Self-Assembly of Amyloid Fibrils into 3D Gel Clusters versus 2D Sheets
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein and Chemicals
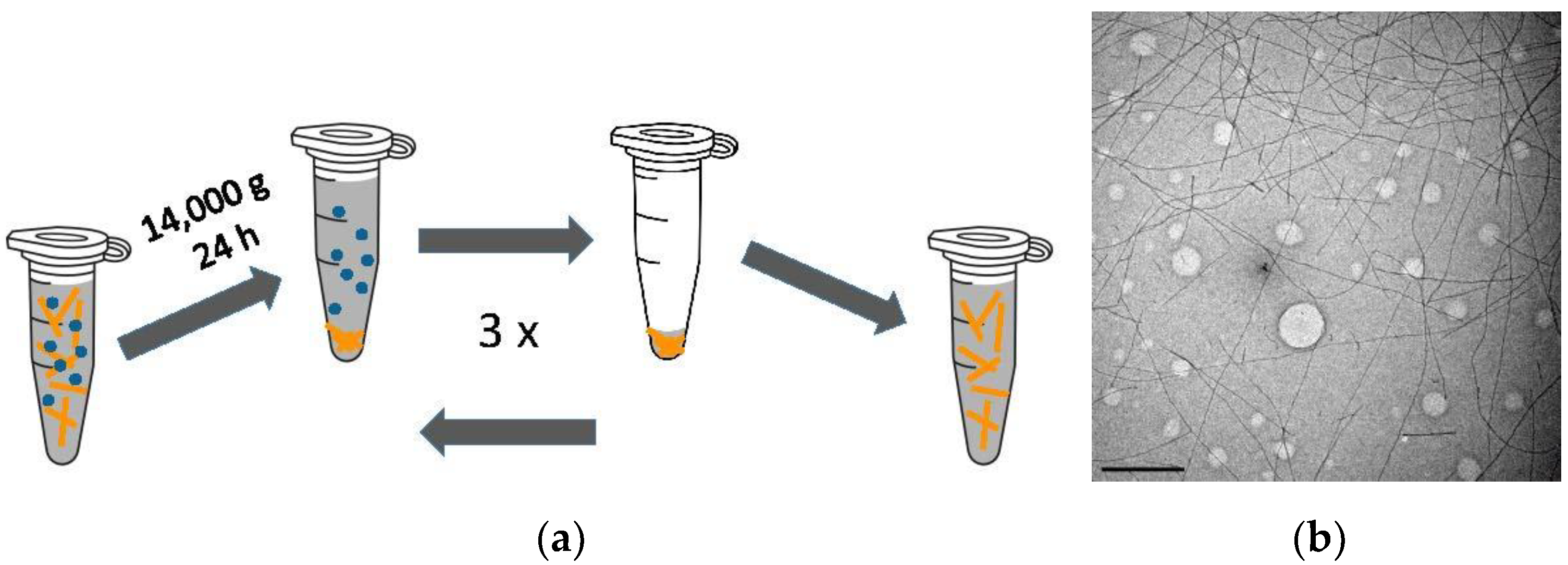
2.2. Growth and Separation of Lysozyme Amyloid Fibrils
2.3. Salt Mediated Fibril Assembly at pH 2
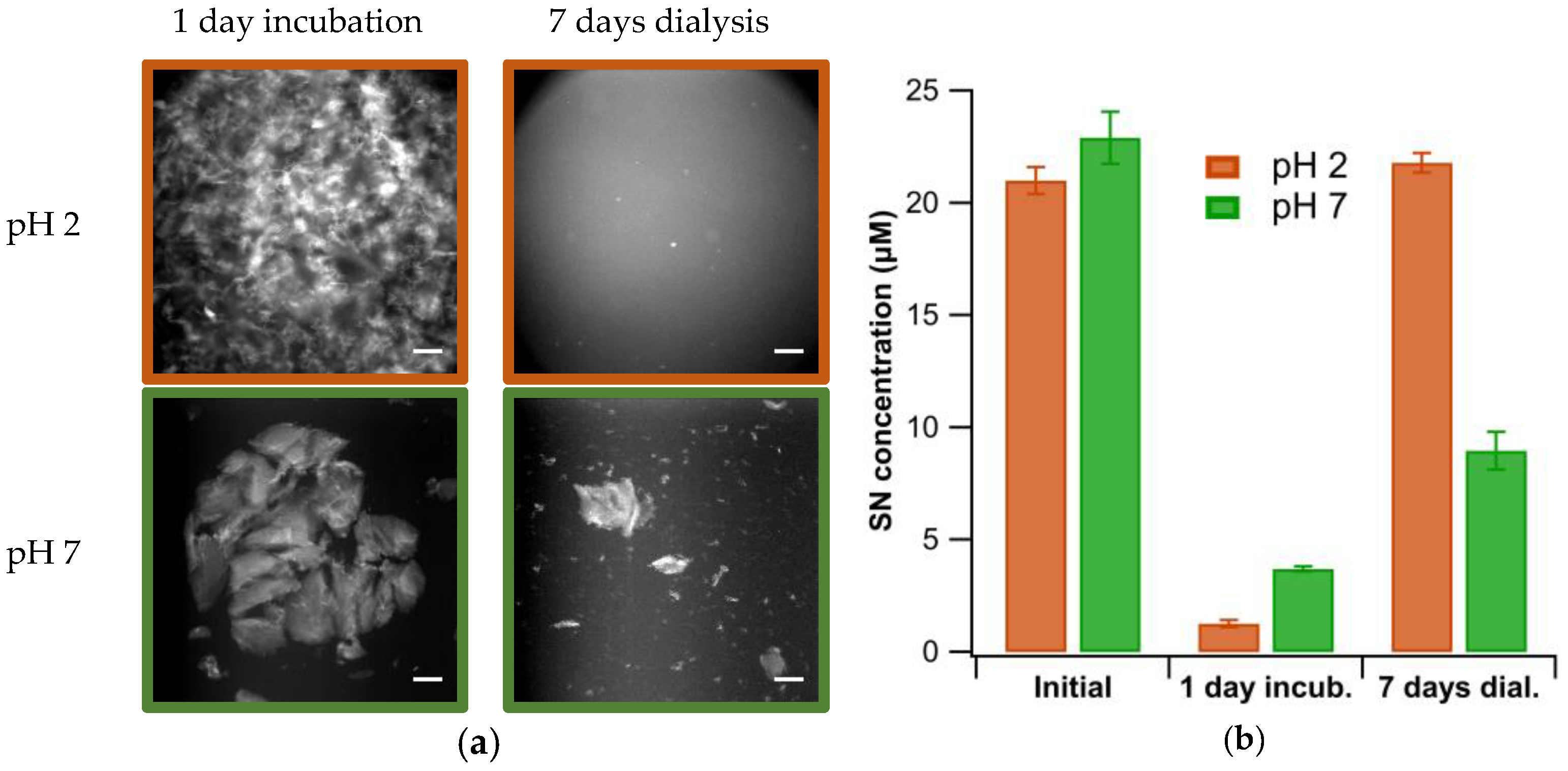
2.4. Visualization of Aggregate Morphology using Fluorescence Microscopy
2.5. Quantification of Fibril Aggregation Using Centrifugation
2.6. Salt-Mediated Fibril Assembly after Transfer to pH 7
2.7. Dialysis Protocol
2.8. Transmission Electron Microscopy
2.9. Dynamic Light Scattering (DLS)
3. Results
3.1. Growth and Characterization of Isolated Lysozyme Amyloid Fibrils
3.2. Salt-Induced Precipitation of Isolated Amyloid Fibrils at pH 2
3.3. Plaque Formation after Transfer to pH 7
3.4. Stability of Gels vs. Sheets
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chiti, F.; Dobson, C.M. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.J.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef]
- Eisenberg, D.; Jucker, M. The Amyloid State of Proteins in Human Diseases. Cell 2012, 148, 1188–1203. [Google Scholar] [CrossRef] [PubMed]
- Buxbaum, J.N. The systemic amyloidoses. Curr. Opin. Rheumatol. 2004, 16, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M. Protein misfolding, evolution and disease. Trends Biochem. Sci. 1999, 24, 329–332. [Google Scholar] [CrossRef]
- Jahn, T.R.; Makin, O.S.; Morris, K.L.; Marshall, K.E.; Tian, P.; Sikorski, P.; Serpell, L.C. The Common Architecture of Cross-b Amyloid. J. Mol. Biol. 2010, 395, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Fändrich, M.; Dobson, C.M. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 2002, 21, 5682–5690. [Google Scholar] [CrossRef] [PubMed]
- Cline, E.N.; Bicca, M.A.; Viola, K.L.; Klein, W.L. The Amyloid-β Oligomer Hypothesis: Beginning of the Third Decade. J. Alzheimer’s Dis. 2018, 64, S567–S610. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, U.; Nilson, A.N.; Kayed, R. The Role of Amyloid-β Oligomers in Toxicity, Propagation, and Immunotherapy. eBioMedicine 2016, 6, 42–49. [Google Scholar] [CrossRef]
- Kayed, R.; Lasagna-Reeves, C.A. Molecular Mechanisms of Amyloid Oligomers Toxicity. J. Alzheimer’s Dis. 2013, 33, S67–S78. [Google Scholar] [CrossRef]
- Pickett, E.K.; Koffie, R.M.; Wegmann, S.; Henstridge, C.M.; Herrmann, A.G.; Colom-Cadena, M.; Lleo, A.; Kay, K.R.; Vaught, M.; Soberman, R.; et al. Non-Fibrillar Oligomeric Amyloid-b within Synapses. J. Alzheimer’s Dis. 2016, 53, 787–800. [Google Scholar] [CrossRef] [PubMed]
- Hefti, F.; Goure, W.F.; Jerecic, J.; Iverson, K.S.; Walicke, P.A.; Krafft, G.A. The case for soluble Aβ oligomers as a drug target in Alzheimer’s disease. Trends Pharmacol. Sci. 2013, 34, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Koffie, R.M.; Meyer-Luehmann, M.; Hashimoto, T.; Adams, K.W.; Mielke, M.L.; Garcia-Alloza, M.; Micheva, K.D.; Smith, S.J.; Kim, M.L.; Lee, V.M.; et al. Oligomeric amyloid β associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc. Natl. Acad. Sci. USA 2009, 106, 4012–4017. [Google Scholar] [CrossRef] [PubMed]
- Reixach, N.; Deechongkit, S.; Jiang, X.; Kelly, J.W.; Buxbaum, J.N. Tissue damage in the amyloidoses: Transthyretin monomers and nonnative oligomers are the major cytotoxic species in tissue culture. Proc. Natl. Acad. Sci. USA 2004, 101, 2817–2822. [Google Scholar] [CrossRef]
- Pepys, M.B. Amyloidosis. Annu. Rev. Med. 2006, 57, 223–241. [Google Scholar] [CrossRef] [PubMed]
- Schreml, S.; Szeimies, R.-M.; Vogt, T.; Landthaler, M.; Schroeder, J.; Babilas, P. Cutaneous amyloidoses and systemic amyloidoses with cutaneous involvement. Eur. J. Dermatol. 2010, 20, 152–160. [Google Scholar] [CrossRef]
- Stine, W.B., Jr.; Dahlgren, K.N.; Krafft, G.A.; LaDu, M.J. In Vitro Characterization of Conditions for Amyloid-beta Peptide Oligomerization and Fibrillogenesis. J. Biol. Chem. 2003, 278, 11612–11622. [Google Scholar] [CrossRef]
- Kayed, R.; Glabe, C.G. Conformation-Dependent Anti-Amyloid Oligomer Antibodies. In Methods Enzymol; Academic Press: Cambridge, MA, USA, 2006; Volume 413, pp. 326–344. [Google Scholar]
- Teplow, D.B.; Noel, L.D.; Bitan, G.; Bernstein, S.; Wyttenbach, T.; Bowers, M.T.; Baumketner, A.; Shea, J.-E.; Urbanc, B.; Cruz, L.; et al. Elucidating Amyloid b-Protein Folding and Assembly: A Multidisciplinary Approach. Acc. Chem. Res. 2006, 39, 635–645. [Google Scholar] [CrossRef]
- Harper, J.D.; Wong, S.S.; Lieber, C.M.; Lansbury, P.T. Observation of metastable Ab amyloid protofibrils by atomic force microscopy. Chem. Biol. 1997, 4, 119–125. [Google Scholar] [CrossRef]
- Jia, Z.; Beugelsdijk, A.; Chen, J.; Schmit, J.D. The Levinthal Problem in Amyloid Aggregation: Sampling of a Flat Reaction Space. J. Phys. Chem. B 2017, 121, 1576–1586. [Google Scholar] [CrossRef]
- Meisl, G.; Rajah, L.; Cohen, S.A.I.; Pfammatter, M.; Saric, A.; Hellstrand, E.; Buell, A.K.; Aguzzi, A.; Linse, S.; Vendruscolo, M.; et al. Scaling behaviour and rate-determining steps in filamentous self-assembly. Chem. Sci. 2017, 8, 7087–7097. [Google Scholar] [CrossRef] [PubMed]
- Iadanza, M.G.; Jackson, M.P.; Hewitt, E.W.; Ranson, N.A.; Radford, S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 755–773. [Google Scholar] [CrossRef]
- Rizevsky, S.; Kurouski, D. Nanoscale Structural Organization of Insulin Fibril Polymorphs Revealed by Atomic Force Microscopy–Infrared Spectroscopy (AFM-IR). ChemBioChem 2020, 21, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, F.; Maiti, P.; Bitan, G. Photo-induced cross-linking of unmodified proteins (PICUP) applied to amyloidogenic peptides. J. Vis. Exp. 2009, 23, e1071. [Google Scholar] [CrossRef] [PubMed]
- Psonka-Antonczyk, K.M.; Hammarström, P.; Johansson, L.B.G.; Lindgren, M.; Stokke, B.T.; Nilsson, K.P.R.; Nyström, S. Nanoscale Structure and Spectroscopic Probing of Aβ1-40 Fibril Bundle Formation. Front. Chem. 2016, 4, 44. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Kollmer, M.; Markx, D.; Claus, S.; Walther, P.; Fändrich, M. Amyloid plaque structure and cell surface interactions of β-amyloid fibrils revealed by electron tomography. Sci. Rep. 2017, 7, 43577. [Google Scholar] [CrossRef] [PubMed]
- Joachim, C.L.; Morris, J.H.; Selkoe, D.J. Diffuse Senile Plaques Occur Commonly in the Cerebellum in Alzheimer’s Disease. Am. J. Pathol. 1989, 135, 309–319. [Google Scholar]
- Lemere, C.A.; Blusztajn, J.K.; Yamaguchi, H.; Wisniewski, T.; Saido, T.C.; Selkoe, D.J. Sequence of Deposition of Heterogeneous Amyloid β-Peptides and APO E in Down Syndrome: Implications for Initial Events in Amyloid Plaque Formation. Neurobiol. Dis. 1996, 3, 16–32. [Google Scholar] [CrossRef]
- Lemke, G.; Huang, Y. The dense-core plaques of Alzheimer’s disease are granulomas. J. Exp. Med. 2022, 219, e20212477. [Google Scholar] [CrossRef]
- Fowler, D.M.; Koulov, A.V.; Alory-Jost, C.; Marks, M.S.; Balch, W.E.; Kelly, J.W. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006, 4, e6. [Google Scholar] [CrossRef]
- Jang, H.; Arce, F.T.; Mustata, M.; Ramachandran, S.; Capone, R.; Nussinov, R.; Lal, R. Antimicrobial protegrin-1 forms amyloid-like fibrils with rapid kinetics suggesting a functional link. Biophys. J. 2011, 100, 1775–1783. [Google Scholar] [CrossRef] [PubMed]
- Hervás, R.; Li, L.; Majumdar, A.; Fernández-Ramírez, M.d.C.; Unruh, J.R.; Slaughter, B.D.; Galera-Prat, A.; Santana, E.; Suzuki, M.; Nagai, Y.; et al. Molecular Basis of Orb2 Amyloidogenesis and Blockade of Memory Consolidation. PLoS Biol. 2016, 14, e1002361. [Google Scholar] [CrossRef] [PubMed]
- Roan, N.R.; Sandi-Monroy, N.; Kohgadai, N.; Usmani, S.M.; Hamil, K.G.; Neidleman, J.; Montano, M.; Ständker, L.; Röcker, A.; Cavrois, M.; et al. Semen amyloids participate in spermatozoa selection and clearance. eLife 2017, 6, e24888. [Google Scholar] [CrossRef] [PubMed]
- Otzen, D. Functional amyloid: Turning swords into plowshares. Prion 2010, 4, 256–264. [Google Scholar] [CrossRef]
- Wei, G.; Su, Z.; Reynolds, N.P.; Arosio, P.; Hamley, I.W.; Gazit, E.; Mezzenga, R. Self-assembling peptide and protein amyloids: From structure to tailored function in nanotechnology. Chem. Soc. Rev. 2017, 46, 4661–4708. [Google Scholar] [CrossRef]
- Reynolds, N.P.; Charnley, M.; Mezzenga, R.; Hartley, P.G. Engineered Lysozyme Amyloid Fibril Networks Support Cellular Growth and Spreading. Biomacromolecules 2014, 15, 599–608. [Google Scholar] [CrossRef]
- Jacob, R.S.; Ghosh, D.; Singh, P.K.; Basu, S.K.; Jha, N.N.; Das, S.; Sukul, P.K.; Patil, S.; Sathaye, S.; Kumar, A.; et al. Self healing hydrogels composed of amyloid nano fibrils for cell culture and stem cell differentiation. Biomaterials 2015, 54, 97–105. [Google Scholar] [CrossRef]
- Woodard, D.; Bell, D.; Tipton, D.; Durrance, S.; Cole, L.; Li, B.; Xu, S. Gel Formation in Protein Amyloid Aggregation: A Physical Mechanism for Cytotoxicity. PLoS ONE 2014, 9, e94789. [Google Scholar] [CrossRef]
- Kumari, A.; Ahmad, B. The physical basis of fabrication of amyloid-based hydrogels by lysozyme. RSC Adv. 2019, 9, 37424–37435. [Google Scholar] [CrossRef]
- Hasecke, F.; Niyangoda, C.; Borjas, G.; Pan, J.; Matthews, G.; Muschol, M.; Hoyer, W. Protofibril-Fibril Interactions Inhibit Amyloid Fibril Assembly by Obstructing Secondary Nucleation. Angew. Chem. Int. Ed. 2021, 60, 3016–3021. [Google Scholar] [CrossRef]
- Miti, T.; Mulaj, M.; Schmit, J.D.; Muschol, M. Stable, Metastable and Kinetically Trapped Amyloid Aggregate Phases. Biomacromolecules 2015, 16, 326–335. [Google Scholar] [CrossRef]
- Hill, S.E.; Miti, T.; Richmond, T.; Muschol, M. Spatial Extent of Charge Repulsion Regulates Assembly Pathways for Lysozyme Amyloid Fibrils. PLoS ONE 2011, 6, e18171. [Google Scholar] [CrossRef]
- Niyangoda, C.; Barton, J.; Bushra, N.; Karunarathne, K.; Strauss, G.; Fakhre, F.; Koria, P.; Muschol, M. Origin, toxicity and characteristics of two amyloid oligomer polymorphs. RSC Chem. Biol. 2021, 2, 1631–1642. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Sörgjerd, K.; Nyström, S.; Nordigården, A.; Yu, Y.-C.; Hammarström, P. Lysozyme Amyloidogenesis Is Accelerated by Specific Nicking and Fragmentation but Decelerated by Intact Protein Binding and Conversion. J. Mol. Biol. 2007, 366, 1029–1044. [Google Scholar] [CrossRef] [PubMed]
- Spassov, V.Z.; Yan, L. A fast and accurate computational approach to protein ionization. Protein Sci. 2008, 17, 1955–1970. [Google Scholar] [CrossRef] [PubMed]
- Kuehner, D.E.; Engmann, J.; Fergg, F.; Wernick, M.; Blanch, H.W.; Prausnitz, J.M. Lysozyme Net Charge and Ion Binding in Concentrated Aqueous Electrolyte Solutions. J. Phys. Chem. B 1999, 103, 1368–1374. [Google Scholar] [CrossRef]
- Hu, B.; Shen, Y.; Adamcik, J.; Fischer, P.; Schneider, M.; Loessner, M.J.; Mezzenga, R. Polyphenol-Binding Amyloid Fibrils Self-Assemble into Reversible Hydrogels with Antibacterial Activity. ACS Nano 2018, 12, 3385–3396. [Google Scholar] [CrossRef]
- Shimanovich, U.; Efimov, I.; Mason, T.O.; Flagmeier, P.; Buell, A.K.; Gedanken, A.; Linse, S.; Åkerfeldt, K.S.; Dobson, C.M.; Weitz, D.A.; et al. Protein Microgels from Amyloid Fibril Networks. ACS Nano 2014, in press. [CrossRef]
- Yan, H.; Saiani, A.; Gough, J.E.; Miller, A.F. Thermoreversible Protein Hydrogel as Cell Scaffold. Biomacromolecules 2006, 7, 2776–2782. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karunarathne, K.; Bushra, N.; Williams, O.; Raza, I.; Tirado, L.; Fakhre, D.; Fakhre, F.; Muschol, M. Self-Assembly of Amyloid Fibrils into 3D Gel Clusters versus 2D Sheets. Biomolecules 2023, 13, 230. https://doi.org/10.3390/biom13020230
Karunarathne K, Bushra N, Williams O, Raza I, Tirado L, Fakhre D, Fakhre F, Muschol M. Self-Assembly of Amyloid Fibrils into 3D Gel Clusters versus 2D Sheets. Biomolecules. 2023; 13(2):230. https://doi.org/10.3390/biom13020230
Chicago/Turabian StyleKarunarathne, Kanchana, Nabila Bushra, Olivia Williams, Imad Raza, Laura Tirado, Diane Fakhre, Fadia Fakhre, and Martin Muschol. 2023. "Self-Assembly of Amyloid Fibrils into 3D Gel Clusters versus 2D Sheets" Biomolecules 13, no. 2: 230. https://doi.org/10.3390/biom13020230
APA StyleKarunarathne, K., Bushra, N., Williams, O., Raza, I., Tirado, L., Fakhre, D., Fakhre, F., & Muschol, M. (2023). Self-Assembly of Amyloid Fibrils into 3D Gel Clusters versus 2D Sheets. Biomolecules, 13(2), 230. https://doi.org/10.3390/biom13020230





