Functions of Representative Terpenoids and Their Biosynthesis Mechanisms in Medicinal Plants
Abstract
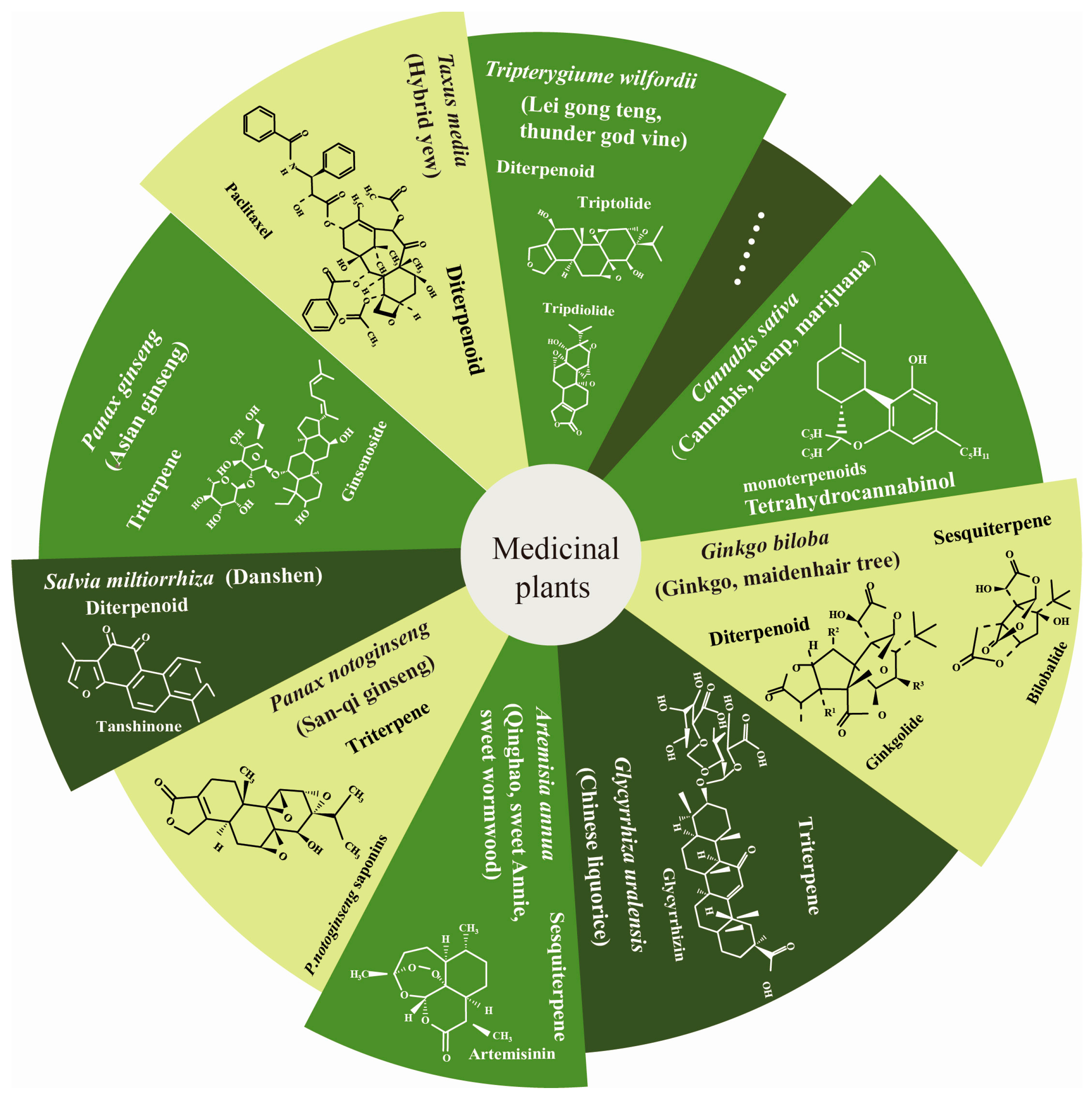
:1. Introduction
2. Function of Terpenoids in Human Health
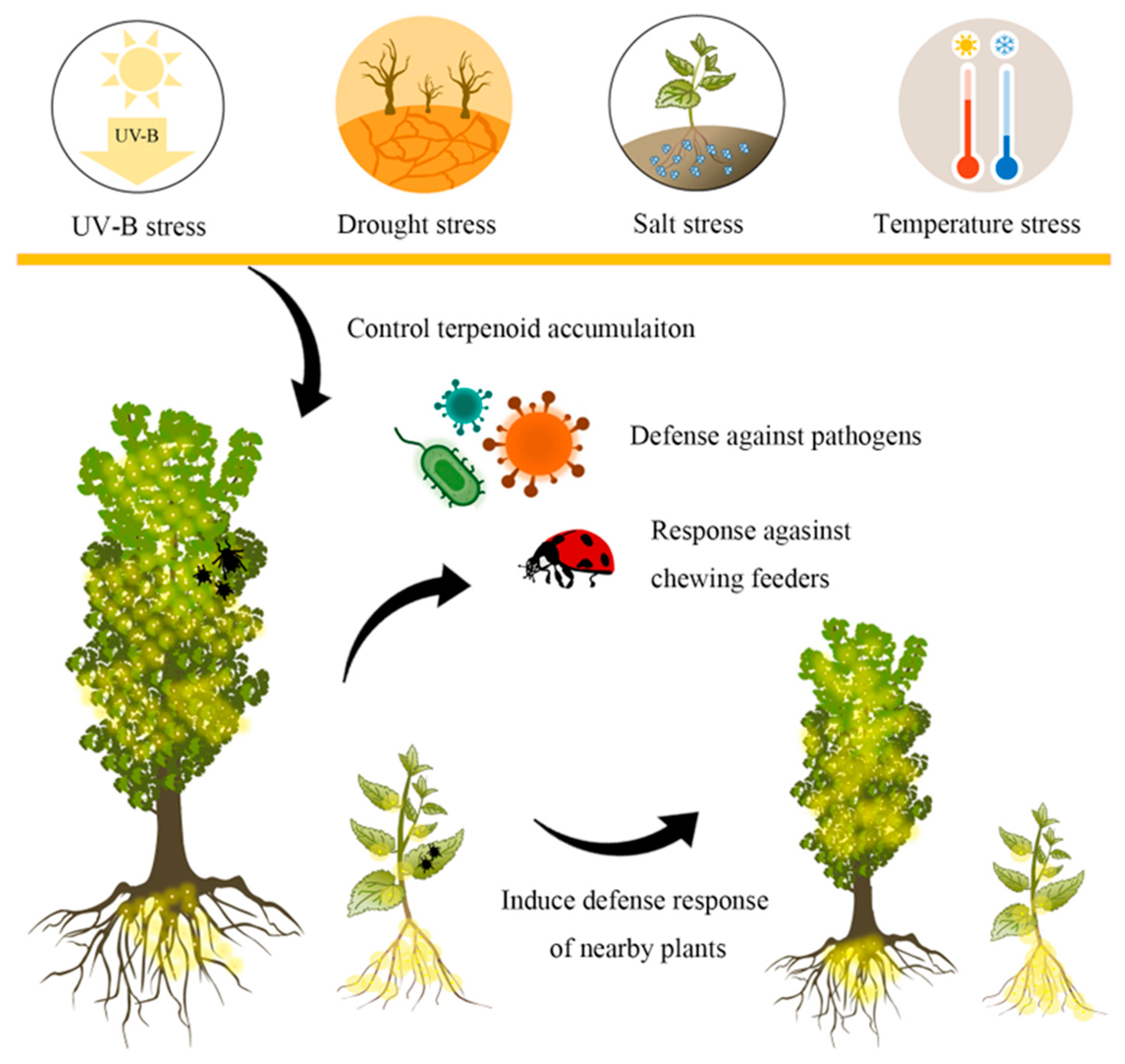
3. Functions of Terpenoids in Biotic Stress
4. Effects of Environmental Factors on Terpenoid Accumulation
5. Accumulation Characteristics of Terpenoids and Their Transport
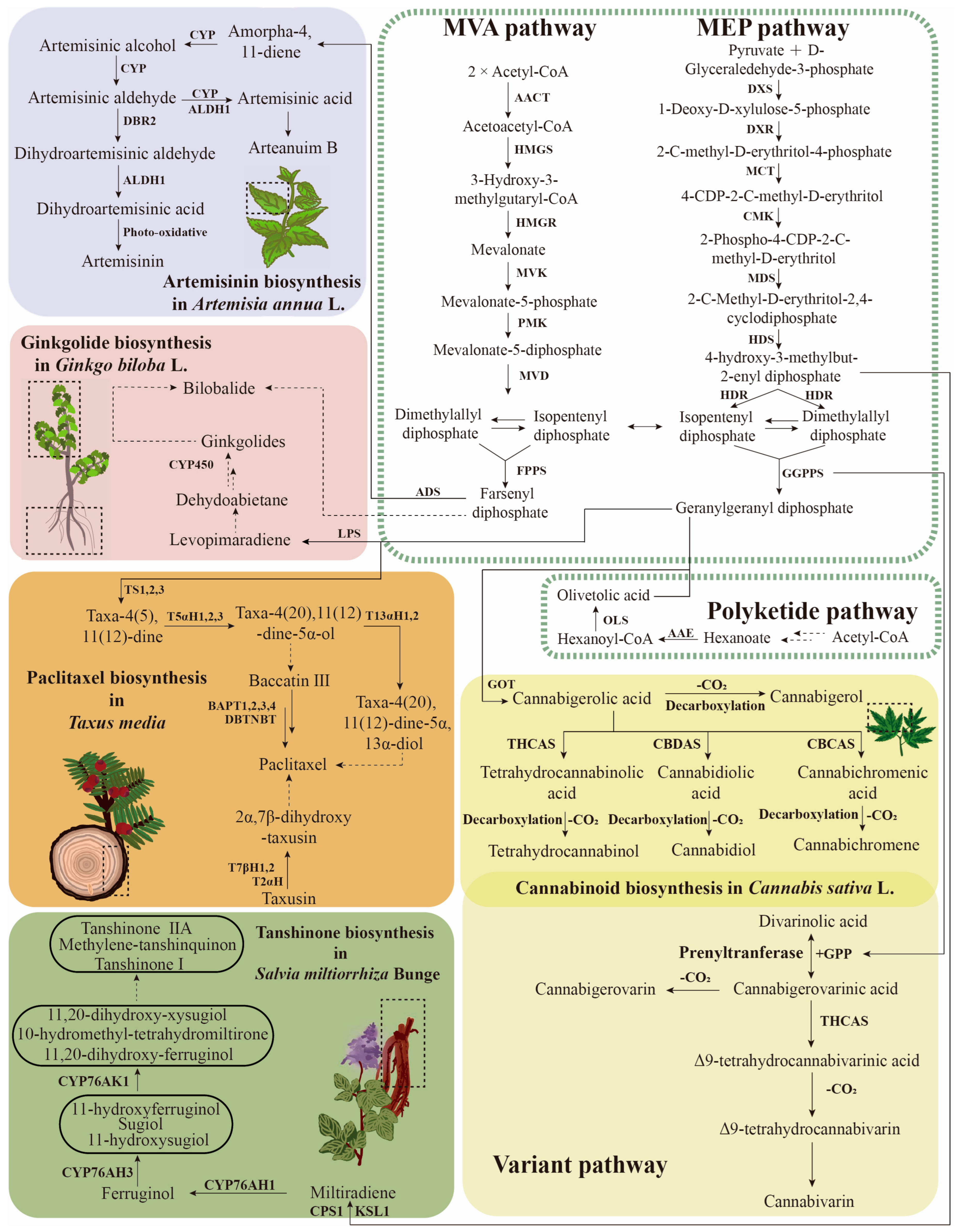
6. Terpenoid Biosynthesis Pathway and Regulation
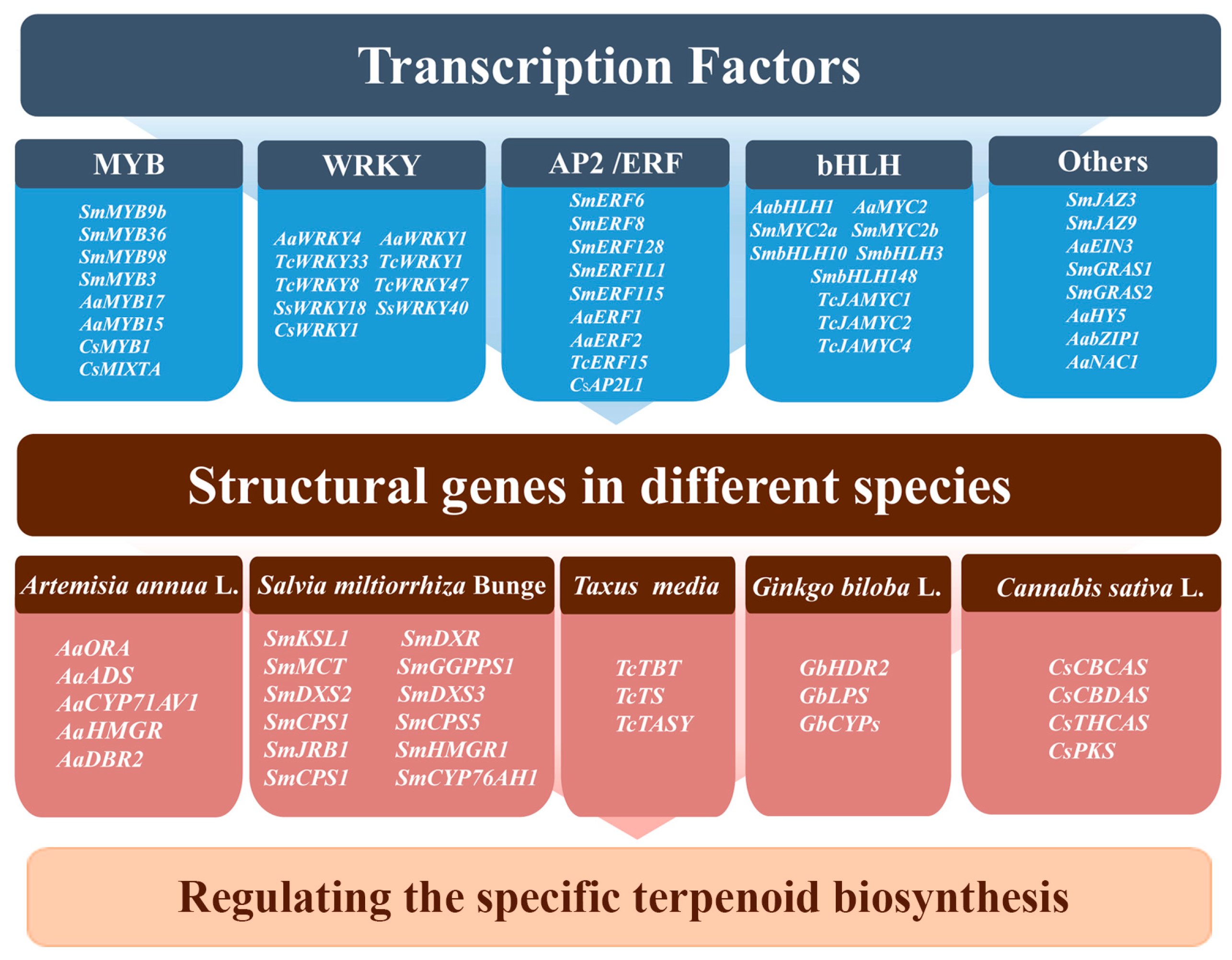
7. Regulation of Terpenoid Biosynthesis by Transcription Factors
8. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Ahad, B.; Shahri, W.; Rasool, H.; Reshi, Z.; Rasool, S.; Hussain, T. Medicinal plants and herbal drugs: An overview. In Medicinal and Aromatic Plants: Healthcare and Industrial Applications; Springer: Berlin/Heidelberg, Germany, 2021; pp. 1–40. [Google Scholar]
- Li, J.; Li, B.; Luo, L.; Cao, F.; Yang, B.; Gao, J.; Yan, Y.; Zhang, G.; Peng, L.; Hu, B. Increased phenolic acid and tanshinone production and transcriptional responses of biosynthetic genes in hairy root cultures of Salvia przewalskii Maxim. treated with methyl jasmonate and salicylic acid. Mol. Biol. Rep. 2020, 47, 8565–8578. [Google Scholar] [CrossRef]
- Bailly, C.; Vergoten, G. Glycyrrhizin: An alternative drug for the treatment of COVID-19 infection and the associated respiratory syndrome? Pharmacol. Ther. 2020, 214, 107618. [Google Scholar] [CrossRef]
- Frank, L.; Wenig, M.; Ghirardo, A.; van der Krol, A.; Vlot, A.C.; Schnitzler, J.P.; Rosenkranz, M. Environment. Isoprene and β-caryophyllene confer plant resistance via different plant internal signalling pathways. Plant Cell Environ. 2021, 44, 1151–1164. [Google Scholar] [CrossRef]
- Zhou, Z.; Tan, H.; Li, Q.; Li, Q.; Wang, Y.; Bu, Q.; Li, Y.; Wu, Y.; Chen, W.; Zhang, L. Trichome and Artemisinin Regulator 2 positively regulates trichome development and artemisinin biosynthesis in Artemisia annua. New Phytol. 2020, 228, 932–945. [Google Scholar] [CrossRef] [PubMed]
- Nagegowda, D.A.; Gupta, P. Advances in biosynthesis, regulation, and metabolic engineering of plant specialized terpenoids. Plant Sci. 2020, 294, 110457. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Jayakodi, M.; Lee, S.C.; Choi, B.S.; Jang, W.; Lee, J.; Kim, H.H.; Waminal, N.E.; Lakshmanan, M.; van Nguyen, B. Genome and evolution of the shade-requiring medicinal herb Panax ginseng. Plant Biotechnol. J. 2018, 16, 1904–1917. [Google Scholar] [CrossRef]
- Lu, S. Biosynthesis and regulatory mechanisms of bioactive compounds in Salvia miltiorrhiza, a model system for medicinal plant biology. Crit. Rev. Plant Sci. 2021, 40, 243–283. [Google Scholar] [CrossRef]
- Shen, Q.; Zhang, L.; Liao, Z.; Wang, S.; Yan, T.; Shi, P.; Liu, M.; Fu, X.; Pan, Q.; Wang, Y. The genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynthesis. Mol. Plant 2018, 11, 776–788. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Gou, J.; Liao, Q.; Li, Y.; Zhou, Q.; Bi, G.; Li, C.; Du, R.; Wang, X.; Sun, T. The Taxus genome provides insights into paclitaxel biosynthesis. Nat. Plants 2021, 7, 1026–1036. [Google Scholar] [CrossRef]
- Zhao, Y.-P.; Fan, G.; Yin, P.-P.; Sun, S.; Li, N.; Hong, X.; Hu, G.; Zhang, H.; Zhang, F.-M.; Han, J.-D. Resequencing 545 ginkgo genomes across the world reveals the evolutionary history of the living fossil. Nat. Commun. 2019, 10, 4201. [Google Scholar] [CrossRef]
- Halder, M.; Jha, S. Medicinal Plants and Bioactive Phytochemical Diversity: A Fountainhead of Potential Drugs Against Human Diseases. In Medicinal Plants: Biodiversity, Biotechnology and Conservation; Springer Nature: Singapore, 2023; pp. 39–93. [Google Scholar]
- Wendel, J.F.; Jackson, S.A.; Meyers, B.C.; Wing, R.A. Evolution of plant genome architecture. Genome Biol. 2016, 17, 37. [Google Scholar] [CrossRef]
- Meng, F.; Tang, Q.; Chu, T.; Li, X.; Lin, Y.; Song, X.; Chen, W. TCMPG: An integrative database for traditional Chinese medicine plant genomes. Hortic. Res. 2022, 9, uhac060. [Google Scholar] [CrossRef] [PubMed]
- Masyita, A.; Sari, R.M.; Astuti, A.D.; Yasir, B.; Rumata, N.R.; Emran, T.B.; Nainu, F.; Simal-Gandara, J. Terpenes and terpenoids as main bioactive compounds of essential oils, their roles in human health and potential application as natural food preservatives. Food Chem. X 2022, 13, 100217. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.K.; Patel, K.H.; Moochhala, S.M. Gut microbiota intervention strategies using active components from medicinal herbs to evaluate clinical efficacy of type 2 diabetes–A review. Clin. Transl. Discov. 2023, 3, e170. [Google Scholar] [CrossRef]
- Briskin, D.P. Medicinal plants and phytomedicines. Linking plant biochemistry and physiology to human health. Plant Physiol. 2000, 124, 507–514. [Google Scholar] [CrossRef]
- Marchev, A.S.; Vasileva, L.V.; Amirova, K.M.; Savova, M.S.; Balcheva-Sivenova, Z.P.; Georgiev, M.I. Metabolomics and health: From nutritional crops and plant-based pharmaceuticals to profiling of human biofluids. Cell. Mol. Life Sci. 2021, 78, 6487–6503. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, L.; Wang, L.-P.; Wan, Q. Therapeutic targets of neuroprotection and neurorestoration in ischemic stroke: Applications for natural compounds from medicinal herbs. Biomed. Pharmacother. 2022, 148, 112719. [Google Scholar] [CrossRef]
- Alexander, J.C.; Joshi, G.P. A review of the anesthetic implications of marijuana use. Bayl. Univ. Med. Cent. Proc. 2019, 3, 364–371. [Google Scholar] [CrossRef]
- Zhao, T.; Zhang, T.; Zhang, Y.; Zhou, B.; Lu, X. Paclitaxel resistance modulated by the interaction between TRPS1 and AF178030. 2 in triple-negative breast cancer. Evid.-Based Complement. Altern. Med. 2022, 2022, 6019975. [Google Scholar]
- Guo, Y.; Zhang, T.; Zhong, J.; Ba, T.; Xu, T.; Zhang, Q.; Sun, M. Identification of the volatile compounds and observation of the glandular trichomes in Opisthopappus taihangensis and four species of Chrysanthemum. Plants 2020, 9, 855. [Google Scholar] [CrossRef]
- Boateng, I.D. Ginkgols and bilobols in Ginkgo biloba L. A review of their extraction and bioactivities. Phytother. Res. 2023, 37, 3211–3223. [Google Scholar] [CrossRef]
- Andre, C.M.; Hausman, J.-F.; Guerriero, G. Cannabis sativa: The plant of the thousand and one molecules. Front. Plant Sci. 2016, 7, 19. [Google Scholar] [CrossRef]
- Alharbi, K.S.; Fuloria, N.K.; Fuloria, S.; Rahman, S.B.; Al-Malki, W.H.; Shaikh, M.A.J.; Thangavelu, L.; Singh, S.K.; Allam, V.S.R.R.; Jha, N.K. Nuclear factor-kappa B and its role in inflammatory lung disease. Chem.-Biol. Interact. 2021, 345, 109568. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and mechanism of jasmonic acid in plant responses to abiotic and biotic stresses. Int. J. Mol. Sci. 2021, 22, 8568. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.S.; Li, J.; Wang, Z.; Mi, C.; Ma, J.; Piao, L.X.; Xu, G.H.; Li, X.; Jin, X. Artemisinin inhibits inflammatory response via regulating NF-κB and MAPK signaling pathways. Immunopharmacol. Immunotoxicol. 2017, 39, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Xie, L.; Liu, C.; Fu, C.; Ye, W.; Liu, H.; Zhang, B. Tanshinone IIA improves hypoxic ischemic encephalopathy through TLR-4-mediated NF-κB signal pathway. Mol. Med. Rep. 2018, 18, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.-B.; Lu, X.-M.; Wang, H.-Y.; Liu, H.-L.; Wu, Q.-Y.; Liao, P.; Li, S.; Long, Z.-Y.; Wang, Y.-T. Effects and mechanisms of Tanshinone IIA on PTSD-like symptoms. Phytomedicine 2023, 120, 155032. [Google Scholar] [CrossRef]
- Fang, J.; Chen, Q.; He, B.; Cai, J.; Yao, Y.; Cai, Y.; Xu, S.; Rengasamy, K.R.; Gowrishankar, S.; Pandian, S.K. Tanshinone IIA attenuates TNF-α induced PTX3 expression and monocyte adhesion to endothelial cells through the p38/NF-κB pathway. Food Chem. Toxicol. 2018, 121, 622–630. [Google Scholar] [CrossRef]
- Kozela, E.; Pietr, M.; Juknat, A.; Rimmerman, N.; Levy, R.; Vogel, Z. Cannabinoids Δ9-tetrahydrocannabinol and cannabidiol differentially inhibit the lipopolysaccharide-activated NF-κB and interferon-β/STAT proinflammatory pathways in BV-2 microglial cells. J. Biol. Chem. 2010, 285, 1616–1626. [Google Scholar] [CrossRef]
- Zhu, P.-C.; Tong, Q.; Zhuang, Z.; Wang, Z.-H.; Deng, L.-H.; Zheng, G.-q.; Wang, Y. Ginkgolide B for myocardial ischemia/reperfusion injury: A preclinical systematic review and meta-analysis. Front. Physiol. 2019, 10, 1292. [Google Scholar] [CrossRef]
- Zhao, S.; Tang, Y.; Wang, R.; Najafi, M. Mechanisms of cancer cell death induction by paclitaxel: An updated review. Apoptosis 2022, 27, 647–667. [Google Scholar] [CrossRef] [PubMed]
- Silva, H.; Martins, F.G. Cardiovascular Activity of Ginkgo Biloba—An Insight from Healthy Subjects. Biology 2022, 12, 15. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Li, L.; Su, J.; Li, S.; Duncan, S.E.; Liu, Z.; Fan, G. Pharmacological activity and mechanism of tanshinone IIA in related diseases. Drug Des. Dev. Ther. 2020, 14, 4735–4748. [Google Scholar] [CrossRef]
- Martins, J.N.; Silva, S.R. Use of Infrared Thermography to Assess Body Temperature as a Physiological Stress Indicator in Horses during Ridden and Lunging Sessions. Animals 2022, 12, 3255. [Google Scholar] [CrossRef] [PubMed]
- Vladu, A.F.; Ficai, D.; Ene, A.G.; Ficai, A. Combination Therapy Using Polyphenols: An Efficient Way to Improve Antitumoral Activity and Reduce Resistance. Int. J. Mol. Sci. 2022, 23, 10244. [Google Scholar] [CrossRef]
- Bergman, M.E.; Davis, B.; Phillips, M.A. Medically Useful Plant Terpenoids: Biosynthesis, Occurrence, and Mechanism of Action. Molecules 2019, 24, 3961. [Google Scholar] [CrossRef]
- Li, Y.-I.; Elmer, G.; LeBoeuf, R.C. Tanshinone IIA reduces macrophage death induced by hydrogen peroxide by upregulating glutathione peroxidase. Life Sci. 2008, 83, 557–562. [Google Scholar] [CrossRef]
- Kopustinskiene, D.M.; Masteikova, R.; Lazauskas, R.; Bernatoniene, J. Cannabis sativa L. Bioactive compounds and their protective role in oxidative stress and inflammation. Antioxidants 2022, 11, 660. [Google Scholar] [CrossRef]
- Russo, E.B. Taming THC: Potential cannabis synergy and phytocannabinoid—Terpenoid entourage effects. Br. J. Pharmacol. 2011, 163, 1344–1364. [Google Scholar] [CrossRef]
- Kostanda, E.; Khatib, S. Biotic stress caused by Tetranychus urticae mites elevates the quantity of secondary metabolites, cannabinoids and terpenes, in Cannabis sativa L. Ind. Crops Prod. 2022, 176, 114331. [Google Scholar] [CrossRef]
- Lv, Z.; Wang, S.; Zhang, F.; Chen, L.; Hao, X.; Pan, Q.; Fu, X.; Li, L.; Sun, X.; Tang, K. Overexpression of a novel NAC domain-containing transcription factor gene (AaNAC1) enhances the content of artemisinin and increases tolerance to drought and Botrytis cinerea in Artemisia annua. Plant Cell Physiol. 2016, 57, 1961–1971. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, F.; Chen, S.; Guan, Z.; Jiang, J.; Fang, W.; Chen, F. Effects of aphid herbivory on volatile organic compounds of Artemisia annua and Chrysanthemum morifolium. Biochem. Syst. Ecol. 2015, 60, 225–233. [Google Scholar] [CrossRef]
- Xu, M.; Jiang, Y.; Chen, S.; Chen, F.; Chen, F. Herbivory-induced emission of volatile terpenes in Chrysanthemum morifolium functions as an indirect defense against Spodoptera litura larvae by attracting natural enemies. J. Agric. Food Chem. 2021, 69, 9743–9753. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Wang, X.; Meng, F.; Liu, Q.; Li, X.; Zhang, Y.; Piao, S. Three-dimensional change in temperature sensitivity of northern vegetation phenology. Glob. Change Biol. 2020, 26, 5189–5201. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Zhao, M.; Jing, T.; Zhang, M.; Lu, M.; Yu, G.; Wang, J.; Guo, D.; Pan, Y.; Hoffmann, T.D. Volatile compound-mediated plant–plant interactions under stress with the tea plant as a model. Hortic. Res. 2023, 10, uhad143. [Google Scholar] [CrossRef]
- Magagnini, G.; Grassi, G.; Kotiranta, S. The effect of light spectrum on the morphology and cannabinoid content of Cannabis sativa L. Med. Cannabis Cannabinoids 2018, 1, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Danziger, N.; Bernstein, N. Light matters: Effect of light spectra on cannabinoid profile and plant development of medical cannabis (Cannabis sativa L.). Ind. Crops Prod. 2021, 164, 113351. [Google Scholar] [CrossRef]
- Hawley, D. The Influence of Spectral Quality of Light on Plant Secondary Metabolism and Photosynthetic Acclimation to Light Quality; University of Guelph: Guelph, ON, Canada, 2018. [Google Scholar]
- Namdar, D.; Charuvi, D.; Ajjampura, V.; Mazuz, M.; Ion, A.; Kamara, I.; Koltai, H. LED lighting affects the composition and biological activity of Cannabis sativa secondary metabolites. Ind. Crops Prod. 2019, 132, 177–185. [Google Scholar] [CrossRef]
- Hawley, D.; Graham, T.; Stasiak, M.; Dixon, M. Improving Cannabis Bud Quality and Yield with Subcanopy Lighting. HortScience 2018, 53, 1593–1599. [Google Scholar] [CrossRef]
- Morello, V.; Brousseau, V.D.; Wu, N.; Wu, B.S.; MacPherson, S.; Lefsrud, M. Light quality impacts vertical growth rate, phytochemical yield and cannabinoid production efficiency in Cannabis sativa. Plants 2022, 11, 2982. [Google Scholar] [CrossRef]
- Arora, A.S.; Yun, C.M. Dynamic spectrum lighting impact on plant morphology and cannabinoid profile of medical and recreational cannabis—A novel leapfrog strategy towards shaping the future of horticulture lighting. Ind. Crops Prod. 2023, 199, 116799. [Google Scholar] [CrossRef]
- Demkura, P.V.; Ballare, C.L. UVR8 mediates UV-B-induced Arabidopsis defense responses against Botrytis cinerea by controlling sinapate accumulation. Mol. Plant 2012, 5, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Trancoso, I.; de Souza, G.A.; dos Santos, P.R.; dos Santos, K.D.; de Miranda, R.M.d.S.N.; da Silva, A.L.P.M.; Santos, D.Z.; García-Tejero, I.F.; Campostrini, E. Cannabis sativa L.: Crop management and abiotic factors that affect phytocannabinoid production. Agronomy 2022, 12, 1492. [Google Scholar] [CrossRef]
- Zhang, C.; Xing, B.; Yang, D.; Ren, M.; Guo, H.; Yang, S.; Liang, Z. SmbHLH3 acts as a transcription repressor for both phenolic acids and tanshinone biosynthesis in Salvia miltiorrhiza hairy roots. Phytochemistry 2020, 169, 112183. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Shi, C.; Alamgir, K.; Kwon, S.; Pan, L.; Zhu, Y.; Yang, X. Global assessment of the distribution and conservation status of a key medicinal plant (Artemisia annua L.): The roles of climate and anthropogenic activities. Sci. Total Environ. 2022, 821, 153378. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Zhang, C.; Lv, Z.; Shen, C. Pre-and post-harvest exposure to stress influence quality-related metabolites in fresh tea leaves (Camellia sinensis). Sci. Hortic. 2021, 281, 109984. [Google Scholar] [CrossRef]
- Wallaart, T.E.; Pras, N.; Beekman, A.C.; Quax, W.J. Seasonal variation of artemisinin and its biosynthetic precursors in plants of Artemisia annua of different geographical origin: Proof for the existence of chemotypes. Planta Medica 2000, 66, 57–62. [Google Scholar] [CrossRef]
- Naeem, M.; Sadiq, Y.; Jahan, A.; Nabi, A.; Aftab, T.; Khan, M.M.A. Salicylic acid restrains arsenic induced oxidative burst in two varieties of Artemisia annua L. by modulating antioxidant defence system and artemisinin production. Ecotoxicol. Environ. Saf. 2020, 202, 110851. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, X.; Fu, M.; Zeng, H.; Ye, J.; Zhang, W.; Liao, Y.; Xu, F. Effects of different stress treatments on the total terpene trilactone content and expression levels of key genes in Ginkgo biloba leaves. Plant Mol. Biol. Rep. 2020, 38, 521–530. [Google Scholar] [CrossRef]
- Vashisth, D.; Kumar, R.; Rastogi, S.; Patel, V.K.; Kalra, A.; Gupta, M.M.; Gupta, A.K.; Shasany, A.K. Transcriptome changes induced by abiotic stresses in Artemisia annua. Sci. Rep. 2018, 8, 3423. [Google Scholar] [CrossRef]
- Rebey, I.B.; Bourgou, S.; Rahali, F.Z.; Msaada, K.; Ksouri, R.; Marzouk, B. Relation between salt tolerance and biochemical changes in cumin (Cuminum cyminum L.) seeds. J. Food Drug Anal. 2017, 25, 391–402. [Google Scholar] [CrossRef]
- Yep, B.; Gale, N.V.; Zheng, Y. Comparing hydroponic and aquaponic rootzones on the growth of two drug-type Cannabis sativa L. cultivars during the flowering stage. Ind. Crops Prod. 2020, 157, 112881. [Google Scholar] [CrossRef]
- Caser, M.; Chitarra, W.; D’Angiolillo, F.; Perrone, I.; Demasi, S.; Lovisolo, C.; Pistelli, L.; Pistelli, L.; Scariot, V. Drought stress adaptation modulates plant secondary metabolite production in Salvia dolomitica Codd. Ind. Crops Prod. 2019, 129, 85–96. [Google Scholar] [CrossRef]
- Haberstroh, S.; Kreuzwieser, J.; Lobo-do-Vale, R.; Caldeira, M.C.; Dubbert, M.; Werner, C. Terpenoid emissions of two Mediterranean woody species in response to drought stress. Front. Plant Sci. 2018, 9, 1071. [Google Scholar] [CrossRef] [PubMed]
- Morshedloo, M.R.; Craker, L.E.; Salami, A.; Nazeri, V.; Sang, H.; Maggi, F. Effect of prolonged water stress on essential oil content, compositions and gene expression patterns of mono-and sesquiterpene synthesis in two oregano (Origanum vulgare L.) subspecies. Plant Physiol. Biochem. 2017, 111, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Caplan, D.; Dixon, M.; Zheng, Y. Increasing inflorescence dry weight and cannabinoid content in medical cannabis using controlled drought stress. HortScience 2019, 54, 964–969. [Google Scholar] [CrossRef]
- Wei, T.; Deng, K.; Liu, D.; Gao, Y.; Liu, Y.; Yang, M.; Zhang, L.; Zheng, X.; Wang, C.; Song, W. Ectopic expression of DREB transcription factor, AtDREB1A, confers tolerance to drought in transgenic Salvia miltiorrhiza. Plant Cell Physiol. 2016, 57, 1593–1609. [Google Scholar] [CrossRef]
- Liao, B.; Shen, X.; Xiang, L.; Guo, S.; Chen, S.; Meng, Y.; Liang, Y.; Ding, D.; Bai, J.; Zhang, D. Allele-aware chromosome-level genome assembly of Artemisia annua reveals the correlation between ADS expansion and artemisinin yield. Mol. Plant 2022, 15, 1310–1328. [Google Scholar] [CrossRef]
- Kennedy, D.O. Plants and the Human Brain; Oxford University Press: Oxford, MS, USA, 2014. [Google Scholar]
- Ryz, N.R.; Remillard, D.J.; Russo, E.B. Cannabis roots: A traditional therapy with future potential for treating inflammation and pain. Cannabis Cannabinoid Res. 2017, 2, 210–216. [Google Scholar] [CrossRef]
- Zhan, X.; Qiu, T.; Zhang, H.; Hou, K.; Liang, X.; Chen, C.; Wang, Z.; Wu, Q.; Wang, X.; Li, X.-l. Mass spectrometry imaging and single-cell transcriptional profiling reveal the tissue-specific regulation of bioactive ingredient biosynthesis in Taxus leaves. Plant Commun. 2023, 4, 100630. [Google Scholar] [CrossRef] [PubMed]
- Soliman, S.S.; Raizada, M.N. Sites of biosynthesis and storage of Taxol in Taxus media (Rehder) plants: Mechanism of accumulation. Phytochemistry 2020, 175, 112369. [Google Scholar] [CrossRef]
- Cartayrade, A.; Neau, E.; Sohier, C.; Balz, J.-P.; Carde, J.-P.; Walter, J. Ginkgolide and bilobalide biosynthesis in Ginkgo biloba. I: Sites of synthesis, translocation and accumulation of ginkgolides and bilobalide. Plant Physiol. Biochem. 1997, 35, 859–868. [Google Scholar]
- Liu, X.-G.; Lu, X.; Gao, W.; Li, P.; Yang, H. Structure, synthesis, biosynthesis, and activity of the characteristic compounds from Ginkgo biloba L. Nat. Prod. Rep. 2022, 39, 474–511. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Yang, H.; Liu, X.; Shen, Q.; Wang, N.; Qi, L.-w.; Li, P. Combining metabolic profiling and gene expression analysis to reveal the biosynthesis site and transport of ginkgolides in Ginkgo biloba L. Front. Plant Sci. 2017, 8, 872. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Lin, C.; Xing, P.; Fen, Y.; Jin, H.; Zhou, C.; Gu, Y.Q.; Wang, J.; Li, X. A high-quality reference genome sequence of Salvia miltiorrhiza provides insights into tanshinone synthesis in its red rhizomes. Plant Genome 2020, 13, e20041. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Peters, R.J.; Weirather, J.; Luo, H.; Liao, B.; Zhang, X.; Zhu, Y.; Ji, A.; Zhang, B.; Hu, S. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 2015, 82, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Wang, Y.; Tian, W.; Cui, X.; Tu, P.; Li, J.; Shi, S.; Liu, X. Biosynthesis Investigations of Terpenoid, Alkaloid, and Flavonoid Antimicrobial Agents Derived from Medicinal Plants. Antibiotics 2022, 11, 1380. [Google Scholar] [CrossRef]
- Verrier, P.J.; Bird, D.; Burla, B.; Dassa, E.; Forestier, C.; Geisler, M.; Klein, M.; Kolukisaoglu, Ü.; Lee, Y.; Martinoia, E. Plant ABC proteins–a unified nomenclature and updated inventory. Trends Plant Sci. 2008, 13, 151–159. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, J.; Chen, H.; Luo, H. Genome-wide analysis of ATP-binding cassette transporter provides insight to genes related to bioactive metabolite transportation in Salvia miltiorrhiza. BMC Genom. 2021, 22, 315. [Google Scholar] [CrossRef]
- Crouzet, J.; Roland, J.; Peeters, E.; Trombik, T.; Ducos, E.; Nader, J.; Boutry, M. NtPDR1, a plasma membrane ABC transporter from Nicotiana tabacum, is involved in diterpene transport. Plant Mol. Biol. 2013, 82, 181–192. [Google Scholar] [CrossRef]
- Zhang, R.; Huang, J.; Zhu, J.; Xie, X.; Tang, Q.; Chen, X.; Luo, J.; Luo, Z. Isolation and characterization of a novel PDR-type ABC transporter gene PgPDR3 from Panax ginseng CA Meyer induced by methyl jasmonate. Mol. Biol. Rep. 2013, 40, 6195–6204. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Shi, P.; He, Q.; Shen, Q.; Tang, Y.; Pan, Q.; Ma, Y.; Yan, T.; Chen, M.; Hao, X. AaPDR3, a PDR transporter 3, is involved in sesquiterpene β-caryophyllene transport in Artemisia annua. Front. Plant Sci. 2017, 8, 723. [Google Scholar] [CrossRef]
- Huang, H.; Xing, S.; Tang, K.; Jiang, W. AaWRKY4 upregulates artemisinin content through boosting the expressions of key enzymes in artemisinin biosynthetic pathway. Plant Cell Tissue Organ Cult. (PCTOC) 2021, 146, 97–105. [Google Scholar] [CrossRef]
- Mostafa, S.; Wang, Y.; Zeng, W.; Jin, B. Floral scents and fruit aromas: Functions, compositions, biosynthesis, and regulation. Front. Plant Sci. 2022, 13, 860157. [Google Scholar] [CrossRef]
- Huang, Y.; Xie, F.; Cao, X.; Li, M. Research progress in biosynthesis and regulation of plant terpenoids. Biotechnol. Biotechnol. Equip. 2021, 35, 1799–1808. [Google Scholar] [CrossRef]
- Huang, J.Q.; Fang, X. Amorpha-4, 11-diene synthase: A key enzyme in artemisinin biosynthesis and engineering. Abiotech 2021, 2, 276–288. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, L.; Zhang, F.; Jiang, W.; Shen, Q.; Zhang, L.; Lv, Z.; Wang, G.; Tang, K. AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol. 2013, 198, 1191–1202. [Google Scholar] [CrossRef]
- Maeda, H.A.; Fernie, A.R. Evolutionary history of plant metabolism. Annu. Rev. Plant Biol. 2021, 72, 185–216. [Google Scholar] [CrossRef]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.; Tai, A.; Main, A.; Eng, D. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Chen, Y.; Wang, T.; Wu, S.; Lu, X.; Zhang, L.; Zhang, F.; Jiang, W.; Wang, G.; Tang, K. Overexpression of the cytochrome P450 monooxygenase (cyp71av1) and cytochrome P450 reductase (cpr) genes increased artemisinin content in Artemisia annua (Asteraceae). Genet. Mol. Res. 2012, 11, 3298–3309. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.-Q.; Li, D.-M.; Tian, X.; Lin, J.-L.; Yang, L.; Xu, J.-J.; Fang, X. Side products of recombinant amorpha-4, 11-diene synthase and their effect on microbial artemisinin production. J. Agric. Food Chem. 2021, 69, 2168–2178. [Google Scholar] [CrossRef] [PubMed]
- Wani, K.I.; Choudhary, S.; Zehra, A.; Naeem, M.; Weathers, P.; Aftab, T. Enhancing artemisinin content in and delivery from Artemisia annua: A review of alternative, classical, and transgenic approaches. Planta 2021, 254, 29. [Google Scholar] [CrossRef]
- Yuan, Y.; Liu, W.; Zhang, Q.; Xiang, L.; Liu, X.; Chen, M.; Lin, Z.; Wang, Q.; Liao, Z. Overexpression of artemisinic aldehyde Δ11 (13) reductase gene–enhanced artemisinin and its relative metabolite biosynthesis in transgenic Artemisia annua L. Biotechnol. Appl. Biochem. 2015, 62, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shen, Q.; Wang, Y.; Wang, T.; Wu, S.; Zhang, L.; Lu, X.; Zhang, F.; Jiang, W.; Qiu, B. The stacked over-expression of FPS, CYP71AV1 and CPR genes leads to the increase of artemisinin level in Artemisia annua L. Plant Biotechnol. Rep. 2013, 7, 287–295. [Google Scholar] [CrossRef]
- Liu, W.; Wang, H.; Chen, Y.; Zhu, S.; Chen, M.; Lan, X.; Chen, G.; Liao, Z. Cold stress improves the production of artemisinin depending on the increase in endogenous jasmonate. Biotechnol. Appl. Biochem. 2017, 64, 305–314. [Google Scholar] [CrossRef]
- Yin, L.; Zhao, C.; Huang, Y.; Yang, R.; Zeng, Q. Abiotic stress-induced expression of artemisinin biosynthesis genes in Artemisia annua L. Chin. J. Appl. Environ. Biology. 2008, 14, 1. [Google Scholar]
- Jing, F.; Zhang, L.; Li, M.; Tang, Y.; Wang, Y.; Wang, Y.; Wang, Q.; Pan, Q.; Wang, G.; Tang, K. Abscisic acid (ABA) treatment increases artemisinin content in Artemisia annua by enhancing the expression of genes in artemisinin biosynthetic pathway. Biologia 2009, 64, 319–323. [Google Scholar] [CrossRef]
- Pan, W.S.; Zheng, L.P.; Tian, H.; Li, W.Y.; Wang, J.W. Transcriptome responses involved in artemisinin production in Artemisia annua L. under UV-B radiation. J. Photochem. Photobiol. B Biol. 2014, 140, 292–300. [Google Scholar] [CrossRef]
- Yadav, R.K.; Sangwan, R.S.; Srivastava, A.K.; Sangwan, N.S. Prolonged exposure to salt stress affects specialized metabolites-artemisinin and essential oil accumulation in Artemisia annua L.: Metabolic acclimation in preferential favour of enhanced terpenoid accumulation accompanying vegetative to reproductive phase transition. Protoplasma 2017, 254, 505–522. [Google Scholar]
- Cui, G.; Duan, L.; Jin, B.; Qian, J.; Xue, Z.; Shen, G.; Snyder, J.H.; Song, J.; Chen, S.; Huang, L. Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza. Plant Physiol. 2015, 169, 1607–1618. [Google Scholar] [CrossRef]
- Wei, X.; Cao, P.; Wang, G.; Han, J. Microbial inoculant and garbage enzyme reduced cadmium (Cd) uptake in Salvia miltiorrhiza (Bge.) under Cd stress. Ecotoxicol. Environ. Saf. 2020, 192, 110311. [Google Scholar] [CrossRef]
- Xu, H.; Song, J.; Luo, H.; Zhang, Y.; Li, Q.; Zhu, Y.; Xu, J.; Li, Y.; Song, C.; Wang, B. Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Mol. Plant 2016, 9, 949–952. [Google Scholar] [CrossRef]
- Ma, Y.; Cui, G.; Chen, T.; Ma, X.; Wang, R.; Jin, B.; Yang, J.; Kang, L.; Tang, J.; Lai, C. Expansion within the CYP71D subfamily drives the heterocyclization of tanshinones synthesis in Salvia miltiorrhiza. Nat. Commun. 2021, 12, 685. [Google Scholar] [CrossRef]
- Contreras, A.; Leroy, B.; Mariage, P.-A.; Wattiez, R. Proteomic analysis reveals novel insights into tanshinones biosynthesis in Salvia miltiorrhiza hairy roots. Sci. Rep. 2019, 9, 5768. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Peters, R.J. Tanshinones: Leading the way into Lamiaceae labdane-related diterpenoid biosynthesis. Curr. Opin. Plant Biol. 2022, 66, 102189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ji, A.; Xu, Z.; Luo, H.; Song, J. The AP2/ERF transcription factor SmERF128 positively regulates diterpenoid biosynthesis in Salvia miltiorrhiza. Plant Mol. Biol. 2019, 100, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, I.I.; Pramastya, H.; Van Merkerk, R.; Sukrasno; Quax, W.J. Metabolic engineering of Bacillus subtilis toward taxadiene biosynthesis as the first committed step for taxol production. Front. Microbiol. 2019, 10, 218. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Mutanda, I.; Wang, K.; Yang, L.; Wang, J.; Wang, Y. Chloroplastic metabolic engineering coupled with isoprenoid pool enhancement for committed taxanes biosynthesis in Nicotiana benthamiana. Nat. Commun. 2019, 10, 4850. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Xu, W.; Huang, W.; Wang, B.; Li, S.; Zhang, J.; Chang, L. Importation of taxadiene synthase into chloroplast improves taxadiene production in tobacco. Planta 2021, 253, 107. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Wang, X.; Liu, X.; Zhu, X.; Li, Z.; Chu, H.; Wang, Q.; Lou, Q.; Cai, B.; Yang, Y. Chromosome-level genome of Himalayan yew provides insights into the origin and evolution of the paclitaxel biosynthetic pathway. Mol. Plant 2021, 14, 1199–1209. [Google Scholar] [CrossRef]
- Jiang, L.; Zhang, K.; Lü, X.; Yang, L.; Wang, S.; Chen, D.; Yang, Y.; Qiu, D. Characterization and expression analysis of genes encoding Taxol biosynthetic enzymes in Taxus spp. J. For. Res. 2021, 32, 2507–2515. [Google Scholar] [CrossRef]
- Schepmann, H.G.; Pang, J.; Matsuda, S.P. Cloning and characterization of Ginkgo biloba levopimaradiene synthase, which catalyzes the first committed step in ginkgolide biosynthesis. Arch. Biochem. Biophys. 2001, 392, 263–269. [Google Scholar] [CrossRef]
- Forman, V.; Luo, D.; Geu-Flores, F.; Lemcke, R.; Nelson, D.R.; Kampranis, S.C.; Staerk, D.; Møller, B.L.; Pateraki, I. A gene cluster in Ginkgo biloba encodes unique multifunctional cytochrome P450s that initiate ginkgolide biosynthesis. Nat. Commun. 2022, 13, 5143. [Google Scholar] [CrossRef]
- Neau, E.; Cartayrade, A.; Balz, J.-P.; Carde, J.-P.; Walter, J. Ginkgolide and bilobalide biosynthesis in Ginkgo biloba. II: Identification of a possible intermediate compound by using inhibitors of cytochrome p-450-dependent oxygenases. Plant Physiol. Biochem. 1997, 35, 869–879. [Google Scholar]
- Lange, B.M.; Zager, J.J. Comprehensive inventory of cannabinoids in Cannabis sativa L.: Can we connect genotype and chemotype? Phytochem. Rev. 2022, 21, 1273–1313. [Google Scholar] [CrossRef]
- Jalali, S.; Salami, S.A.; Sharifi, M.; Sohrabi, S. Signaling compounds elicit expression of key genes in cannabinoid pathway and related metabolites in cannabis. Ind. Crops Prod. 2019, 133, 105–110. [Google Scholar] [CrossRef]
- Grassa, C.J.; Weiblen, G.D.; Wenger, J.P.; Dabney, C.; Poplawski, S.G.; Timothy Motley, S.; Michael, T.P.; Schwartz, C. A new Cannabis genome assembly associates elevated cannabidiol (CBD) with hemp introgressed into marijuana. New Phytol. 2021, 230, 1665–1679. [Google Scholar] [CrossRef]
- Hao, X.; Pu, Z.; Cao, G.; You, D.; Zhou, Y.; Deng, C.; Shi, M.; Nile, S.H.; Wang, Y.; Zhou, W. Tanshinone and salvianolic acid biosynthesis are regulated by SmMYB98 in Salvia miltiorrhiza hairy roots. J. Adv. Res. 2020, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, L.; Zheng, X.; Zhang, J.; Yang, L.; Tan, R.; Zhao, S. Overexpression of SmMYB9b enhances tanshinone concentration in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 2017, 36, 1297–1309. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, D.; Zhou, L.; Yu, X.; Yan, X.; Zhang, Q.; Li, B.; Liu, Y.; Zhou, W.; Cao, X. JA-responsive transcription factor SmMYB97 promotes phenolic acid and tanshinone accumulation in Salvia miltiorrhiza. J. Agric. Food Chem. 2020, 68, 14850–14862. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Luo, X.; Zhang, C.; Xu, X.; Huang, J.; Chen, Y.; Feng, S.; Zhan, X.; Zhang, L.; Yuan, H. Tissue-specific study across the stem of Taxus media identifies a phloem-specific TmMYB3 involved in the transcriptional regulation of paclitaxel biosynthesis. Plant J. 2020, 103, 95–110. [Google Scholar] [CrossRef]
- Qin, W.; Xie, L.; Li, Y.; Liu, H.; Fu, X.; Chen, T.; Hassani, D.; Li, L.; Sun, X.; Tang, K. An R2R3-MYB transcription factor positively regulates the glandular secretory trichome initiation in Artemisia annua L. Front. Plant Sci. 2021, 12, 657156. [Google Scholar] [CrossRef]
- Wu, Z.; Li, L.; Liu, H.; Yan, X.; Ma, Y.; Li, Y.; Chen, T.; Wang, C.; Xie, L.; Hao, X. AaMYB15, an R2R3-MYB TF in Artemisia annua, acts as a negative regulator of artemisinin biosynthesis. Plant Sci. 2021, 308, 110920. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, P.; Cai, S.; Haughn, G.; Page, J.E. Three novel transcription factors involved in cannabinoid biosynthesis in Cannabis sativa L. Plant Mol. Biol. 2021, 106, 49–65. [Google Scholar] [CrossRef]
- Haiden, S.R.; Apicella, P.V.; Ma, Y.; Berkowitz, G.A. Overexpression of CsMIXTA, a Transcription Factor from Cannabis sativa, Increases Glandular Trichome Density in Tobacco Leaves. Plants 2022, 11, 1519. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Pu, G.; Lei, C.; Ma, L.; Wang, H.; Guo, Y.; Chen, J.; Du, Z.; Wang, H.; Li, G. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4, 11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 2009, 50, 2146–2161. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, Y.; Nie, L.; Jin, X.; Liao, W.; Zhao, S.; Fu, C.; Yu, L. Transcriptome-wide identification and screening of WRKY factors involved in the regulation of taxol biosynthesis in Taxus chinensis. Sci. Rep. 2018, 8, 5197. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, H.; Zhang, M.; Zhang, W.; Ou, Z.; Peng, Z.; Fu, C.; Zhao, C.; Yu, L. Salicylic acid-responsive factor TcWRKY33 positively regulates taxol biosynthesis in Taxus chinensis in direct and indirect ways. Front. Plant Sci. 2021, 12, 697476. [Google Scholar] [CrossRef]
- Cao, W.; Wang, Y.; Shi, M.; Hao, X.; Zhao, W.; Wang, Y.; Ren, J.; Kai, G. Transcription factor SmWRKY1 positively promotes the biosynthesis of tanshinones in Salvia miltiorrhiza. Front. Plant Sci. 2018, 9, 554. [Google Scholar] [CrossRef]
- Deng, C.; Hao, X.; Shi, M.; Fu, R.; Wang, Y.; Zhang, Y.; Zhou, W.; Feng, Y.; Makunga, N.P.; Kai, G. Tanshinone production could be increased by the expression of SmWRKY2 in Salvia miltiorrhiza hairy roots. Plant Sci. 2019, 284, 1–8. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Li, W.; Jia, Y.; Yue, Z.; Jiao, J.; Huang, W.; Xia, P.; Liang, Z. The ethylene response factor SmERF6 co-regulates the transcription of SmCPS1 and SmKSL1 and is involved in tanshinone biosynthesis in Salvia miltiorrhiza hairy roots. Planta 2018, 248, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Li, J.; Yang, C.; Hu, W.; Wang, L.; Chen, X. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 2012, 5, 353–365. [Google Scholar] [CrossRef]
- Huang, Q.; Sun, M.; Yuan, T.; Wang, Y.; Shi, M.; Lu, S.; Tang, B.; Pan, J.; Wang, Y.; Kai, G. The AP2/ERF transcription factor SmERF1L1 regulates the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Food Chem. 2019, 274, 368–375. [Google Scholar] [CrossRef]
- Zhang, M.; Li, S.; Nie, L.; Chen, Q.; Xu, X.; Yu, L.; Fu, C. Two jasmonate-responsive factors, TcERF12 and TcERF15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in Taxus chinensis. Plant Mol. Biol. 2015, 89, 463–473. [Google Scholar] [CrossRef]
- Ji, Y.; Xiao, J.; Shen, Y.; Ma, D.; Li, Z.; Pu, G.; Li, X.; Huang, L.; Liu, B.; Ye, H. Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol. 2014, 55, 1592–1604. [Google Scholar] [CrossRef]
- Xing, B.; Yang, D.; Yu, H.; Zhang, B.; Yan, K.; Zhang, X.; Han, R.; Liang, Z. Overexpression of SmbHLH10 enhances tanshinones biosynthesis in Salvia miltiorrhiza hairy roots. Plant Sci. 2018, 276, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Xing, B.; Liang, L.; Liu, L.; Hou, Z.; Yang, D.; Yan, K.; Zhang, X.; Liang, Z. Overexpression of SmbHLH148 induced biosynthesis of tanshinones as well as phenolic acids in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 2018, 37, 1681–1692. [Google Scholar] [CrossRef] [PubMed]
- Lenka, S.K.; Nims, N.E.; Vongpaseuth, K.; Boshar, R.A.; Roberts, S.C.; Walker, E.L. Jasmonate-responsive expression of paclitaxel biosynthesis genes in Taxus cuspidata cultured cells is negatively regulated by the bHLH transcription factors TcJAMYC1, TcJAMYC2, and TcJAMYC4. Front. Plant Sci. 2015, 6, 115. [Google Scholar] [CrossRef]
- Zhang, F.; Fu, X.; Lv, Z.; Lu, X.; Shen, Q.; Zhang, L.; Zhu, M.; Wang, G.; Sun, X.; Liao, Z. A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Mol. Plant 2015, 8, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Sun, T.-H.; Zhao, L.; Pan, X.-W.; Lu, S. The bZIP transcription factor HY5 interacts with the promoter of the monoterpene synthase gene QH6 in modulating its rhythmic expression. Front. Plant Sci. 2015, 6, 304. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Zhou, W.; Zhang, J.; Huang, S.; Wang, H.; Kai, G. Methyl jasmonate induction of tanshinone biosynthesis in Salvia miltiorrhiza hairy roots is mediated by Jasmonate Zim-Domain repressor proteins. Sci. Rep. 2016, 6, 20919. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Bai, Z.; Pei, T.; Yang, D.; Mao, R.; Zhang, B.; Liu, C.; Liang, Z. SmGRAS1 and SmGRAS2 regulate the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Front. Plant Sci. 2019, 10, 1367. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Zhao, X.; Jiang, Y.; Jin, B.; Wang, L. Functions of Representative Terpenoids and Their Biosynthesis Mechanisms in Medicinal Plants. Biomolecules 2023, 13, 1725. https://doi.org/10.3390/biom13121725
Wang Q, Zhao X, Jiang Y, Jin B, Wang L. Functions of Representative Terpenoids and Their Biosynthesis Mechanisms in Medicinal Plants. Biomolecules. 2023; 13(12):1725. https://doi.org/10.3390/biom13121725
Chicago/Turabian StyleWang, Qingjie, Xiya Zhao, Yang Jiang, Biao Jin, and Li Wang. 2023. "Functions of Representative Terpenoids and Their Biosynthesis Mechanisms in Medicinal Plants" Biomolecules 13, no. 12: 1725. https://doi.org/10.3390/biom13121725
APA StyleWang, Q., Zhao, X., Jiang, Y., Jin, B., & Wang, L. (2023). Functions of Representative Terpenoids and Their Biosynthesis Mechanisms in Medicinal Plants. Biomolecules, 13(12), 1725. https://doi.org/10.3390/biom13121725


_Kwok.png)


