Identification of a Prognostic Signature for Ovarian Cancer Based on Ubiquitin-Related Genes Suggesting a Potential Role for FBXO9
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Specimens
2.2. Immunohistochemistry (IHC)
2.3. Data Extraction and Patient Information Precondition
2.4. Screening and Annotation of Ubiquitin-Related Genes
2.5. Construction and Validation of a Prognostic Signature Based on Ubiquitin-Related Genes
2.6. Construction of Calibration Curves and Nomograms
2.7. Analysis of Tumor Immune Microenvironments and Somatic Mutations
2.8. Predicting the Susceptibility to Chemotherapy Agents
2.9. Analysis of the FBXO9 and UBD Interaction Network and Functional Enrichment
2.10. Cell Lines
2.11. RNA Extraction and Gene Expression Analysis
2.12. Western Blot (WB) Assay
2.13. Statistical Analysis
3. Results
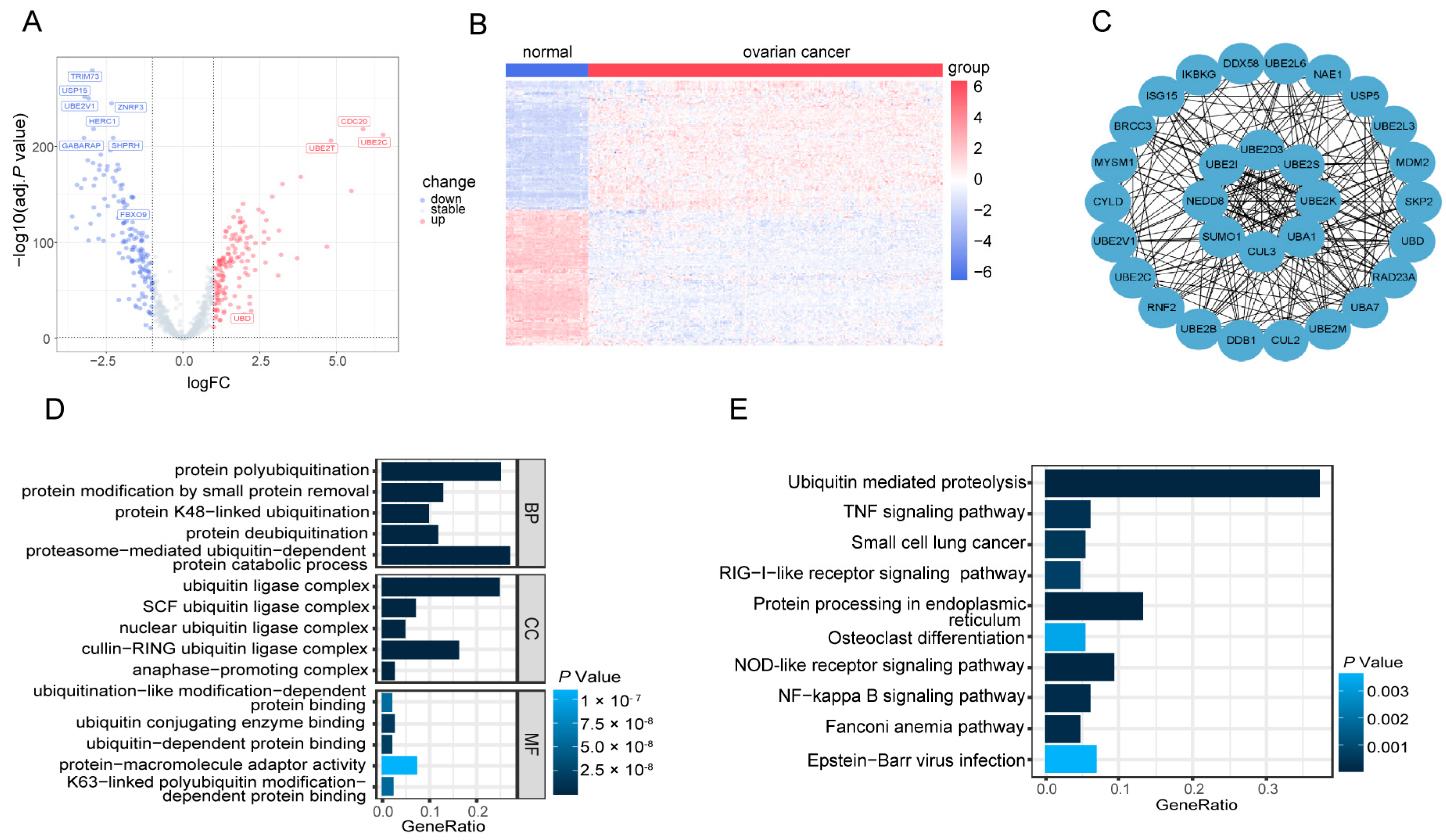
3.1. K48-Linked Ubiquitination was Suppressed in OV
3.2. Identification of 287 DE-UbRGs Involved in OV
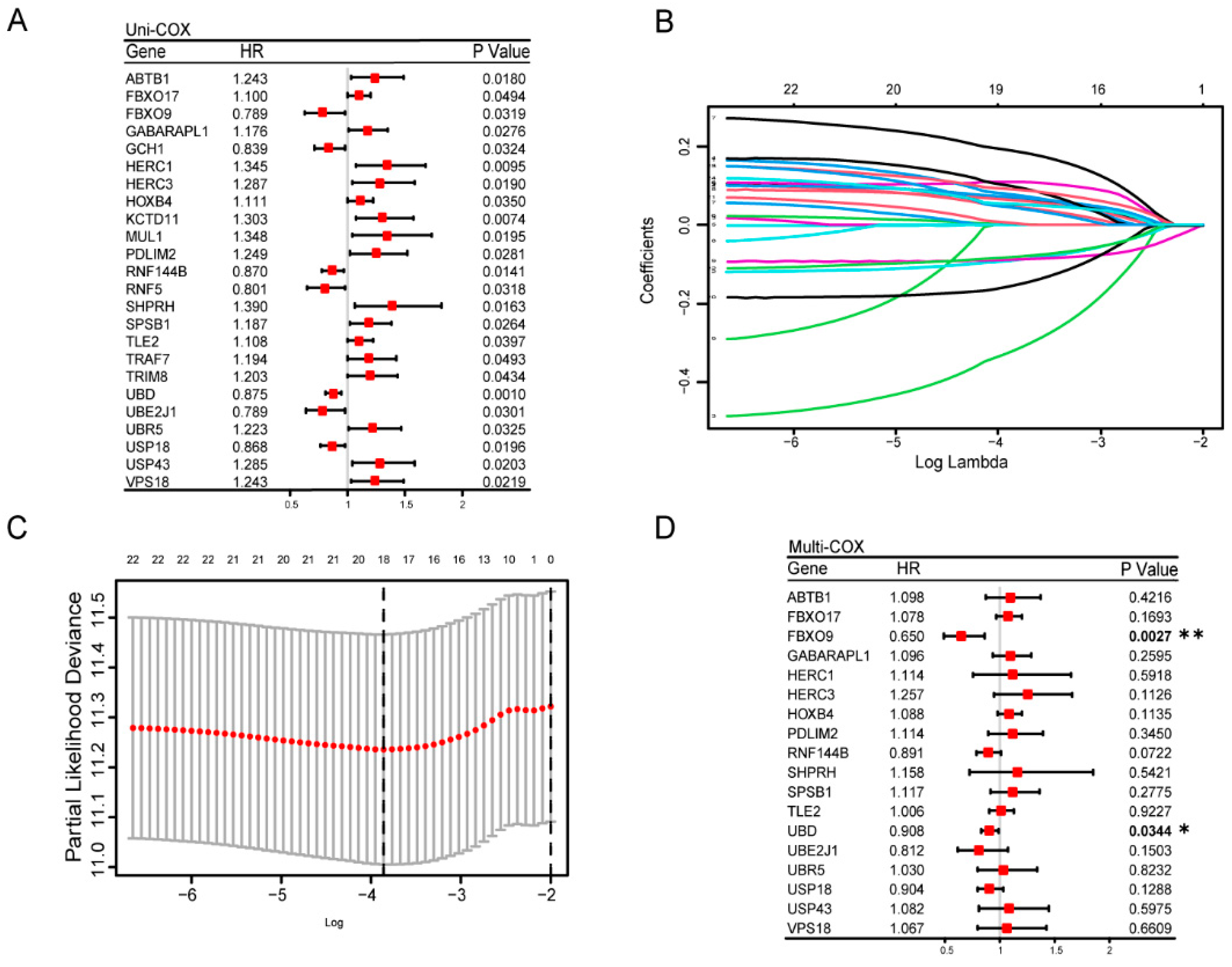
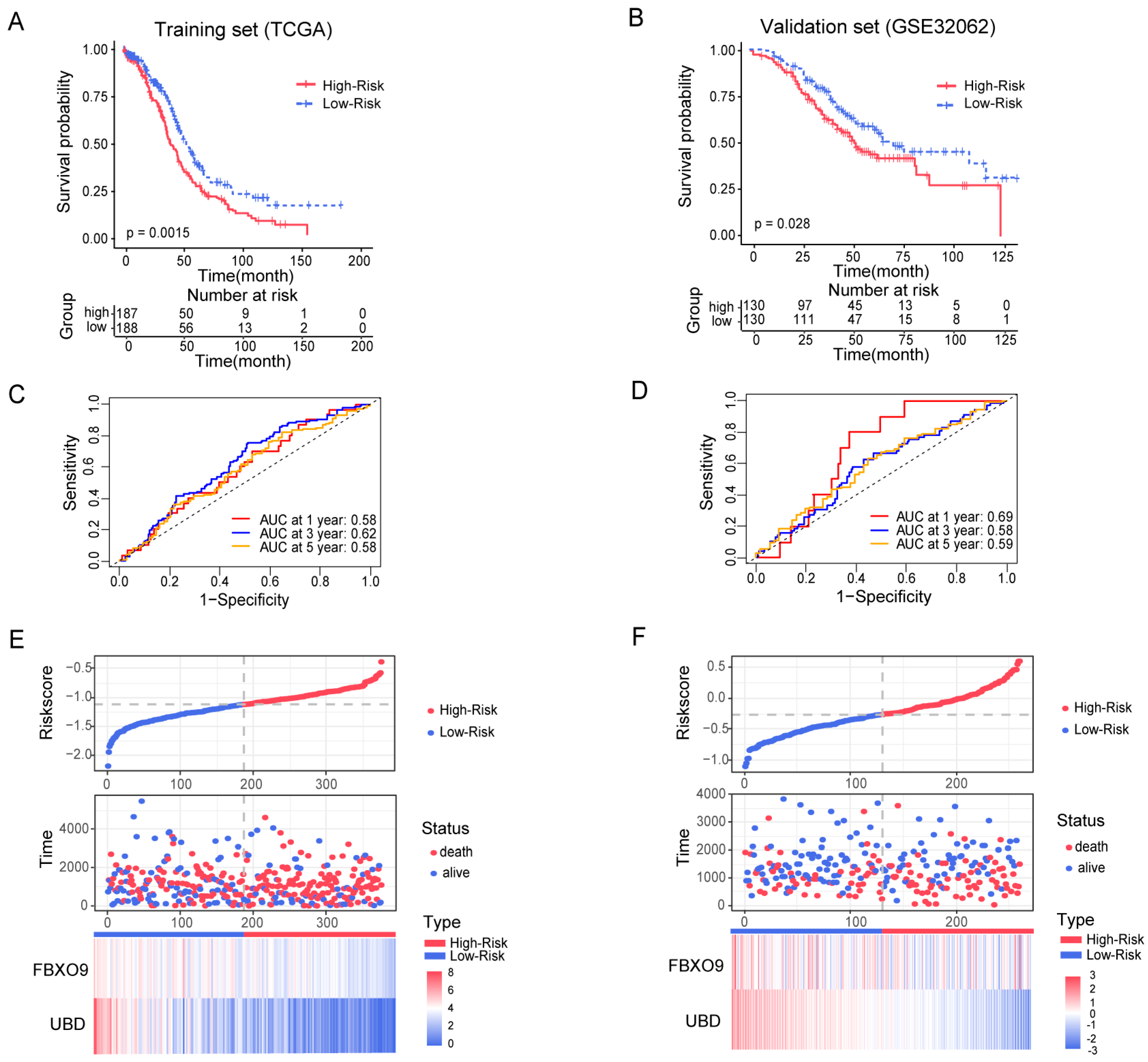
3.3. Construction and Estimation of a Prognostic Signature of UbRGs for OV
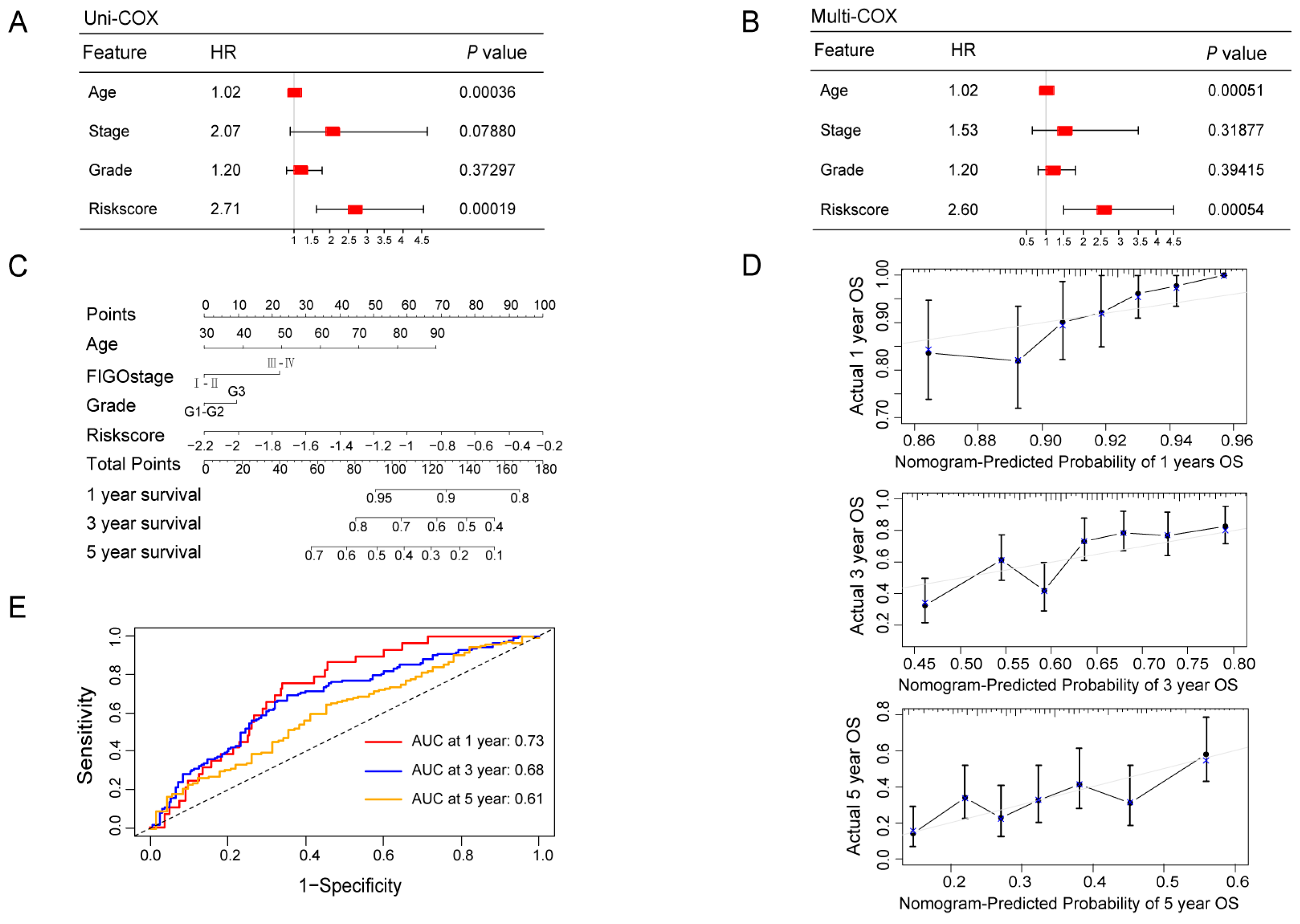
3.4. Construction and Validation of a Ubiquitin-Related Prognostic Nomogram
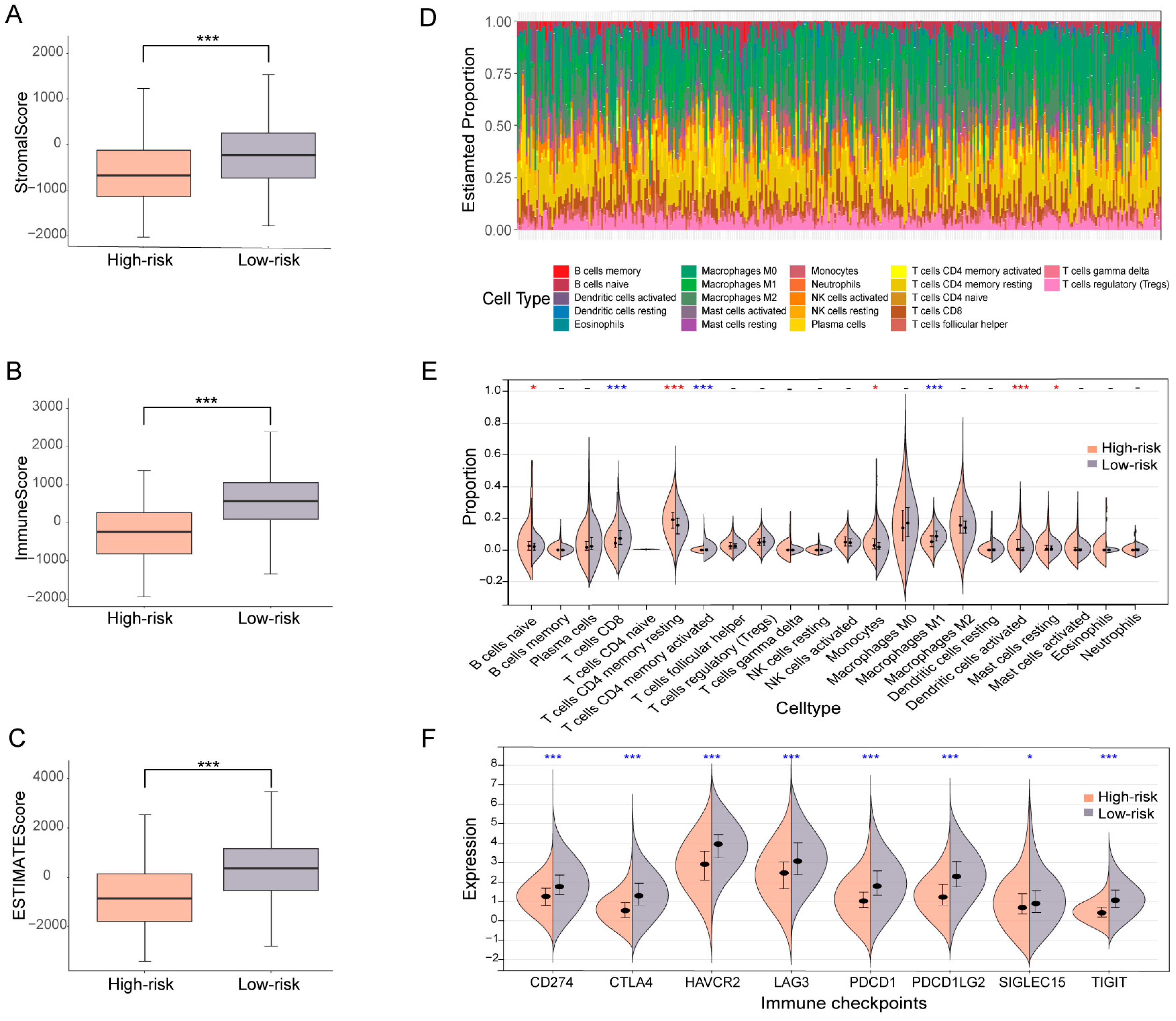
3.5. OV patients Subgrouped Using the Ubiquitin-Related Signature Demonstrate Distinct TME Characteristics
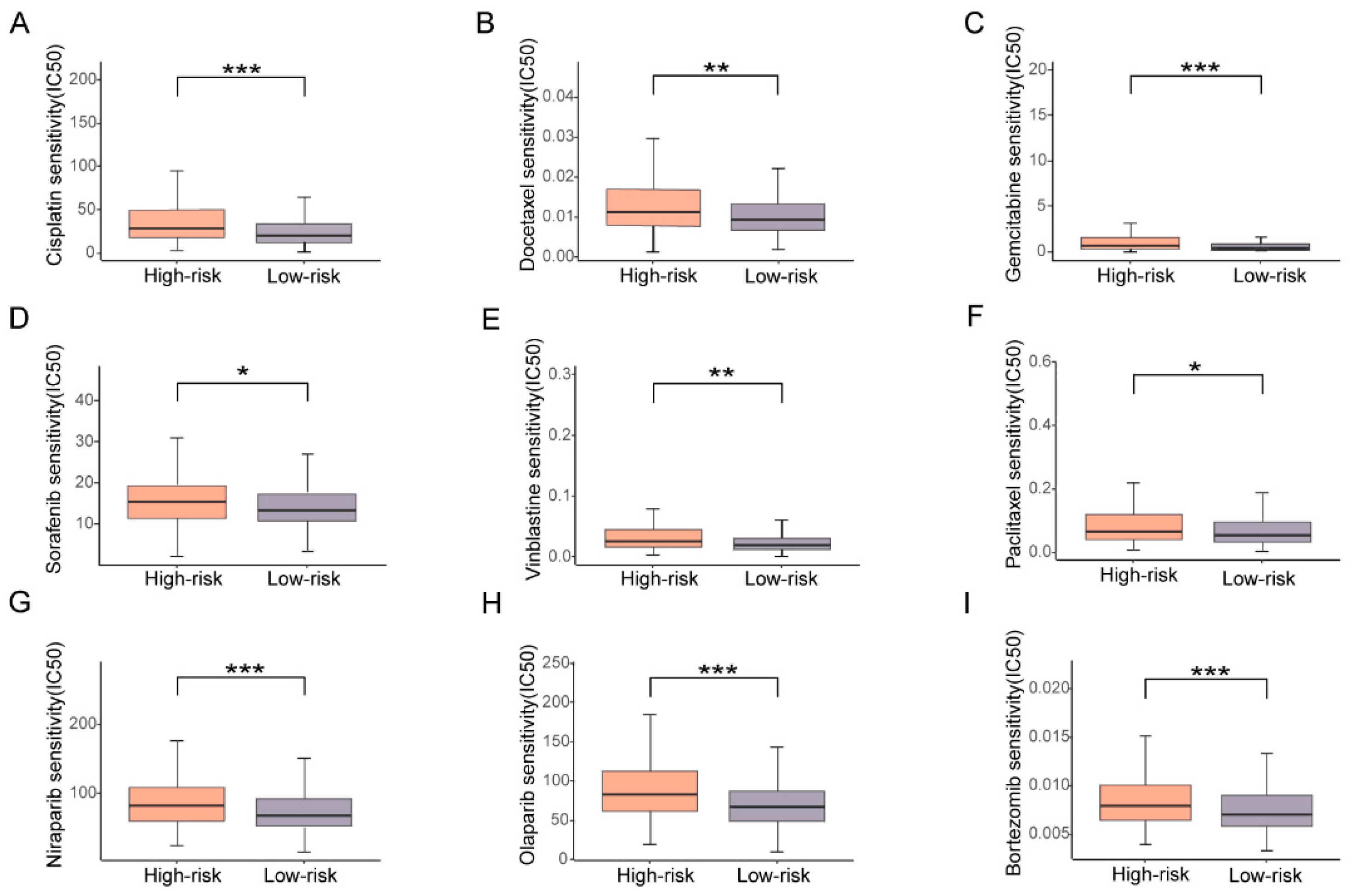
3.6. High Risk Based on the Prognostic Signature Corresponds with High Drug Resistance
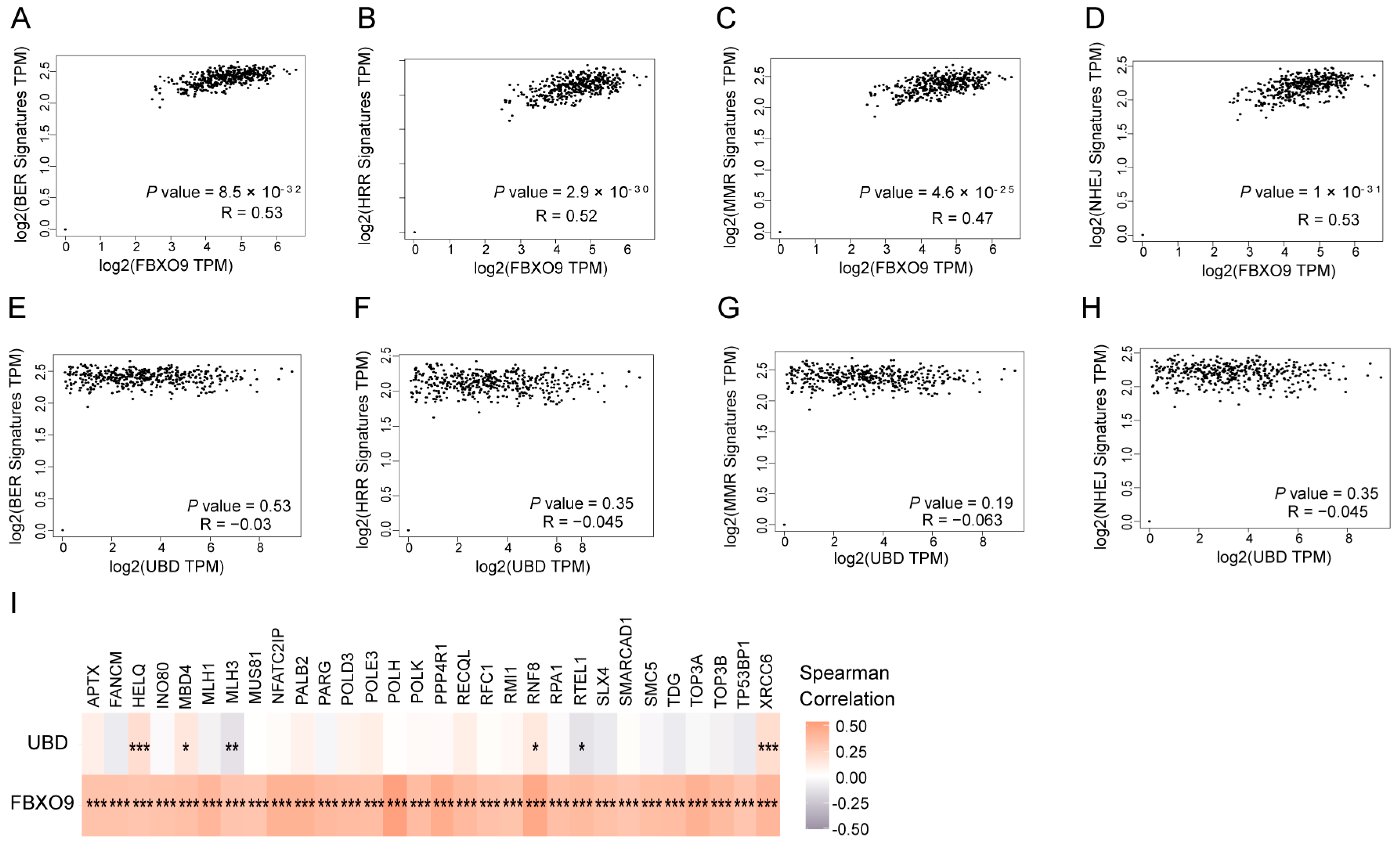
3.7. FBXO9 Is Involved in DNA Damage Repair
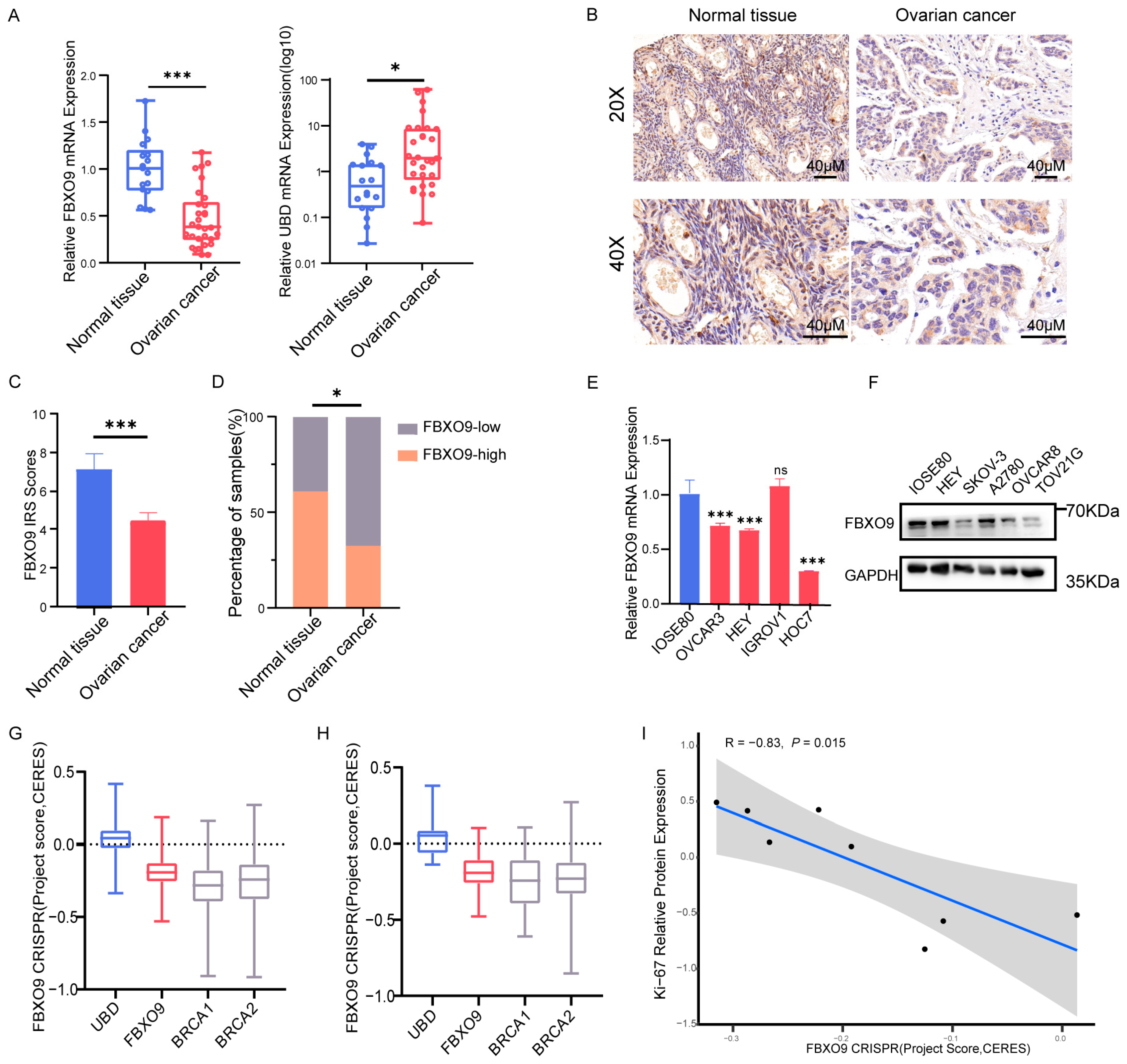
3.8. Altered Expression Level of FBXO9 in OV
4. Discussion
4.1. Advances of the UbRGs Signature for Prognostic Prediction in OV
4.2. Potential Mechanisms Underlying Ubiquitination-Mediated OV Risk
4.3. Potential Functions of FBXO9 in OV
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OV | Ovarian cancer |
| DE-UbRGs | Differentially expressed ubiquitin-related genes |
| DUB | Deubiquitinating enzymes |
| GTEx | Genotype-Tissue Expression Project |
| TCGA | The Cancer Genome Atlas |
| GEO | Gene Expression Omnibus |
| TME | Tumor microenvironment |
| IRS | Immunoreactivity scores |
| TPM | Transcripts Per Million |
| GSEA | Gene set enrichment analysis |
| GDSC | Genomics of Drug Sensitivity in Cancer |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| ROC | Receiver operating characteristic |
| UbRGs | Ubiquitin-related genes |
| PPI | Protein–protein interaction |
| OS | Overall survival |
| mRNA | Message RNA |
| IHC | Immunohistochemistry |
| TMB | Tumor mutation burden |
| AUC | Area under the curve |
| IC50 | Half-maximal inhibitory concentration |
| DDR | DNA damage repair |
| BER | Base excision repair |
| HRR | Homologous Recombination repair |
| MMR | Mismatch repair |
| NHEJ | Non-homologous end joining |
| LncRNAs | long noncoding RNAs |
| LASSO | least absolute shrinkage and selection operator |
| K-M | Kaplan–Meier |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CCLE | Cancer Cell Line Encyclopedia |
| KO | Knowout |
| HE | Hematoxylin and Eosin |
| BP | Biological Process |
| CC | Cellular Component |
| MF | Molecular Function |
References
- Ledermann, J.A.; Raja, F.A.; Fotopoulou, C.; Gonzales-Martín, A.; Colombo, N.; Sessa, C. ESMO Guidelines Working Group. Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2013, 24 (Suppl. S6), vi24–vi32. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Chen, W.; Zheng, R.; Zhang, S.; Ji, J.S.; Zou, X.; Xia, C.; Sun, K.; Yang, Z.; Li, H.; et al. Changing cancer survival in China during 2003–15: A pooled analysis of 17 population-based cancer registries. Lancet Glob. Health 2018, 6, e555–e567. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Weissman, A.M. Themes and Variations on Ubiquitylation. Nat. Rev. Mol. Cell Biol. 2001, 2, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Cockram, P.E.; Kist, M.; Prakash, S.; Chen, S.-H.; Wertz, I.E.; Vucic, D. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 2021, 28, 591–605. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Chen, Z.J.; Sun, L.J. Nonproteolytic Functions of Ubiquitin in Cell Signaling. Mol. Cell 2009, 33, 275–286. [Google Scholar] [CrossRef]
- Dang, F.; Nie, L.; Wei, W. Ubiquitin signaling in cell cycle control and tumorigenesis. Cell Death Differ. 2020, 28, 427–438. [Google Scholar] [CrossRef]
- Han, S.; Wang, R.; Zhang, Y.; Li, X.; Gan, Y.; Gao, F.; Rong, P.; Wang, W.; Li, W. The role of ubiquitination and deubiquitination in tumor invasion and metastasis. Int. J. Biol. Sci. 2022, 18, 2292–2303. [Google Scholar] [CrossRef]
- Çetin, G.; Klafack, S.; Studencka-Turski, M.; Krüger, E.; Ebstein, F. The Ubiquitin–Proteasome System in Immune Cells. Biomolecules 2021, 11, 60. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Sun, S.-C. Targeting ubiquitin signaling for cancer immunotherapy. Signal Transduct. Target. Ther. 2021, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Gavali, S.; Liu, J.; Li, X.; Paolino, M. Ubiquitination in T-Cell Activation and Checkpoint Inhibition: New Avenues for Targeted Cancer Immunotherapy. Int. J. Mol. Sci. 2021, 22, 10800. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.; Park, H.-B.; Baek, K.-H.; Hwang, S. Cellular Functions of Deubiquitinating Enzymes in Ovarian Adenocarcinoma. Genes 2023, 14, 886. [Google Scholar] [CrossRef]
- Vriend, J.; Nachtigal, M.W. Ubiquitin Proteasome Pathway Transcriptome in Epithelial Ovarian Cancer. Cancers 2021, 13, 2659. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, Y.; Yang, X.; Ren, X.; Huang, S.; Gong, S.; Tan, X.; Li, J.; He, S.; Li, Y.; et al. USP44 regulates irradiation-induced DNA double-strand break repair and suppresses tumorigenesis in nasopharyngeal carcinoma. Nat. Commun. 2022, 13, 501. [Google Scholar] [CrossRef]
- Zhou, J.; Xu, Y.; Lin, S.; Guo, Y.; Deng, W.; Zhang, Y.; Guo, A.; Xue, Y. iUUCD 2.0: An update with rich annotations for ubiquitin and ubiquitin-like conjugations. Nucleic Acids Res. 2018, 46, D447–D453. [Google Scholar] [CrossRef]
- Xiang, G.; Dong, X.; Xu, T.; Feng, Y.; He, Z.; Ke, C.; Xiao, J.; Weng, Y.-M. A Nomogram for Prediction of Postoperative Pneumonia Risk in Elderly Hip Fracture Patients. Risk Manag. Health Policy 2020, 13, 1603–1611. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Liu, H.; Yu, Y.; Xu, L. TRIM9 overexpression promotes uterine leiomyoma cell proliferation and inhibits cell apoptosis via NF-κB signaling pathway. Life Sci. 2020, 257, 118101. [Google Scholar] [CrossRef]
- Ye, B.; Liu, B.; Hao, L.; Zhu, X.; Yang, L.; Wang, S.; Xia, P.; Du, Y.; Meng, S.; Huang, G.; et al. Klf4 glutamylation is required for cell reprogramming and early embryonic development in mice. Nat. Commun. 2018, 9, 1261. [Google Scholar] [CrossRef]
- Peake, J.D.; Noguchi, E. Fanconi anemia: Current insights regarding epidemiology, cancer, and DNA repair. Hum. Genet. 2022, 141, 1811–1836. [Google Scholar] [CrossRef]
- Lee, K.W.; Kwak, S.H.; Koo, Y.D.; Cho, Y.-K.; Lee, H.M.; Jung, H.S.; Cho, Y.M.; Park, Y.J.; Chung, S.S.; Park, K.S. F-box only protein 9 is an E3 ubiquitin ligase of PPARγ. Exp. Mol. Med. 2016, 48, e234. [Google Scholar] [CrossRef]
- Aichem, A.; Groettrup, M. The ubiquitin-like modifier FAT10—Much more than a proteasome-targeting signal. J. Cell Sci. 2020, 133, jcs246041. [Google Scholar] [CrossRef]
- Richardson, P.G.; Hideshima, T.; Anderson, K.C. Bortezomib (PS-341): A Novel, First-in-Class Proteasome Inhibitor for the Treatment of Multiple Myeloma and Other Cancers. Cancer Control. 2003, 10, 361–369. [Google Scholar] [CrossRef]
- Xiong, W.; Gao, X.; Zhang, T.; Jiang, B.; Hu, M.-M.; Bu, X.; Gao, Y.; Zhang, L.-Z.; Xiao, B.-L.; He, C.; et al. USP8 inhibition reshapes an inflamed tumor microenvironment that potentiates the immunotherapy. Nat. Commun. 2022, 13, 1700. [Google Scholar] [CrossRef]
- Groelly, F.J.; Fawkes, M.; Dagg, R.A.; Blackford, A.N.; Tarsounas, M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer 2023, 23, 78–94. [Google Scholar] [CrossRef] [PubMed]
- Isildak, M.; Guven, G.S.; Kekilli, M.; Beyazit, Y.; Erman, M. Ovarian carcinoma in two patients with chronic liver disease. World J. Gastroenterol. 2005, 11, 4445–4446. [Google Scholar] [CrossRef]
- Ji, J.; Shen, J.; Xu, Y.; Xie, M.; Qian, Q.; Qiu, T.; Shi, W.; Ren, D.; Ma, J.; Liu, W.; et al. FBXO2 targets glycosylated SUN2 for ubiquitination and degradation to promote ovarian cancer development. Cell Death Dis. 2022, 13, 442. [Google Scholar] [CrossRef] [PubMed]
- Richardson, P.G.; Barlogie, B.; Berenson, J.; Singhal, S.; Jagannath, S.; Irwin, D.; Rajkumar, S.V.; Srkalovic, G.; Alsina, M.; Alexanian, R.; et al. A Phase 2 Study of Bortezomib in Relapsed, Refractory Myeloma. N. Engl. J. Med. 2003, 348, 2609–2617. [Google Scholar] [CrossRef] [PubMed]
- Che, Y.; Jiang, D.; Xu, L.; Sun, Y.; Wu, Y.; Liu, Y.; Chang, N.; Fan, J.; Xi, H.; Qiu, D.; et al. The Clinical Prediction Value of the Ubiquitination Model Reflecting the Immune Traits in LUAD. Front. Immunol. 2022, 13, 846402. [Google Scholar] [CrossRef]
- Li, Y.; An, L.; Jia, Z.; Li, J.; Zhou, E.; Wu, F.; Yin, Z.; Geng, W.; Liao, T.; Xiao, W.; et al. Identification of Ubiquitin-Related Gene-Pair Signatures for Predicting Tumor Microenvironment Infiltration and Drug Sensitivity of Lung Adenocarcinoma. Cancers 2022, 14, 3478. [Google Scholar] [CrossRef]
- Ni, W.; Bian, S.; Zhu, M.; Song, Q.; Zhang, J.; Xiao, M.; Zheng, W. Identification and Validation of Ubiquitin-Specific Proteases as a Novel Prognostic Signature for Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 629327. [Google Scholar] [CrossRef]
- Tang, Q.; Chen, Z.; Xie, J.; Mo, C.; Lu, J.; Zhang, Q.; Wang, Z.; Wu, W.; Wang, H. Transcriptome Analysis and Single-Cell Sequencing Analysis Constructed the Ubiquitination-Related Signature in Glioma and Identified USP4 as a Novel Biomarker. Front. Immunol. 2022, 13, 915709. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, J.; Du, X.; Zhao, Y.; Zou, C.; Zhou, H.; Li, W.; Yan, X. Construction and validation of a novel aging-related gene signature and prognostic nomogram for predicting the overall survival in ovarian cancer. Cancer Med. 2021, 10, 9097–9114. [Google Scholar] [CrossRef]
- Yang, S.; Ji, J.; Wang, M.; Nie, J.; Wang, S. Construction of Ovarian Cancer Prognostic Model Based on the Investigation of Ferroptosis-Related lncRNA. Biomolecules 2023, 13, 306. [Google Scholar] [CrossRef]
- Mo, J.; Ruan, S.; Yang, B.; Jin, Y.; Liu, K.; Luo, X.; Jiang, H. A novel defined risk signature of endoplasmic reticulum stress-related genes for predicting the prognosis and immune infiltration status of ovarian cancer. J. Zhejiang Univ. B 2023, 24, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Geng, R.; Chen, T.; Zhong, Z.; Ni, S.; Bai, J.; Liu, J. The m6A-Related Long Noncoding RNA Signature Predicts Prognosis and Indicates Tumor Immune Infiltration in Ovarian Cancer. Cancers 2022, 14, 4056. [Google Scholar] [CrossRef] [PubMed]
- Romeo, E.; Caserta, C.A.; Rumio, C.; Marcucci, F. The Vicious Cross-Talk between Tumor Cells with an EMT Phenotype and Cells of the Immune System. Cells 2019, 8, 460. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.; Zhu, M.; Wang, L.; Liu, Y.; Tian, H.; Zheng, Y.; Tan, F.; Sun, N.; Li, C.; He, J. Immune profile of the tumor microenvironment and the identification of a four-gene signature for lung adenocarcinoma. Aging 2020, 13, 2397–2417. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Hu, R.; Zeng, Y.; Zhang, W.; Zhou, H.-H. Tumour immune cell infiltration and survival after platinum-based chemotherapy in high-grade serous ovarian cancer subtypes: A gene expression-based computational study. EBioMedicine 2020, 51, 102602. [Google Scholar] [CrossRef]
- Wouters, M.C.A.; Nelson, B.H. Prognostic Significance of Tumor-Infiltrating B Cells and Plasma Cells in Human Cancer. Clin. Cancer Res. 2018, 24, 6125–6135. [Google Scholar] [CrossRef]
- Arians, N.; Kieser, M.; Benner, L.; Rochet, N.; Katayama, S.; Sterzing, F.; Herfarth, K.; Schubert, K.; Schröder, L.; Leitzen, C.; et al. Adjuvant Intensity Modulated Whole-Abdominal Radiation Therapy for High-Risk Patients With Ovarian Cancer (International Federation of Gynecology and Obstetrics Stage III): First Results of a Prospective Phase 2 Study. Int. J. Radiat. Oncol. 2017, 99, 912–920. [Google Scholar] [CrossRef]
- Dias, M.P.; Moser, S.C.; Ganesan, S.; Jonkers, J. Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 2021, 18, 773–791. [Google Scholar] [CrossRef]
- Chou, C.-L.; Chen, T.-J.; Li, W.-S.; Lee, S.-W.; Yang, C.-C.; Tian, Y.-F.; Lin, C.-Y.; He, H.-L.; Wu, H.-C.; Shiue, Y.-L.; et al. Upregulated Ubiquitin D is a Favorable Prognostic Indicator for Rectal Cancer Patients Undergoing Preoperative Concurrent Chemoradiotherapy. OncoTargets Ther. 2022, 15, 1171–1181. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, X.; Zhou, L.; Zhao, X.; Ge, C.; Zhao, F.; Xie, H.; Chen, T.; Tian, H.; Li, H.; et al. FBXO9 Mediates the Cancer-Promoting Effects of ZNF143 by Degrading FBXW7 and Facilitates Drug Resistance in Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 930220. [Google Scholar] [CrossRef]
- Hussain, S.; Dong, J.; Ma, X.; Li, J.; Chen, S.; Clement, A.; Liu, H. F-box only protein 9 and its role in cancer. Mol. Biol. Rep. 2022, 49, 1537–1544. [Google Scholar] [CrossRef]
- Liu, J.A.; Tai, A.; Hong, J.; Cheung, M.P.L.; Sham, M.H.; Cheah, K.S.E.; Cheung, C.W.; Cheung, M. Fbxo9 functions downstream of Sox10 to determine neuron-glial fate choice in the dorsal root ganglia through Neurog2 destabilization. Proc. Natl. Acad. Sci. USA 2020, 117, 4199–4210. [Google Scholar] [CrossRef] [PubMed]
- Foulkes, W.; Ragoussis, J.; Stamp, G.; Allan, G.; Trowsdale, J. Frequent loss of heterozygosity on chromosome 6 in human ovarian carcinoma. Br. J. Cancer 1993, 67, 551–559. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fernández-Sáiz, V.; Targosz, B.-S.; Lemeer, S.; Eichner, R.; Langer, C.; Bullinger, L.; Reiter, C.; Slotta-Huspenina, J.; Schroeder, S.; Knorn, A.-M.; et al. SCFFbxo9 and CK2 direct the cellular response to growth factor withdrawal via Tel2/Tti1 degradation and promote survival in multiple myeloma. Nature 2013, 15, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, Y.; Xu, S.; Huang, S.; Wu, M.; Chen, G.; Wang, Y. Endoplasmic Reticulum Stress-Related Ten-Biomarker Risk Classifier for Survival Evaluation in Epithelial Ovarian Cancer and TRPM2: A Potential Therapeutic Target of Ovarian Cancer. Int. J. Mol. Sci. 2023, 24, 14010. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, X.; Wang, Y.; Zhang, H.; Chen, G.; Sheng, J.; Tian, X.; Xue, R.; Wang, Y. Identification of a Prognostic Signature for Ovarian Cancer Based on Ubiquitin-Related Genes Suggesting a Potential Role for FBXO9. Biomolecules 2023, 13, 1724. https://doi.org/10.3390/biom13121724
Luo X, Wang Y, Zhang H, Chen G, Sheng J, Tian X, Xue R, Wang Y. Identification of a Prognostic Signature for Ovarian Cancer Based on Ubiquitin-Related Genes Suggesting a Potential Role for FBXO9. Biomolecules. 2023; 13(12):1724. https://doi.org/10.3390/biom13121724
Chicago/Turabian StyleLuo, Xiaomei, Yingjie Wang, Hao Zhang, Guangquan Chen, Jindan Sheng, Xiu Tian, Renhao Xue, and Yu Wang. 2023. "Identification of a Prognostic Signature for Ovarian Cancer Based on Ubiquitin-Related Genes Suggesting a Potential Role for FBXO9" Biomolecules 13, no. 12: 1724. https://doi.org/10.3390/biom13121724
APA StyleLuo, X., Wang, Y., Zhang, H., Chen, G., Sheng, J., Tian, X., Xue, R., & Wang, Y. (2023). Identification of a Prognostic Signature for Ovarian Cancer Based on Ubiquitin-Related Genes Suggesting a Potential Role for FBXO9. Biomolecules, 13(12), 1724. https://doi.org/10.3390/biom13121724





