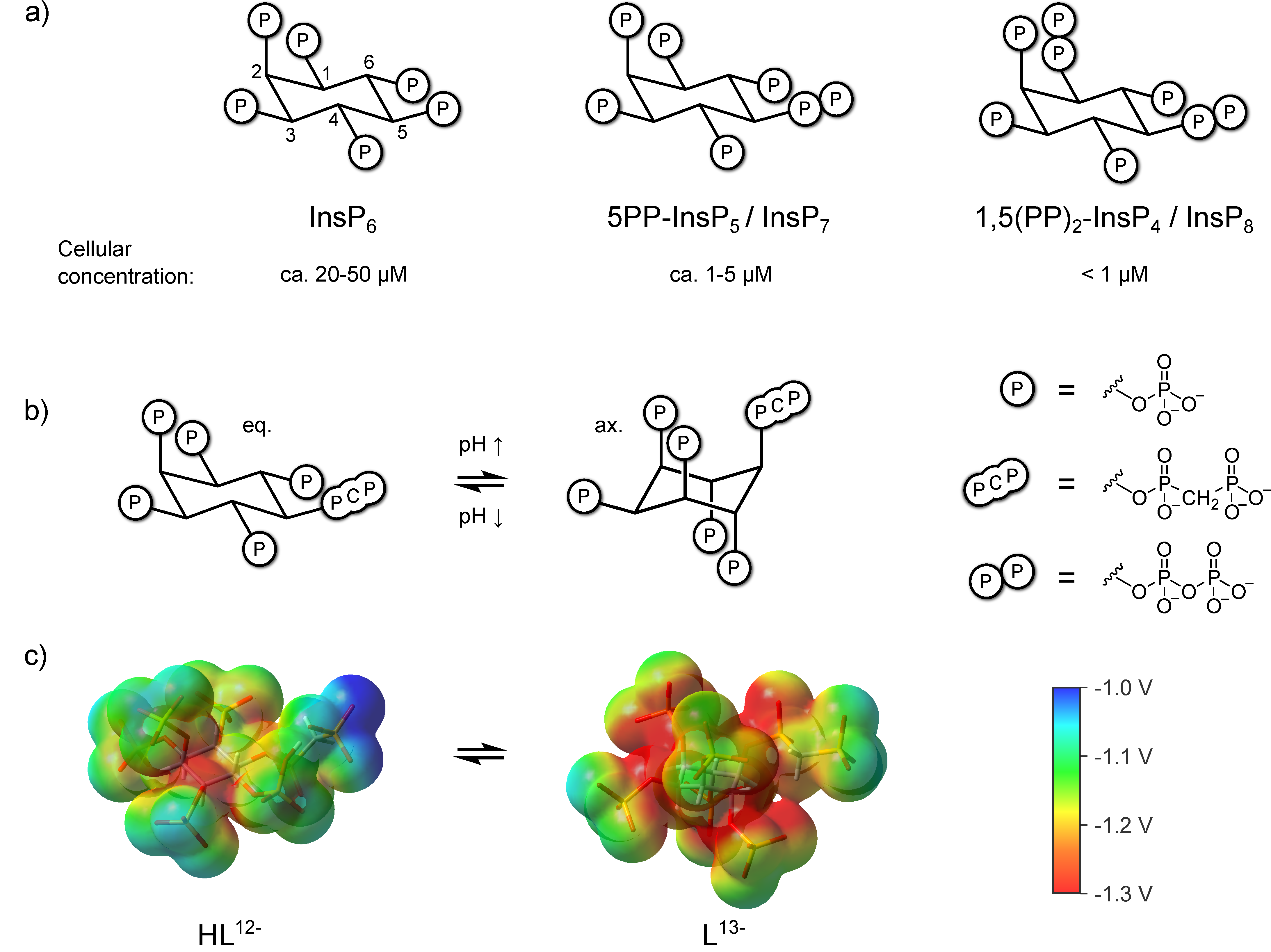
3. Results
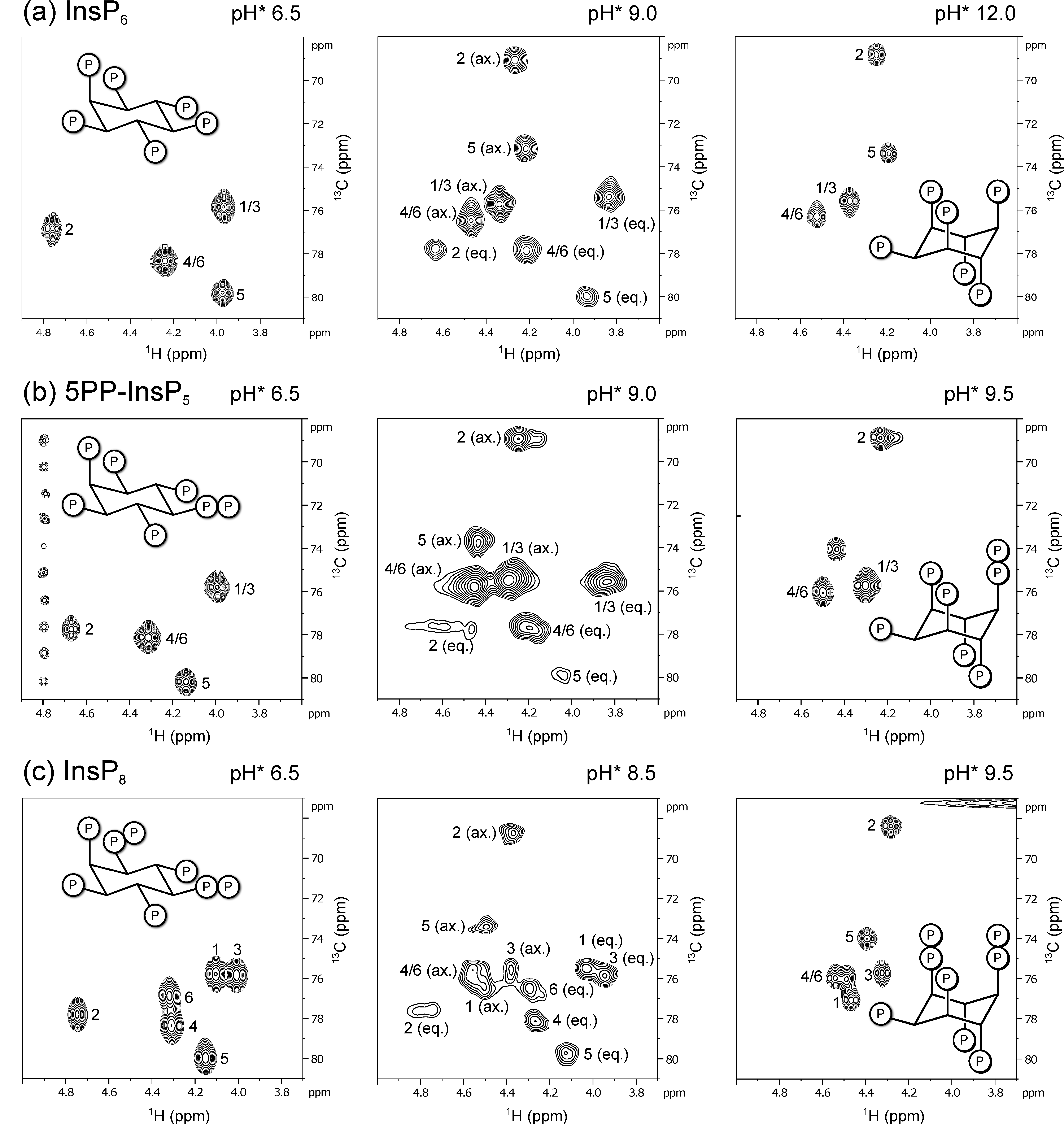
3.1. 1H,13C-HMQC NMR Spectra of PP-InsPs Can Show Two Conformations Simultaneously
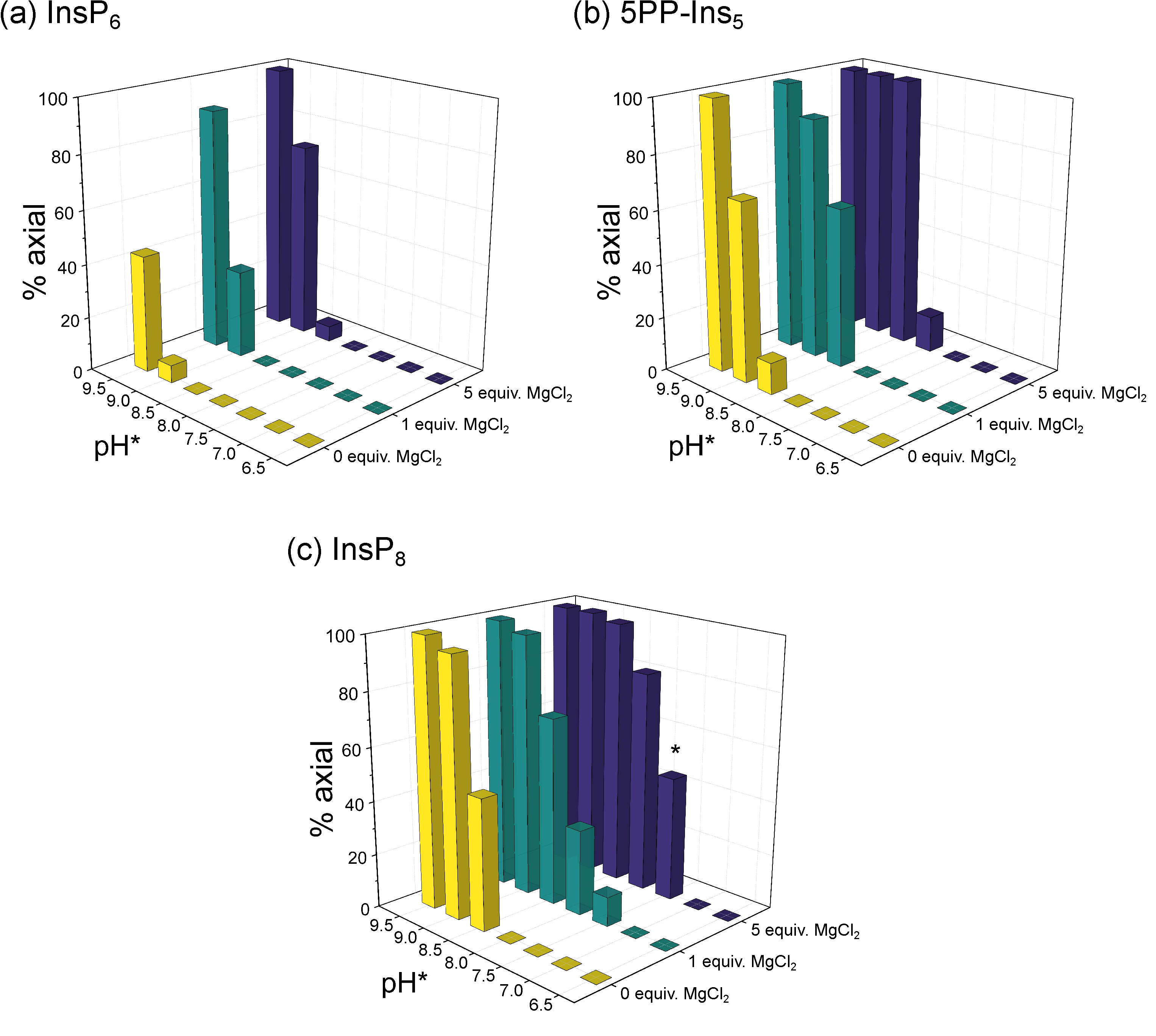
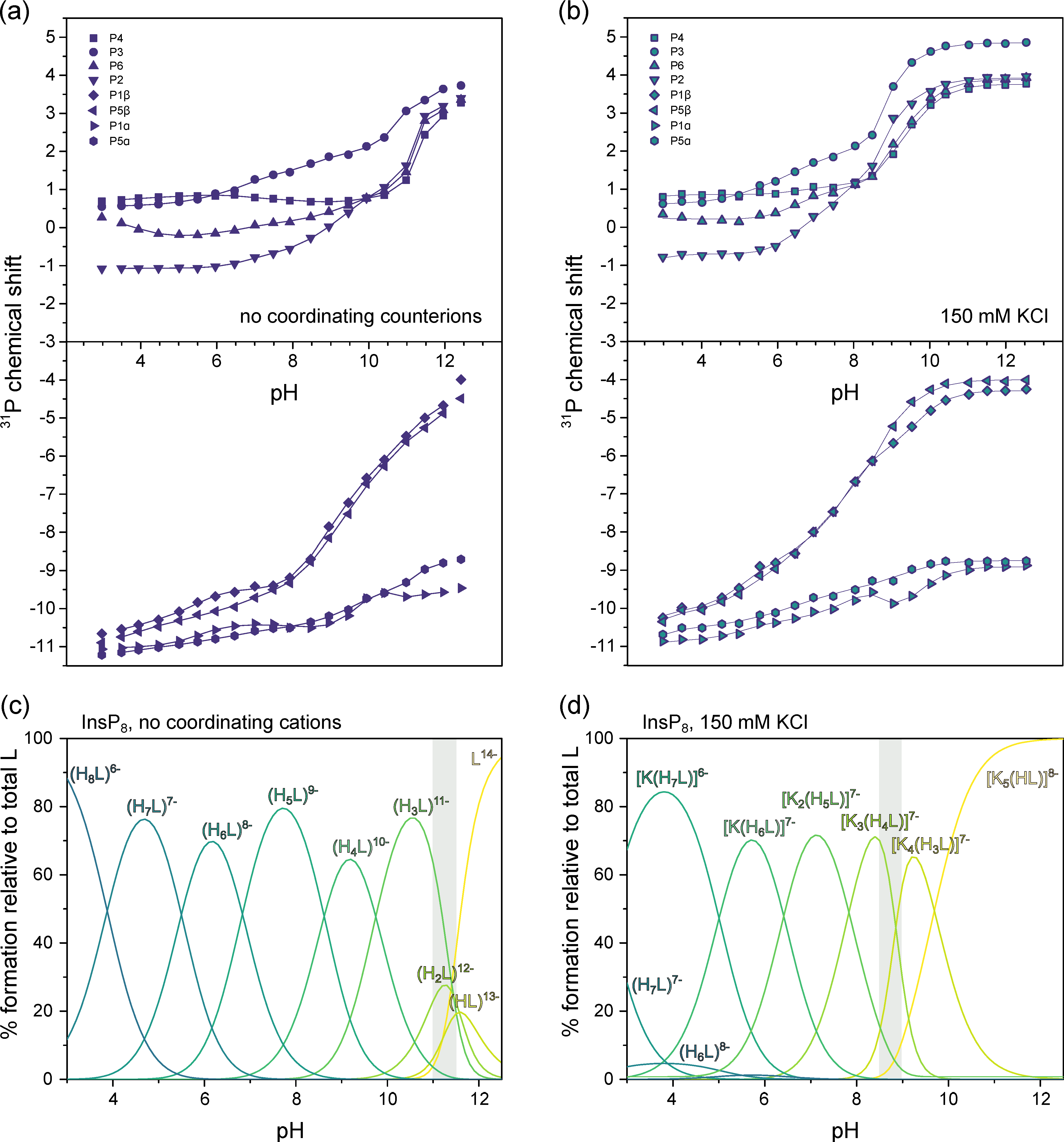
3.2. Conformational Equilibria of InsP6 and PP-InsPs Are Sensitive to pH and Ionic Composition
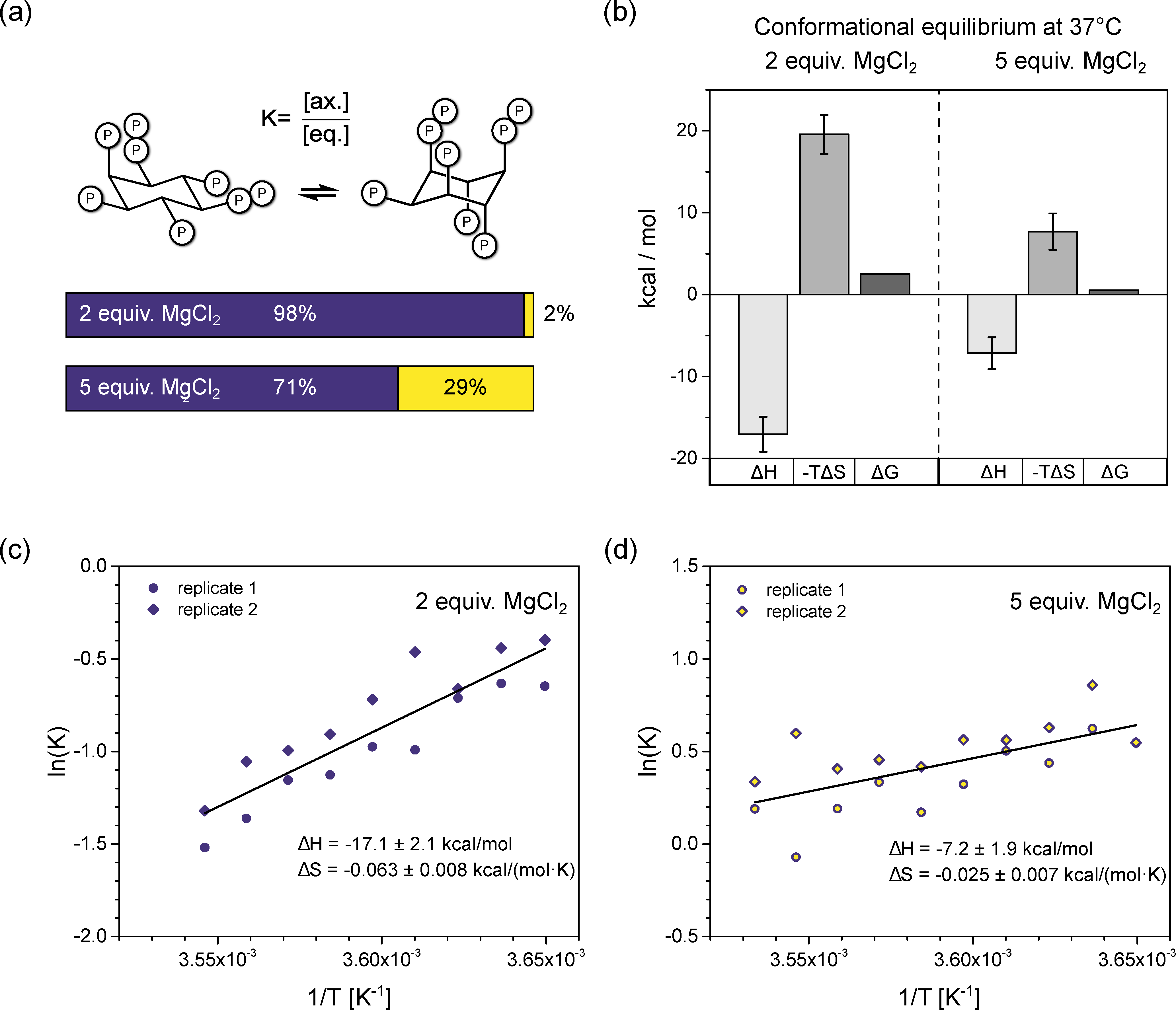
3.3. InsP8 Is Present in Both Conformations under Near-Physiological Conditions
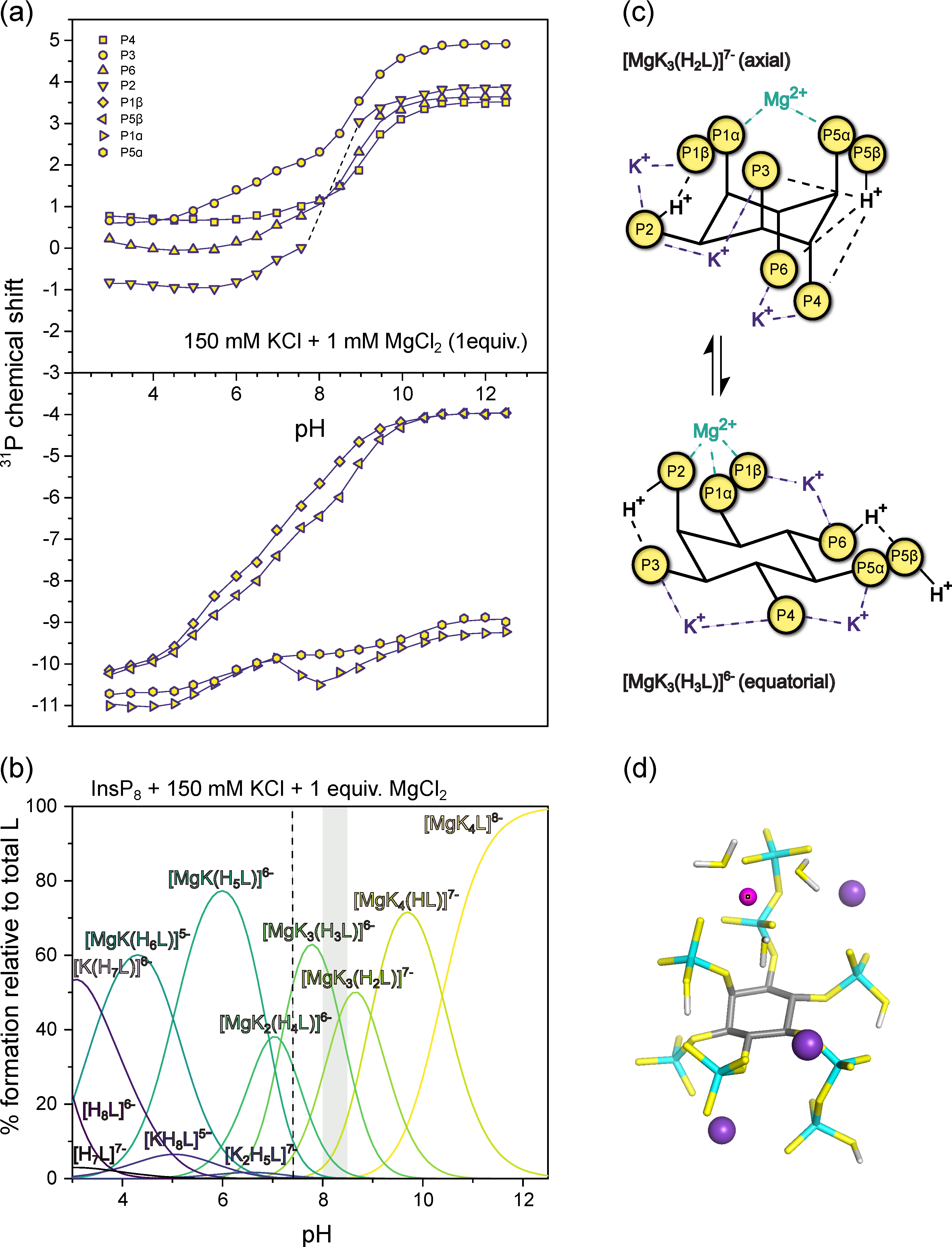
3.4. InsP8 Forms Strong Complexes with Potassium and Magnesium Ions
3.5. Complex Speciation of InsP8 Affects Protein Binding
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chatree, S.; Thongmaen, N.; Tantivejkul, K.; Sitticharoon, C.; Vucenik, I. Role of Inositols and Inositol Phosphates in Energy Metabolism. Molecules 2020, 25, 5079. [Google Scholar] [CrossRef]
- Shears, S.B. Inositol Pyrophosphates: Why so Many Phosphates? Adv. Biol. Regul. 2015, 57, 203–216. [Google Scholar] [CrossRef]
- Nguyen Trung, M.; Furkert, D.; Fiedler, D. Versatile Signaling Mechanisms of Inositol Pyrophosphates. Curr. Opin. Chem. Biol. 2022, 70, 102177. [Google Scholar] [CrossRef]
- Gu, C.; Nguyen, H.N.; Hofer, A.; Jessen, H.J.; Dai, X.; Wang, H.; Shears, S.B. The Significance of the Bifunctional Kinase/Phosphatase Activities of Diphosphoinositol Pentakisphosphate Kinases (PPIP5Ks) for Coupling Inositol Pyrophosphate Cell Signaling to Cellular Phosphate Homeostasis. J. Biol. Chem. 2017, 292, 4544–4555. [Google Scholar] [CrossRef]
- Benjamin, B.; Garg, A.; Jork, N.; Jessen, H.J.; Schwer, B.; Shuman, S. Activities and Structure-Function Analysis of Fission Yeast Inositol Pyrophosphate (IPP) Kinase-Pyrophosphatase Asp1 and Its Impact on Regulation of Pho1 Gene Expression. MBio 2022, 13, e0103422. [Google Scholar] [CrossRef]
- Pascual-Ortiz, M.; Walla, E.; Fleig, U.; Saiardi, A. The PPIP5K Family Member Asp1 Controls Inorganic Polyphosphate Metabolism in S. Pombe. J. Fungi 2021, 7, 626. [Google Scholar] [CrossRef]
- Zhu, J.; Lau, K.; Puschmann, R.; Harmel, R.K.; Zhang, Y.; Pries, V.; Gaugler, P.; Broger, L.; Dutta, A.K.; Jessen, H.J.; et al. Two Bifunctional Inositol Pyrophosphate Kinases/Phosphatases Control Plant Phosphate Homeostasis. Elife 2019, 8, e43582. [Google Scholar] [CrossRef]
- Dong, J.; Ma, G.; Sui, L.; Wei, M.; Satheesh, V.; Zhang, R.; Ge, S.; Li, J.; Zhang, T.E.; Wittwer, C.; et al. Inositol Pyrophosphate InsP8 Acts as an Intracellular Phosphate Signal in Arabidopsis. Mol. Plant 2019, 12, 1463–1473. [Google Scholar] [CrossRef]
- Li, X.; Gu, C.; Hostachy, S.; Sahu, S.; Wittwer, C.; Jessen, H.J.; Fiedler, D.; Wang, H.; Shears, S.B. Control of XPR1-Dependent Cellular Phosphate Efflux by InsP8 Is an Exemplar for Functionally-Exclusive Inositol Pyrophosphate Signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 3568–3574. [Google Scholar] [CrossRef]
- Wilson, M.S.; Jessen, H.J.; Saiardi, A. The Inositol Hexakisphosphate Kinases IP6K1 and -2 Regulate Human Cellular Phosphate Homeostasis, Including XPR1-Mediated Phosphate Export. J. Biol. Chem. 2019, 294, 11597–11608. [Google Scholar] [CrossRef]
- Legati, A.; Giovannini, D.; Nicolas, G.; López-Sánchez, U.; Quintáns, B.; Oliveira, J.R.M.; Sears, R.L.; Ramos, E.M.; Spiteri, E.; Sobrido, M.-J.; et al. Mutations in XPR1 Cause Primary Familial Brain Calcification Associated with Altered Phosphate Export. Nat. Genet. 2015, 47, 579–581. [Google Scholar] [CrossRef]
- Yousaf, R.; Gu, C.; Ahmed, Z.M.; Khan, S.N.; Friedman, T.B.; Riazuddin, S.; Shears, S.B.; Riazuddin, S. Mutations in Diphosphoinositol-Pentakisphosphate Kinase PPIP5K2 Are Associated with Hearing Loss in Human and Mouse. PLoS Genet. 2018, 14, e1007897. [Google Scholar] [CrossRef]
- Khaled, M.L.; Bykhovskaya, Y.; Gu, C.; Liu, A.; Drewry, M.D.; Chen, Z.; Mysona, B.A.; Parker, E.; McNabb, R.P.; Yu, H.; et al. PPIP5K2 and PCSK1 Are Candidate Genetic Contributors to Familial Keratoconus. Sci. Rep. 2019, 9, 19406. [Google Scholar] [CrossRef]
- Szijgyarto, Z.; Garedew, A.; Azevedo, C.; Saiardi, A. Influence of Inositol Pyrophosphates on Cellular Energy Dynamics. Science 2011, 334, 802–805. [Google Scholar] [CrossRef]
- Qin, N.; Li, L.; Ji, X.; Pereira, R.; Chen, Y.; Yin, S.; Li, C.; Wan, X.; Qiu, D.; Jiang, J.; et al. Flux Regulation through Glycolysis and Respiration Is Balanced by Inositol Pyrophosphates in Yeast. Cell 2023, 186, 748–763.e15. [Google Scholar] [CrossRef]
- Illies, C.; Gromada, J.; Fiume, R.; Leibiger, B.; Yu, J.; Juhl, K.; Yang, S.-N.; Barma, D.K.; Falck, J.R.; Saiardi, A.; et al. Requirement of Inositol Pyrophosphates for Full Exocytotic Capacity in Pancreatic b Cells. Science 2007, 318, 1299–1302. [Google Scholar] [CrossRef]
- Zhang, X.; Li, N.; Zhang, J.; Zhang, Y.; Yang, X.; Luo, Y.; Zhang, B.; Xu, Z.; Zhu, Z.; Yang, X.; et al. 5-IP7 Is a GPCR Messenger Mediating Neural Control of Synaptotagmin-Dependent Insulin Exocytosis and Glucose Homeostasis. Nat. Metab. 2021, 3, 1400–1414. [Google Scholar] [CrossRef]
- Chakraborty, A.; Koldobskiy, M.A.; Bello, N.T.; Maxwell, M.; Potter, J.J.; Juluri, K.R.; Maag, D.; Kim, S.; Huang, A.S.; Dailey, M.J.; et al. Inositol Pyrophosphates Inhibit Akt Signaling, Thereby Regulating Insulin Sensitivity and Weight Gain. Cell 2010, 143, 897–910. [Google Scholar] [CrossRef]
- Pavlovic, I.; Thakor, D.T.; Vargas, J.R.; McKinlay, C.J.; Hauke, S.; Anstaett, P.; Camunã, R.C.; Bigler, L.; Gasser, G.; Schultz, C.; et al. Cellular Delivery and Photochemical Release of a Caged Inositol-Pyrophosphate Induces PH-Domain Translocation in Cellulo. Nat. Commun. 2016, 7, 10622. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, C.; Liu, B.; Liang, D.; Qin, X.; Li, X.; Zhang, R.; Li, C.; Wang, H.; Sun, D.; et al. Inositol Pyrophosphates Mediate the Effects of Aging on Bone Marrow Mesenchymal Stem Cells by Inhibiting Akt Signaling. Stem Cell Res. Ther. 2014, 5, 33. [Google Scholar] [CrossRef]
- Zhu, Q.; Ghoshal, S.; Rodrigues, A.; Gao, S.; Asterian, A.; Kamenecka, T.M.; Barrow, J.C.; Chakraborty, A. Adipocyte-Specific Deletion of Ip6k1 Reduces Diet-Induced Obesity by Enhancing AMPK-Mediated Thermogenesis. J. Clin. Investig. 2016, 126, 4273–4288. [Google Scholar] [CrossRef]
- Moritoh, Y.; Abe, S.-I.; Akiyama, H.; Kobayashi, A.; Koyama, R.; Hara, R.; Kasai, S.; Watanabe, M. The Enzymatic Activity of Inositol Hexakisphosphate Kinase Controls Circulating Phosphate in Mammals. Nat. Commun. 2021, 12, 4847. [Google Scholar] [CrossRef]
- Gerasimaite, R.; Pavlovic, I.; Capolicchio, S.; Hofer, A.; Schmidt, A.; Jessen, H.J.; Mayer, A. Inositol Pyrophosphate Specificity of the SPX-Dependent Polyphosphate Polymerase VTC. ACS Chem. Biol. 2017, 12, 648–653. [Google Scholar] [CrossRef]
- Veiga, N.; Torres, J.; Godage, H.Y.; Riley, A.M.; Domínguez, S.; Potter, B.V.L.; Díaz, A.; Kremer, C. The Behaviour of Inositol 1,3,4,5,6-Pentakisphosphate in the Presence of the Major Biological Metal Cations. J. Biol. Inorg. Chem. 2009, 14, 1001–1013. [Google Scholar] [CrossRef]
- Veiga, N.; Torres, J.; Cerdá, F.; González, G.; Gómez, K.; Mansell, D.; Freeman, S.; Domínguez, S.; Kremer, C. Redox and Structural Aspects on Iron Inositol 1,2,3-Trisphosphate Interaction: An Experimental and Computational Approach. J. Mol. Struct. 2011, 994, 343–349. [Google Scholar] [CrossRef]
- Veiga, N.; Torres, J.; MacHo, I.; Gómez, K.; González, G.; Kremer, C. Coordination, Microprotonation Equilibria and Conformational Changes of Myo-Inositol Hexakisphosphate with Pertinence to Its Biological Function. Dalt. Trans. 2014, 43, 16238–16251. [Google Scholar] [CrossRef]
- Riley, A.M.; Trusselle, M.; Kuad, P.; Borkovec, M.; Cho, J.; Choi, J.H.; Qian, X.; Shears, S.B.; Spiess, B.; Potter, B.V.L. Scyllo-Inositol Pentakisphosphate as an Analogue of Myo-Inositol 1,3,4,5,6-Pentakisphosphate: Chemical Synthesis, Physicochemistry and Biological Applications. ChemBioChem 2006, 7, 1114–1122. [Google Scholar] [CrossRef]
- Hager, A.; Wu, M.; Wang, H.; Brown, N.W.J.; Shears, S.B.; Veiga, N.; Fiedler, D. Cellular Cations Control Conformational Switching of Inositol Pyrophosphate Analogues. Chemistry 2016, 22, 12406–12414. [Google Scholar] [CrossRef]
- Barker, C.J.; Wright, J.; Hughes, P.J.; Kirk, C.J.; Michell, R.H. Complex Changes in Cellular Inositol Phosphate Complement Accompany Transit through the Cell Cycle. Biochem. J. 2004, 380, 465–473. [Google Scholar] [CrossRef]
- Harmel, R.K.; Puschmann, R.; Nguyen Trung, M.; Saiardi, A.; Schmieder, P.; Fiedler, D. Harnessing (13)C-Labeled Myo-Inositol to Interrogate Inositol Phosphate Messengers by NMR. Chem. Sci. 2019, 10, 5267–5274. [Google Scholar] [CrossRef]
- Qiu, D.; Wilson, M.S.; Eisenbeis, V.B.; Harmel, R.K.; Riemer, E.; Haas, T.M.; Wittwer, C.; Jork, N.; Gu, C.; Shears, S.B.; et al. Analysis of Inositol Phosphate Metabolism by Capillary Electrophoresis Electrospray Ionization Mass Spectrometry. Nat. Commun. 2020, 11, 6035. [Google Scholar] [CrossRef]
- Gu, C.; Wilson, M.S.C.; Jessen, H.J.; Saiardi, A.; Shears, S.B. Inositol Pyrophosphate Profiling of Two HCT116 Cell Lines Uncovers Variation in InsP8 Levels. PLoS ONE 2016, 11, e0165286. [Google Scholar] [CrossRef]
- Puschmann, R.; Harmel, R.K.; Fiedler, D. Scalable Chemoenzymatic Synthesis of Inositol Pyrophosphates. Biochemistry 2019, 58, 3927–3932. [Google Scholar] [CrossRef]
- Findeisen, M.; Berger, T.; Berger, S. A 1H-NMR Thermometer Suitable for Cryoprobes.Pdf. Magn. Reson. Chem. 2007, 45, 175–178. [Google Scholar] [CrossRef]
- Krȩzel, A.; Bal, W. A Formula for Correlating PKa Values Determined in D 2O and H2O. J. Inorg. Biochem. 2004, 98, 161–166. [Google Scholar] [CrossRef]
- Frassineti, C.; Ghelli, S. NMR as a Tool for Determining Protonation Constants of Nutral Polyprotic Bases in Solution. Anal. Biochem. 1995, 231, 374–382. [Google Scholar] [CrossRef]
- Gans, P.; Sabatini, A.; Vacca, A. Investigation of Equilibria in Solution. Determination of Equilibrium Constants with the HYPERQUAD Suite of Programs. Talanta 1996, 43, 1739–1753. [Google Scholar] [CrossRef]
- Alderighi, L.; Gans, P.; Ienco, A.; Peters, D.; Sabatini, A.; Vacca, A. Hyperquad Simulation and Speciation (HySS): A Utility Program for the Investigation of Equilibria Involving Soluble and Partially Soluble Species. Coord. Chem. Rev. 1999, 184, 311–318. [Google Scholar] [CrossRef]
- Couto, D.; Richter, A.; Walter, H.; Furkert, D.; Hothorn, M.; Fiedler, D. Using Biotinylated Myo -Inositol Hexakisphosphate to Investigate Inositol Pyrophosphate—Protein Interactions with Surface-Based Biosensors. Biochemistry 2021, 60, 2739–2748. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision A.02; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Hay, P.J.; Wadt, W.R. Ab Initio Effective Core Potentials for Molecular Calculations. Potentials for K to Au Including the Outermost Core Orbitale. J. Chem. Phys. 1985, 82, 299–310. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef]
- Frost, E.B. Conformational States of Myo-Inositol Hexakisphosphate in Aqueous Solution. A 13C NMR, 31P NMR, and Raman Spectroscopic Investigation. Science 1979, 47, 416–417. [Google Scholar] [CrossRef]
- Barrientos, L.G.; Murthy, P.P.N. Conformational Studies of Myo-Inositol Phosphates. Carbohydr. Res. 1996, 296, 39–54. [Google Scholar] [CrossRef]
- Veiga, N.; Torres, J.; Macho, I.; Gómez, K.; Godage, H.Y.; Riley, A.M.; Potter, B.V.L.; González, G.; Kremer, C. Inframolecular Acid–Base and Coordination Properties towards Na+ and Mg2+ of Myo-Inositol 1,3,4,5,6-Pentakisphosphate: A Structural Approach to Biologically Relevant Species. J. Chem. Soc. Dalt. Trans. 2013, 42, 6021–6032. [Google Scholar] [CrossRef]
- Torres, J.; Domínguez, S.; Cerdá, M.F.; Obal, G.; Mederos, A.; Irvine, R.F.; Díaz, A.; Kremer, C. Solution Behaviour of Myo-Inositol Hexakisphosphate in the Presence of Multivalent Cations. Prediction of a Neutral Pentamagnesium Species under Cytosolic/Nuclear Conditions. J. Inorg. Biochem. 2005, 99, 828–840. [Google Scholar] [CrossRef]
- Torres, J.; Veiga, N.; Gancheff, J.S.; Domínguez, S.; Mederos, A.; Sundberg, M.; Sánchez, A.; Castiglioni, J.; Díaz, A.; Kremer, C. Interaction of Myo-Inositol Hexakisphosphate with Alkali and Alkaline Earth Metal Ions: Spectroscopic, Potentiometric and Theoretical Studies. J. Mol. Struct. 2008, 874, 77–88. [Google Scholar] [CrossRef]
- Wild, R.; Gerasimaite, R.; Jung, J.-Y.; Truffault, V.; Pavlovic, I.; Schmidt, A.; Saiardi, A.; Jessen, H.J.; Poirier, Y.; Hothorn, M.; et al. Control of Eukaryotic Phosphate Homeostasis by Inositol Polyphosphate Sensor Domains. Science 2016, 352, 986–990. [Google Scholar] [CrossRef]
- Reynolds, C.H.; Holloway, M.K. Thermodynamics of Ligand Binding and Efficiency. ACS Med. Chem. Lett. 2011, 2, 433–437. [Google Scholar] [CrossRef]
- Šala, M.; Makuc, D.; Kolar, J.; Plavec, J.; Pihlar, B. Potentiometric and 31P NMR Studies on Inositol Phosphates and Their Interaction with Iron(III) Ions. Carbohydr. Res. 2011, 346, 488–494. [Google Scholar] [CrossRef]
- Veiga, N.; Macho, I.; Gómez, K.; González, G.; Kremer, C.; Torres, J. Potentiometric and Spectroscopic Study of the Interaction of 3d Transition Metal Ions with Inositol Hexakisphosphate. J. Mol. Struct. 2015, 1098, 55–65. [Google Scholar] [CrossRef]
- Asensio, G.; Hernández-Arriaga, A.M.; Martín del Campo, M.; Prieto, A.M.; Rojo, L.; Vázquez-Lasa, B. A Study on Sr/Zn Phytate Complexes: Structural Properties and Antimicrobial Synergistic Effects against Streptococcus Mutans. Sci. Rep. 2022, 12, 20177. [Google Scholar] [CrossRef]
- Romani, A.M.P. Intracellular Magnesium Homeostasis. Arch. Biochem. Biophys. 2011, 512, 13–58. [Google Scholar] [CrossRef]
- Smith, R.M.; Martell, A.E.; Chen, Y. Critical Evaluation of Stability Constants for Nucleotide Complexes with Protons and Metal Ions and the Accompanying Enthalpy Changes. Pure Appl. Chem. 1991, 63, 1015–1080. [Google Scholar] [CrossRef]
- Young, B.P.; Shin, J.J.H.; Orij, R.; Chao, J.T.; Li, S.C.; Guan, X.L.; Loewen, C.J.R. Phosphatidic Acid Is a PH Biosensor That Links Membrane Biogenesis to Metabolism. Science 2010, 329, 1085–1088. [Google Scholar] [CrossRef]
- Orij, R.; Urbanus, M.L.; Vizeacoumar, F.J.; Giaever, G.; Boone, C.; Nislow, C.; Brul, S.; Smits, G.J. Genome-Wide Analysis of Intracellular PH Reveals Quantitative Control of Cell Division Rate by PH(c) in Saccharomyces Cerevisiae. Genome Biol. 2012, 13, R80. [Google Scholar] [CrossRef]
- Shears, S.B. Assessing the Omnipotence of Inositol Hexakisphosphate. Cell. Signal. 2001, 13, 151–158. [Google Scholar] [CrossRef]
- Moedritzer, K. PH Dependence of Phosphorus-31 Chemical Shifts and Coupling Constants of Some Oxyacids of Phosphorus. Inorg. Chem. 1967, 6, 936–939. [Google Scholar] [CrossRef]

| Equilibrium | log K | ||
|---|---|---|---|
| InsP8 | 5PCP-InsP5 | InsP6 | |
| 31P NMR (22 °C) a | 31P NMR (22 °C) b | Potentiometry (37 °C) b | |
| L14− + H+ ↔ HL13− | 11.21(1) | 11.48(1) | 10.8(1) |
| L14− + 2 H+ ↔ H2L12− | 22.78(2) | 22.42(2) | 21.3(1) |
| L14− + 3 H+ ↔ H3L11− | 34.22(2) | 32.26(2) | 31.63(6) |
| L14− + 4 H+ ↔ H4L10− | 43.96(2) | 40.94(2) | 40.42(6) |
| L14− + 5 H+ ↔ H5L9− | 52.58(3) | 47.67(2) | 47.32(6) |
| L14− + 6 H+ ↔ H6L8− | 59.41(8) | 51.93(2) | 53.04(7) |
| L14− + 7 H+ ↔ H7L7− | 64.91(6) | 55.64(2) | 56.14(9) |
| L14− + 8 H+ ↔ H8L6− | 68.79(7) | --- | --- |
| 5 K+ + HL13− ↔ [K5(HL)]8− | 12.47(3) | 6.57(3) | --- |
| 4 K+ + H2L12− ↔ [K4(H2L)]8− | 9.76(3) | 4.61(3) | --- |
| 4 K+ + H3L11− ↔ [K4(H3L)]7− | --- | 4.50(5) | 5.42(5) |
| 3 K+ + H4L10− ↔ [K3(H4L)]7− | 5.446(5) | 3.94(4) | 3.36(5) |
| 2 K+ + H5L9− ↔ [K2(H5L)]7− | 3.820(7) | 2.79(7) | --- |
| K+ + H6L8− ↔ [K(H6L)]7− | 2.58(5) | --- | --- |
| K+ + H7L7− ↔ [K(H7L)]6− | 2.08(3) | --- | --- |
| Mg2+ + L14− + 4 K+ ↔ [MgK4L]8− | 22.4(1) | --- | --- |
| Mg2+ + HL13− + 4 K+ ↔ [MgK4(HL)]7− | 21.6(1) | 11.64(5) | --- |
| Mg2+ + H2L12− + 3 K+ ↔ [MgK3(H2L)]7− | 18.1(1) | 9.75(7) | --- |
| Mg2+ + H3L11− + 3 K+ ↔ [MgK3(H3L)]6− | 15.0(1) | 8.79(7) | --- |
| Mg2+ + H4L10− + 2 K+ ↔ [MgK2(H4L)]6− | 11.6(1) | 6.96(7) | --- |
| Mg2+ + H5L9− + K+ ↔ [MgK(H5L)]6− | 9.1(1) | 5.26(7) | --- |
| Mg2+ + H6L8− + K+ ↔ [MgK(H6L)]5− | 7.3(1) | --- | --- |
| Mg2+ + H6L8− ↔ [Mg(H6L)]6− | --- | 4.71(7) | --- |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurz, L.; Schmieder, P.; Veiga, N.; Fiedler, D. One Scaffold, Two Conformations: The Ring-Flip of the Messenger InsP8 Occurs under Cytosolic Conditions. Biomolecules 2023, 13, 645. https://doi.org/10.3390/biom13040645
Kurz L, Schmieder P, Veiga N, Fiedler D. One Scaffold, Two Conformations: The Ring-Flip of the Messenger InsP8 Occurs under Cytosolic Conditions. Biomolecules. 2023; 13(4):645. https://doi.org/10.3390/biom13040645
Chicago/Turabian StyleKurz, Leonie, Peter Schmieder, Nicolás Veiga, and Dorothea Fiedler. 2023. "One Scaffold, Two Conformations: The Ring-Flip of the Messenger InsP8 Occurs under Cytosolic Conditions" Biomolecules 13, no. 4: 645. https://doi.org/10.3390/biom13040645
APA StyleKurz, L., Schmieder, P., Veiga, N., & Fiedler, D. (2023). One Scaffold, Two Conformations: The Ring-Flip of the Messenger InsP8 Occurs under Cytosolic Conditions. Biomolecules, 13(4), 645. https://doi.org/10.3390/biom13040645